Abstract
Aberrant methylation of promoter CpG islands is causally linked with a number of inherited syndromes and most sporadic cancers, and may provide valuable diagnostic and prognostic biomarkers. In this report, we describe an approach to simultaneous analysis of multiple CpG islands, where methylation-specific oligonucleotide probes are joined by ligation and subsequently amplified by polymerase chain reaction (PCR) when hybridized in juxtaposition on bisulfite-treated DNA. Specificity of the ligation reaction is achieved by (i) using probes containing CpGpCpG (for methylated sequences) or CpApCpA (for unmethylated sequences) at the 3′ ends, (ii) including three or more probes for each target, and (iii) using a thermostable DNA ligase. The external probes carry universal tails to allow amplification of multiple ligation products using a common primer pair. As proof-of-principle applications, we established duplex assays to examine the FMR1 promoter in individuals with fragile-X syndrome and the SNRPN promoter in individuals with Prader-Willi syndrome or Angelman syndrome, and a multiplex assay to simultaneously detect hypermethylation of seven genes (ID4, APC, RASSF1A, CDH1, ESR1, HIN1 and TWIST1) in breast cancer cell lines and tissues. These data show that ligation of oligonucleotide probes hybridized to bisulfite-treated DNA is a simple and cost-effective approach to analysis of CpG methylation.
INTRODUCTION
Cytosine methylation is a modification of DNA that is important for epigenetic regulation of endogenous genes, silencing of transposons and control of genome stability (1,2). In humans, DNA methylation occurs almost exclusively at cytosines within the context of CpG dinucleotides. A large proportion of human gene promoters contain CpG clusters, so-called CpG islands, which may be either unmethylated or hypermethylated. Promoter hypermethylation is often associated with transcriptional inactivation and has been shown to be responsible for the stable monoallelic silencing of imprinted genes and genes on the X chromosome during human development (3,4).
Aberrant methylation of promoter CpG islands has been implicated in a number of inherited disorders, including the fragile X syndrome, Beckwith-Wiedemann syndrome, Prader-Willi syndrome (PWS) and Angelman syndrome (AS) (4,5). Although the genomic abnormalities responsible for these syndromes include deletions, uniparental disomy, imprinting mutations and trinucleotide repeat expansions, the resulting changes in DNA methylation may serve as valuable diagnostic markers. Changes in DNA methylation have also been associated with neoplastic initiation and progression (6,7). The best characterized epigenetic change in tumor cells is transcriptional silencing of tumor suppressor genes caused by increases in promoter methylation. The patterns of aberrant promoter methylation are non-random and tumor-type specific (8–10), and methylation events that frequently occur in one or more tumor types are being developed as biomarkers for risk assessment, early detection and response to therapy (11–13).
The gold standard for high-resolution mapping of DNA methylation is bisulfite genomic sequencing (14). The basis of this method is treatment of genomic DNA with bisulfite, which effectively deaminates unmethylated cytosine residues to uracil, while 5-methylcytosines are resistant to this treatment and remain unchanged (15). Bisulfite-treated DNA can be used as template in a standard PCR, in which uracils (formerly unmethylated cytosines) will be amplified as thymine and only 5-methylcytosines will be amplified as cytosine, to generate a template that is amenable to direct or cloned sequence analysis. Despite the power of sequence analysis to distinguish between cytosine and 5-methylcytosine, this approach may still be impracticable in many research and diagnostic laboratories and may not always be the most rational approach to detect aberrant DNA methylation. A wealth of PCR-based methods have been developed to analyze bisulfite-treated DNA, including methylation-specific PCR (16), methylation-sensitive single-nucleotide primer extension (Ms-SnuPE) (17), MethyLight (18), HeavyMethyl (19), combined bisulfite restriction analysis (COBRA) (20) and methylation-specific melting curve analysis (MS-MCA) (21). For comprehensive reviews of methods for analysis of DNA methylation, see references (22,23).
The different techniques provide information on DNA methylation at different levels and have their own advantages and limitations. Notably, none of the above methods provides a simple means to simultaneously analyze multiple genes in a single reaction. Here, we describe an approach to multiplex analysis of CpG islands, which utilizes oligonucleotide probes that can hybridize to bisulfite-treated DNA and then be ligated into an amplifiable product when perfectly matched to the target sequence. We describe a probe design that, in combination with the use of a thermostable DNA ligase, proved sufficiently robust to discriminate between methylated and unmethylated sequences. As proof-of-principle applications, duplex assays were used to examine the SNRPN and FMR1 promoters in individuals with inherited disorders, and a multiplex assay was used to simultaneously detect aberrant promoter hypermethylation of ID4, APC, RASSF1A, CDH1, ESR1, HIN1 and TWIST1 in breast cancer cell lines and tissues.
MATERIALS AND METHODS
Cell lines, tumor biopsies and DNA
MCF7, CAMA-1, BT-20, HBL-100, HCC1937, HCC1569, MDA-MD-157 and ZR-75-1 breast cancer cell lines were grown in RPMI 1640 medium with Glutamax-1 (GIBCO) and 10% fetal bovine serum (GIBCO). Tissue biopsies from 17 high-risk breast cancer patients (15 ductal invasive carcinomas and two lobular lesions) were obtained and processed as previously described (24). For 10 of these patients, tissue was also available from axillary nodal metastases.
DNA and sodium bisulfite treatment
DNA from cell lines and biopsies was extracted using the NucleoSpin Tissue kit (Macherey-Nagel). Peripheral blood lymphocyte (PBL) DNA from healthy donors and from persons with AS, PWS or fragile X syndrome was previously analyzed using MS-MCA (21,25). Two micrograms of genomic DNA were treated with sodium bisulfite according to standard procedures (15). The bisulfite-modified DNA was resuspended in 20 µl of TE buffer (10 mM Tris–HCl and 1 mM EDTA, pH 8.5) and used immediately or stored at −80°C until use. Bisulfite-treated DNA gave no detectable signal in a TaqMan® assay with primers for a normal, non-converted ACTB sequence (18), which confirms that the bisulfite conversion was complete.
Probes
The sequences of oligonucleotide probes (Sigma-Genosys) used for ligation-based DNA methylation analysis are shown in Table 1. Promoter sequences were obtained from the UCSC Genome Browser, using the accession numbers listed in Table 1. All probes carrying a 5′ phosphate group were ordered purified by polyacrylamide gel electrophoresis.
Table 1.
Oligonucleotide probes for ligation-based detection of CpG methylation
| Number of probes | GenBank accession no. | Probes sequences (5′–3′) | Product length (bp) | ||
|---|---|---|---|---|---|
| SNRPN U | 3 | L32702 | P-TCAAACATCTCCAACAACCACTCCACT-[X] | 122 | |
| P-TCACTAACCACTCCTCAAACAAATACA | |||||
| [Y]-CACAACTAACCTTACCCACTCCATCACA | |||||
| SNRPN M | 3 | L32702 | P-TCAAACATCTCCGACGACCGCT-[X] | 109 | |
| P-TCACTAACCGCTCCTCAAACAAATACG | |||||
| [Y]-ACCTTACCCGCTCCATCGCG | |||||
| FMR1 U | 3 | X61378 | P-CATACACACTACTAAAAACCAACCAAAATACCAAATCAAA-[X] | 138 | |
| P-CCCTCTCTCTTCAAATAACCTAAAAACACACA | |||||
| [Y]-CCCACAAACTCAACCCCTCAACCCCA | |||||
| FMR1M | 3 | X61378 | P-CATACGCGCTACTAAAAACCGACCGAAATAC-[X] | 125 | |
| P-CCCTCTCTCTTCAAATAACCTAAAAACGCGCG | |||||
| [Y]-CGAACTCAACCCCTCGACCCCG | |||||
| ID4 | 3 | NM_001546 | P-AACGAAACCCGCTAAACGCGC-[X] | 104 | |
| P-AACCGAATCGCAACGCGAACCG | |||||
| [Y]-CGAAAACGAAACCAACGCGCG | |||||
| APC | 3 | NM_000038 | P-AACTCCCGACGAAAATAAAAAACGC-[X] | 115 | |
| P-TACGCCCACACCCAACCAATCGACG | |||||
| [Y]-CCAATACAACCACATATCGATCACG | |||||
| RASSF1A | 3 | NM_007182 | P-CCCAACGAATACCAACTCCCGCAACTCAATA-[X] | 126 | |
| P-CTCCAACCGAATACGACCCTTCCCAACGCG | |||||
| [Y]-GCGCGATACGCAACGCGTTAACACG | |||||
| CHD1 | 3 | NM_004360 | P-CCCACCCGACCTCGCATAAACGCGATAACCC-[X] | 140 | |
| P-AACCCCTCCCCAAAACGAAACTAACGACCCG | |||||
| [Y]-AAAAATTCACCTACCGACCACAACCAATCAACAACGCG | |||||
| ESR1 | 3 | NM_000125 | P-ATAAAACCGAACGACCCGACGAAAACAAATACAATCCC-[X] | 154 | |
| P-AACTCTAACCCCGACCCTACCCCGAAAACCTACGAATCCG | |||||
| [Y]-TCAAAAACGACGCAACGCATATCCCGCCGACACGCG | |||||
| PAH | 4 | NM_000277 | P-ATCAATATTCCCTACTACATCCCATAAACC-[X] | 175 | |
| P-CCCCAAATAAAAAATTATTATCACTATTAAATCAAA | |||||
| P-AAAAAAAACTTTAACTTCTCTAATAAACAATACTATAAA | |||||
| [Y]-TTTTAAATAACTATCTTCTCCAACTCCAAA | |||||
| HIN1 | 4 | NM_052863 | P-CCGACCTCGCCCGCGCTCCTAAAAAAACCC-[X] | 193 | |
| P-AACAAAACCACGAAACTTCTTATACCCGATCCTCGCCCCTCCAACG | |||||
| P-AAAACTCGAAACGCGCGAAAAACCTACGACTACCCG | |||||
| [Y]-ACCACGCAAAACCCCAAAAAAACGACGAACTTCATAACGCG | |||||
| TWIST1 | 5 | NM_000474 | P-CGATAACAACCCCATCCGAAATAACTATAACAACAACAATAACAACAA-[X] | 213 | |
| P-AACCCTAACGCAACCCAAAAAACGATCGAAAAAAACTATCCTAACCG | |||||
| P-CCGAAACGTACGAACAACGCCCCCG | |||||
| P-ACGAACGCGAAACGATTTCCTTCCCCG | |||||
| [Y]-CCTTCCCTCCCCGTCGCCTTCCTCCG |
[X] = 5′-ACCCAATTCGCCCTATAATA-3′
[Y] = 5′-TATGTAAAACGACGGCCAGT-3′
Hybridization, ligation and PCR
Hybridization was performed in a Peltier thermal cycler (PTC-200; MJ Research) in total volumes of 8 µl containing bisulfite-treated DNA (5 µl), 1.5 µl of SALSA MLPA buffer (MRC-Holland) and 1.5 µl of a mix containing 0.5–10 fmol of each probe diluted in TE buffer. The samples were heated to 95°C for 1 min and then incubated at 60°C for 16 h. Ligation of the hybridized oligonucleotides was achieved by addition of 32 µl of a ligase mix containing 3 µl of Ligase buffer A (MRC-Holland), 3 µl of Ligase buffer B (MRC-Holland) and 1 U of Taq DNA ligase (New England Biolabs) and incubation at 54°C for 15 min. After inactivation of the ligase at 98°C for 5 min, 6 µl of the ligation reaction was included in a total volume of 25 µl, containing 7.5 pmol of each primer (5′-TATGTAAAACGACGGCCAGT-3′ and 5′-TATTATAGGGCGAATTGGGT-3′), 1 × Multiplex PCR Master Mix (Qiagen) and 1 × Q-solution (Qiagen). PCR conditions were: 95°C for 15 min to activate the enzyme, followed by 40 cycles at 95°C for 30 s, 52°C for 1.5 min and 72°C for 1.5 min, and a final incubation at 72°C for 10 min. The PCR products were resolved by electrophoresis in 4% NuSieve GTG agarose gels (Cambrex) and visualized by ethidium bromide staining.
Methylation-specific PCR and MS-MCA
Methylation-specific PCR (16) was performed using the HotStarTaq Kit (Qiagen) and methylation-specific primers, designed according to previously described principles (23). MS-MCA (21) was carried out using the LightCycler 1.1 instrument (Roche) and the FastStart DNA Master SYBR Green I Kit (Roche). Primer sequences and PCR conditions are available upon request.
RESULTS
Outline of the assay
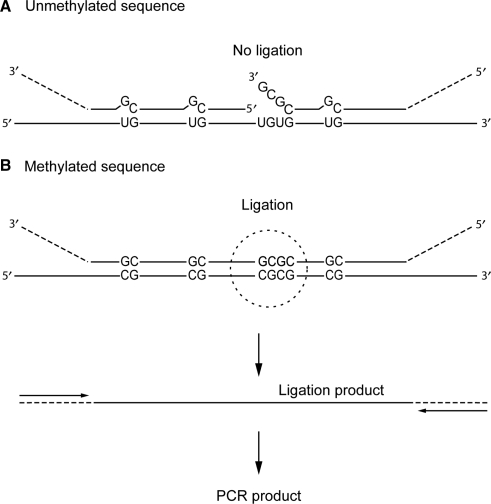
The principle of the assay is shown in Figure 1. For each target, oligonucleotide probes specific for either methylated or unmethylated DNA are allowed to hybridize immediately adjacent to each other on bisulfite-treated DNA and then ligated using a DNA ligase. The probes carry universal tails, which serve as primer binding sites for PCR amplification of the ligation products. Previous work has shown that DNA ligases are sensitive to mispairs present on the 3′ side of the ligase junction (26–28). Therefore, to discriminate between methylated and unmethylated DNA on the basis of the ligation reaction, methylation-specific bases are included at the 3′ end of the probes. When the target sequence contains one or more CpGpCpG sites, the 5′ probe is designed to contain CpGpCpG (for the methylated sequence) or CpApCpA (for the corresponding unmethylated sequence) at the 3′ end, which will introduce a non-ligatable mismatch at the ligation junction when hybridized to sequences that do not match the methylation status. If the promoter contains no CpGpCpG sites, a single CpG site is used as ligation junction.
Figure 1.
Schematic of ligation-based detection of CpG methylation. In this example, two oligonucleotide probes, specific for the methylated target sequence, hybridize to bisulfite-treated DNA. The 5′ probe contains a CpGpCpG at its 3′ end, and both probes carry non-hybridizing tails, which contain binding sites for amplification primers. (A) The probes may hybridize to the unmethylated template despite mismatches, but cannot be ligated due to the mismatch at the ligation junction. (B) On the methylated template, the probes will hybridize in juxtaposition and can be joined by ligation and subsequently amplified by PCR.
In principle, two probes would be sufficient to discriminate between methylated and unmethylated sequences of a specific target. However, some of our assays based on two probes gave unspecific results, particularly when DNA from tumor specimens was used as template, probably because some probes could hybridize unspecifically to other sites and lead to the generation of an amplifiable ligation product. To increase the specificity, another strategy was adopted, which involves three or more adjacent probes for each target sequence. In this setting, generation of an amplifiable ligation product depends on at least two independent ligation reactions. At the same time, it provides a simple means for adjusting the length of the ligation products to allow electrophoretic separation after PCR amplification.
Analysis of the SNRPN CpG island in individuals with PWS and AS
As the first proof-of-principle application, we established an assay to examine SNRPN on chromosome 15q11.2 in individuals with PWS or AS. In healthy individuals, SNRPN is subject to genomic imprinting, with one unmethylated allele of paternal origin and one methylated allele of maternal origin (29). The majority of PWS cases have a deletion of the SNRPN locus on the paternal-origin chromosome 15 or maternal uniparental disomy, and hence carry only the methylated maternal allele. In AS, ∼70% of the cases harbor a deletion of the maternal methylated allele and hence carry only the unmethylated copy (29).
Two sets of three probes were designed, which are specific for either methylated or unmethylated SNRPN. The probes were hybridized to bisulfite-treated PBL DNA from persons with PWS and AS as well as from healthy individuals. Subsequently, the reactions were subjected to ligation and PCR, and the reaction products were resolved on agarose gels. As shown in Figure 2A, healthy individuals showed two bands corresponding to the unmethylated and methylated SNRPN alleles, whereas DNA from individuals with PWS showed only the methylated band, and DNA from individuals with AS showed only the unmethylated band. These experiments showed that ligation-based analysis of bisulfite-treated DNA is sufficiently robust to discriminate between methylated and unmethylated sequences, and can be used to correctly identify individuals with PWS and AS.
Figure 2.
Ligation-based detection of aberrant methylation of the SNRPN and FMR1 CpG islands. (A) Bisulfite-treated DNA from a healthy individual, two individuals with AS and two individuals with PWS were incubated in mixtures containing two sets of three probes each, which are specific for methylated and unmethylated SNRPN, respectively. After hybridization, the probes were ligated and amplified using a primer pair that recognizes both probe sets. (B) Ligation-based methylation analysis of the FMR1 CpG island in two healthy males, one healthy female and two males with fragile X syndrome (FRAXA #1 and #2). IVM, in vitro-methylated DNA; M, 20-bp ladder.
Analysis of the FMR1 promoter CpG island in individuals with fragile X syndrome
As another proof of principle, we designed an assay to examine the methylation status of FMR1 in individuals with fragile X syndrome. This syndrome is associated with the expansion of a naturally occurring CGG trinucleotide tandem repeat in the 5′ UTR of FMR1 at Xq27.3 (30). While most healthy individuals have <55 CGG repeats and an unmethylated, transcriptionally active FMR1 promoter, individuals with fragile X syndrome have >200 repeats (full mutation), which is associated with promoter hypermethylation and transcriptional silencing (30). Because FMR1 is located on the X chromosome, healthy females have both methylated and unmethylated FMR1 due to X-chromosome inactivation, whereas healthy males have only unmethylated FMR1.
Ligation-based analysis of bisulfite-treated DNA from healthy individuals and fragile X males correctly determined the methylation status of the FMR1 promoter. Healthy females showed two bands corresponding to the unmethylated and methylated FMR1 alleles, healthy males showed only the unmethylated band, and fragile X males showed a band corresponding to hypermethylated FMR1 (Figure 2B).
Multiplex analysis of seven breast cancer genes
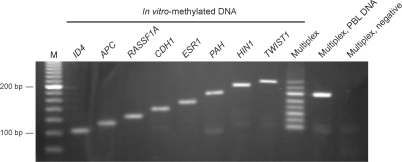
To establish an assay to test for hypermethylation events in breast cancer specimens, we designed a total of seven sets of 3–5 probes specific for the methylated sequences of ID4, APC, RASSF1A, CHD1, ESR1, HIN1 and TWIST1. The promoter CpG islands of these genes have been shown to be frequently hypermethylated in breast carcinomas (31,32). At least one of the probes in each set contained CpGpCpG at the 3′ end, except for the probes for APC and TWIST1, which contained single CpG sites. We also designed an additional set of four probes, which hybridize to a CpG-free region of the phenylalanine hydroxylase gene (PAH). This probe set will generate an amplifiable ligation product on bisulfite-treated DNA in a methylation-independent manner and therefore will serve as a control for DNA integrity as well as for all steps in the procedure, including hybridization, ligation and PCR. The number and length of the probes in the individual sets were chosen to provide ∼10% length increments between the amplification products (from 104 to 213 bp), which would allow resolution by agarose gel electrophoresis.
First, the specificity of the eight probe sets (including the PAH control) was tested by hybridizing them individually to bisulfite-treated, in vitro-methylated DNA, followed by ligation and amplification. All probe sets gave rise to a single band of the expected size (Figure 3). Next, a mix of all 28 probes representing the eight targets was used to analyze in vitro-methylated DNA and PBL DNA. As shown in Figure 3, in vitro-methylated DNA gave rise to a ladder containing all eight bands, whereas PBL DNA showed only one band corresponding to the control gene (PAH). These data show that ligation of probes hybridized to bisulfite-treated DNA can be applied to multiplex analysis of CpG methylation.
Figure 3.
Multiplex ligation-based methylation analysis. Eight probe sets (consisting of 3–5 probes and generating ligation products of 104–213 bp in length) were hybridized to in vitro-methylated DNA in singleplex and multiplex configurations. Multiplex analysis of DNA from peripheral blood lymphocytes (PBL) gave rise to only one band corresponding to the control gene (PAH). M, 20-bp ladder.
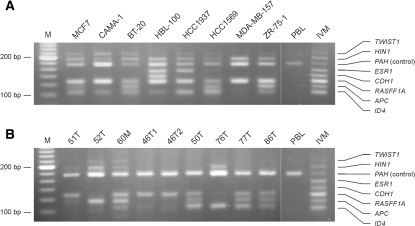
To test the applicability of the multiplex assay to the analysis of breast cancer specimens, we first analyzed DNA from eight breast cancer cell lines. All cell lines gave rise to positive signals for the control gene and at least two of the promoter targets (Figure 4A). MCF7, BT-20 and ZR-75-1 showed similar profiles with hypermethylation of ID4, APC, RASSF1A, HIN1 and TWIST1, whereas the remaining cell lines showed unique profiles (Figure 4A; Supplementary Table 1). The methylation status of the same seven gene promoters was also examined using methylation-specific PCR or MS-MCA, with consistent results (data not shown).
Figure 4.
Multiplex ligation-based analysis of breast cancer specimens. (A) Cell lines. (B) Tumor biopsies. PBL, peripheral blood lymphocytes; IVM, in vitro-methylated DNA; M, 20-bp ladder.
The multiplex assay was then applied to the analysis of 28 tissue biopsies from 17 breast cancer patients (Figure 4B; Supplementary Table 1). Three of the tumors were scored as not informative because all bands were weak, including that for the control gene, which may be ascribed to DNA degradation (see ‘Discussion’). For many of the tumors, the methylation-specific signals were weak compared with the signal from the control gene, which probably reflects tumor heterogeneity and/or contamination of the sample with normal stromal cells. While both tumor cells and normal cells contribute to the control-gene band, only the tumor cells contribute to the methylation-specific bands. The profile of hypermethylated genes showed a high degree of variation among tumors, whereas paired tumors from individual patients showed identical patterns (Figure 4B; Supplementary Table 1). For the 16 independent cases analyzed, the frequencies of promoter hypermethylation were 69% for ID4, 63% for APC, 75% for RASSF1A, 0% for CHD1, 0% for ESR1, 50% for HIN1 and 13% for TWIST1 (Supplementary Table 1). The lack of ESR1 hypermethylation in this series of breast tumors was confirmed using both MS-MCA and a previously validated methylation-specific PCR assay (33). Routine immunohistochemical analysis showed that 22/25 (88%) of these tumors were estrogen receptor positive [(24) and unpublished data].
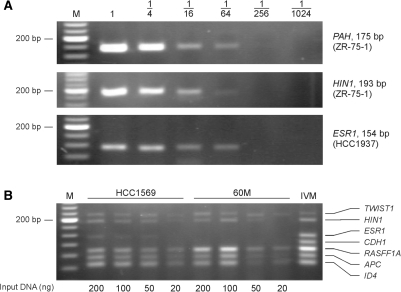
Assay sensitivity
To estimate the limit of detection of the ligation assay, we serially diluted DNA from breast cancer cell lines and analyzed it with relevant probe sets. As shown in Figure 5A, methylated sequences could be detected down to the 1% level. When the same serial dilutions of methylated DNA were mixed with a constant amount of unmethylated DNA, methylated sequences could be detected in the presence of a 100-fold excess of unmethylated DNA, showing that the limit of detection was not influenced by the presence of unmethylated DNA (data not shown).
Figure 5.
Assay sensitivity. (A) Bisulfite-treated DNA from breast cancer cell lines was serially diluted and analyzed using relevant probe sets. (B) Various amounts of DNA from breast cancer cell lines and tissues were mixed with 500 ng of plasmid, treated with sodium bisulfite and analyzed using the multiplex assay. The PAH control was excluded from these analyses. IVM, in vitro-methylated DNA; M, 20-bp ladder.
In the standard protocol for multiplex analysis, each hybridization cocktail contained bisulfite-treated template corresponding to 500 ng of human chromosomal DNA. As the amount of DNA from clinical samples is often limited, we next estimated the amount of starting template needed to obtain reproducible results. DNA from breast cancer cell lines and tissues was mixed with 500 ng of plasmid as carrier prior to bisulfite treatment and then subjected to multiplex analysis. In these experiments, the probe set for the PAH control was excluded to prevent preferential amplification of this target in samples from tumors with a high content of normal tissue. As shown in Figures 4 and 5B, the results were not influenced by the amount of input DNA within the range of 50–500 ng, whereas some of the methylation-specific signals were lost for samples containing <50 ng DNA. None of the samples gave rise to false-positive signals.
DISCUSSION
The recognition that changes in CpG methylation have broad implications for human health has created a strong need for techniques to reliably detect DNA methylation in research and diagnostic settings. We describe a simple assay to simultaneously analyze the methylation status of multiple genes, using bisulfite-treated DNA as template and gene-specific oligonucleotide probes, which bind to either methylated or unmethylated sequences and can be ligated only if perfectly matched in juxtaposition on the target sequence. Although the hybridization step may confer some specificity due to base differences between the methylated and the corresponding unmethylated sequences (Figure 1), the prime source of specificity is the ligation reaction. Wherever possible, CpGpCpG sites were chosen as the ligation junction to create the largest possible mismatch, but discrimination was also possible with a single CpG site, consistent with previous work showing that probe ligation can be used to distinguish between single-base substitutions in genomic DNA (34,35). An additional level of specificity was achieved by including at least three probes for each target, which makes the generation of an amplifiable ligation product dependent on at least two ligation reactions.
The strongest features of this assay are that it is simple to establish, is highly flexible and, in the setting described in this report, requires only inexpensive reagents, a standard block thermal cycler and equipment for agarose gel electrophoresis. It may be adapted to any set of genes in a user-defined configuration, and the lengths of the ligation products can be easily adjusted to suit the preferred separation technique. Furthermore, the addition of a probe set or replacement of one probe set with another in a multiplex assay will require no or only very few adjustments of the assay. In our hands, very little optimization of the assays was needed; all 12 probe sets tested in this study functioned under the same assay conditions despite a large variation in length and Tm of the individual probes, despite extensive multiplexing, and despite the fact that some of the targets required ligation of up to five adjacent probes. It should be emphasized that the assay in its present configuration was not designed for sensitive, quantitative methylation analysis and therefore should not be considered an alternative to Ms-SnuPE (17), MethyLight (18), HeavyMethyl (19) or COBRA (20,36).
One potential limitation of PCR-based methylation analysis using bisulfite-treated DNA as template is PCR bias, which is explained by different amplification efficiencies for the methylated and unmethylated molecules (37). In this respect, ligation-based analysis of bisulfite-treated DNA may be advantageous because of the non-competitive hybridization and ligation reactions. The fact that only the synthetic ligation products are amplified, together with the use of single primer pair common to all targets, will minimize the differences in amplification efficiency among the targets (38).
Another important limitation inherent with the use of bisulfite-treated DNA as template is that most bisulfite protocols involve prolonged heating under acidic conditions, which leads to DNA degradation due to oxidative damage and depurination (39,40). Because hybridization and ligation of probes in juxtaposition require intact template molecules, the extent of DNA degradation will influence the maximum length of the ligation products as well as the limit of detection. Using DNA from breast cancer cell lines and tissues modified by bisulfite under standard conditions, ligation products up to 213 bp were successfully generated and amplified, methylated sequences could be detected down to the 1% level, and reproducible results were obtained using as little as 50 ng of DNA as starting template. Even lower detection levels may be obtained by using high-recovery procedures for bisulfite conversion, which cause less extensive degradation of DNA (41,42). However, analysis of long ligation products may be problematic in samples characterized by a high degree of DNA degradation, such as formalin-fixed, paraffin-embedded tissue and tumors exhibiting high levels of necrosis or apoptosis. A more accurate multiplex analysis of low-quality DNA may require the use of only three short probes for each target, to generate products of <100 bp in length, in combination with a system for higher resolution of length differences.
Recently, Nygren et al. (43) described an assay for multiplex analysis of CpG methylation, called methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA), where plasmid-generated probes are hybridized to genomic DNA, followed by ligation, digestion with a methylation-sensitive restriction enzyme, and amplification using a common primer pair. Apart from the fact that incomplete restriction digests may complicate the analysis, this assay has proven useful for analysis of a defined set of genes in clinical specimens (43). On the basis of the results presented here, ligation of oligonucleotide probes hybridized to bisulfite-treated DNA affords a simple and cost-effective alternative to multiplex analysis of CpG methylation in patient samples.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors are grateful to Vibeke Ahrenkiel for technical assistance. This study was supported by grants from the Danish Cancer Society, the Neye Foundation and the Danish Medical Research Council. Funding to pay the Open Access publication charges for this article was provided by the Danish Medical Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21:461–467. doi: 10.1093/carcin/21.3.461. [DOI] [PubMed] [Google Scholar]
- 2.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 3.Costello JF, Plass C. Methylation matters. J. Med. Genet. 2001;38:285–303. doi: 10.1136/jmg.38.5.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 5.Paulsen M, Ferguson-Smith AC. DNA methylation in genomic imprinting, development, and disease. J. Pathol. 2001;195:97–110. doi: 10.1002/path.890. [DOI] [PubMed] [Google Scholar]
- 6.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 7.Ting AH, McGarvey KM, Baylin SB. The cancer epigenome–components and functional correlates. Genes Dev. 2006;20:3215–3231. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 8.Costello JF, Frühwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomäki P, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat. Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 9.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 10.Adorjan P, Distler J, Lipscher E, Model F, Muller J, Pelet C, Braun A, Florl AR, Gutig D, et al. Tumour class prediction and discovery by microarray-based DNA methylation analysis. Nucleic Acids Res. 2002;30:e21. doi: 10.1093/nar/30.5.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laird PW. The power and the promise of DNA methylation markers. Nat. Rev. Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 12.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 13.Brena RM, Plass C, Costello JF. Mining methylation for early detection of common cancers. PLoS. Med. 2006;3:e479. doi: 10.1371/journal.pmed.0030479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalgo ML, Jones PA. Rapid quantitation of methylation differences at specific sites using methylation-sensitive single nucleotide primer extension (Ms-SNuPE) Nucleic Acids Res. 1997;25:2529–2531. doi: 10.1093/nar/25.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cottrell SE, Distler J, Goodman NS, Mooney SH, Kluth A, Olek A, Schwope I, Tetzner R, Ziebarth H, et al. A real-time PCR assay for DNA-methylation using methylation-specific blockers. Nucleic Acids Res. 2004;32:e10. doi: 10.1093/nar/gnh008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Worm J, Aggerholm A, Guldberg P. In-tube DNA methylation profiling by fluorescence melting curve analysis. Clin. Chem. 2001;47:1183–1189. [PubMed] [Google Scholar]
- 22.Fraga MF, Esteller M. DNA methylation: a profile of methods and applications. Biotechniques. 2002;33:632–649. doi: 10.2144/02333rv01. [DOI] [PubMed] [Google Scholar]
- 23.Dahl C, Guldberg P. DNA methylation analysis techniques. Biogerontology. 2003;4:233–250. doi: 10.1023/a:1025103319328. [DOI] [PubMed] [Google Scholar]
- 24.Cabezon T, Celis JE, Skibshoj I, Klingelhofer J, Grigorian M, Gromov P, Rank F, Myklebust JH, Maelandsmo GM, Lukanidin E, et al. Expression of S100A4 by a variety of cell types present in the tumor microenvironment of human breast cancer. Int. J. Cancer. 2007;121:1433–1444. doi: 10.1002/ijc.22850. [DOI] [PubMed] [Google Scholar]
- 25.Dahl C, Grønskov K, Larsen LA, Guldberg P, Brøndum-Nielsen K. A homogeneous assay for analysis of FMR1 promoter methylation in patients with fragile X syndrome. Clin. Chem. 2007;53:790–793. doi: 10.1373/clinchem.2006.080762. [DOI] [PubMed] [Google Scholar]
- 26.Nilsson M, Barbany G, Antson DO, Gertow K, Landegren U. Enhanced detection and distinction of RNA by enzymatic probe ligation. Nat. Biotechnol. 2000;18:791–793. doi: 10.1038/77367. [DOI] [PubMed] [Google Scholar]
- 27.Bhagwat AS, Sanderson RJ, Lindahl T. Delayed DNA joining at 3' mismatches by human DNA ligases. Nucleic Acids Res. 1999;27:4028–4033. doi: 10.1093/nar/27.20.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu J, Tong J, Feng H, Huang J, Afonso CL, Rock DL, Barany F, Cao W. Unique ligation properties of eukaryotic NAD+-dependent DNA ligase from Melanoplus sanguinipes entomopoxvirus. Biochim. Biophys. Acta. 2004;1701:37–48. doi: 10.1016/j.bbapap.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Horsthemke B, Buiting K. Imprinting defects on human chromosome 15. Cytogenet. Genome Res. 2006;113:292–299. doi: 10.1159/000090844. [DOI] [PubMed] [Google Scholar]
- 30.Warren ST, Sherman SL. The fragile X syndrome. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill; 2001. pp. 1257–1289. [Google Scholar]
- 31.Widschwendter M, Jones PA. DNA methylation and breast carcinogenesis. Oncogene. 2002;21:5462–5482. doi: 10.1038/sj.onc.1205606. [DOI] [PubMed] [Google Scholar]
- 32.Fackler MJ, McVeigh M, Mehrotra J, Blum MA, Lange J, Lapides A, Garrett E, Argani P, Sukumar S. Quantitative multiplex methylation-specific PCR assay for the detection of promoter hypermethylation in multiple genes in breast cancer. Cancer Res. 2004;64:4442–4452. doi: 10.1158/0008-5472.CAN-03-3341. [DOI] [PubMed] [Google Scholar]
- 33.Lapidus RG, Nass SJ, Butash KA, Parl FF, Weitzman SA, Graff JG, Herman JG, Davidson NE. Mapping of ER gene CpG island by methylation-specific polymerase chain reaction. Cancer Res. 1998;58:2515–2519. [PubMed] [Google Scholar]
- 34.Landegren U, Kaiser R, Sanders J, Hood L. A ligase-mediated gene detection technique. Science. 1988;241:1077–1080. doi: 10.1126/science.3413476. [DOI] [PubMed] [Google Scholar]
- 35.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brena RM, Auer H, Kornacker K, Hackanson B, Raval A, Byrd JC, Plass C. Accurate quantification of DNA methylation using combined bisulfite restriction analysis coupled with the Agilent 2100 Bioanalyzer platform. Nucleic Acids Res. 2006;34:e17. doi: 10.1093/nar/gnj017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warnecke PM, Stirzaker C, Melki JR, Millar DS, Paul CL, Clark SJ. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res. 1997;25:4422–4426. doi: 10.1093/nar/25.21.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin Z, Cui X, Li H. Multiplex genotype determination at a large number of gene loci. Proc. Natl Acad. Sci. USA. 1996;93:2582–2587. doi: 10.1073/pnas.93.6.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:E65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munson K, Clark J, Lamparska-Kupsik K, Smith SS. Recovery of bisulfite-converted genomic sequences in the methylation-sensitive QPCR. Nucleic Acids Res. 2007;35:2893–2903. doi: 10.1093/nar/gkm055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olek A, Oswald J, Walter J. A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res. 1996;24:5064–5066. doi: 10.1093/nar/24.24.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyd VL, Zon G. Bisulfite conversion of genomic DNA for methylation analysis: protocol simplification with higher recovery applicable to limited samples and increased throughput. Anal. Biochem. 2004;326:278–280. doi: 10.1016/j.ab.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 43.Nygren AO, Ameziane N, Duarte HM, Vijzelaar RN, Waisfisz Q, Hess CJ, Schouten JP, Errami A. Methylation-specific MLPA (MS-MLPA): simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. Nucleic Acids Res. 2005;33:e128. doi: 10.1093/nar/gni127. [DOI] [PMC free article] [PubMed] [Google Scholar]