Abstract
The activation of a telomere maintenance mechanism is required for cancer development in humans. While most tumors achieve this by expressing the enzyme telomerase, a fraction (5–15%) employs a recombination-based mechanism termed alternative lengthening of telomeres (ALT). Here we show that loss of the single-stranded DNA-binding protein replication protein A (RPA) in human ALT cells, but not in telomerase-positive cells, causes increased exposure of single-stranded G-rich telomeric DNA, cell cycle arrest in G2/M phase, accumulation of single-stranded telomeric DNA within ALT-associated PML bodies (APBs), and formation of telomeric aggregates at the ends of metaphase chromosomes. This study demonstrates differences between ALT cells and telomerase-positive cells in the requirement for RPA in telomere processing and implicates the ALT mechanism in tumor cells as a possible therapeutic target.
INTRODUCTION
Telomeres are specialized protein–DNA complexes at the ends of linear chromosomes. They consist of arrays of the hexameric nucleotide sequence 5′-TTAGGG-3′ and a number of telomere-associated proteins that function in telomere length regulation or protection of the chromosomal ends from degradation or inappropriate recombination events (1).
Several features distinguish telomeres in alternative lengthening of telomeres (ALT) cells from those of normal cells or telomerase-positive tumor cells. First, while telomerase-positive cells contain telomeres of roughly the same length, telomeres in ALT cells exhibit a high degree of length heterogeneity ranging from critically short to abnormally long (2). Second, ALT cells, but not telomerase-positive cells, contain extrachromosomal circular and linear DNA in addition to telomeres at the chromosomal ends. It is not clear whether these extrachromosomal DNA species simply represent by-products of the telomere recombination activities that take place in ALT cells or if they play an active role in telomere elongation (3–6). Finally, ALT cells have been found to contain special promyelocytic leukemia-nuclear bodies (PML-NBs) termed ALT-associated PML bodies (APBs) (7). These structures contain telomeric DNA, telomere-binding proteins and several proteins with known functions in DNA repair and recombination. A recent study revealed that a proportion of the telomeric DNA that was retained in partially purified APBs is of extrachromosomal origin (8). It has not been determined, however, if these structures also contain genomic telomeres.
Replication protein A (RPA) plays crucial roles in DNA metabolic processes such as recombination, replication and repair (9,10). It is also becoming increasingly clear that this protein is both structurally and functionally related to a family of single-stranded telomere-binding proteins which include POT1-TPP1 in mammals, Pot1 in fission yeast, Cdc13-Stn1-Ten1 in budding yeast and TEBP in ciliated protozoan (11,12). For example, similar to the single-stranded telomere-binding proteins, all three RPA subunits (RPA14, RPA32 and RPA70) contain oligonucleotide/oligosaccharide-binding folds (OB-folds) that facilitate single-stranded DNA binding (9). Furthermore, in vitro studies have shown that RPA is able to bind and unfold the telomeric G-rich 3′ overhangs in a manner similar to that of human POT1 (13,14). Moreover, RPA in Saccharomyces cerevisiae has been shown to regulate telomerase-mediated telomere elongation (15), a feature that appears to be common among the telomere capping family of proteins (16–20). Finally, a role of RPA in telomere maintenance is also suggested by studies showing that combined loss of RPA and Ku70 in budding yeast or RPA and Taz1 in fission yeast causes a telomere shortening phenotype (21,22).
ALT cells and telomerase-positive cells may use different proteins for processing and protection of telomere ends. This notion is supported by recent studies demonstrating that the single-stranded telomere-binding protein Cdc13, which is required for survival in S. cerevisiae, becomes dispensable in post-senescence telomerase-negative yeast strains (so-called type II survivors) that use a telomerase maintenance strategy similar to that of human ALT cells (23–25). In agreement with these studies, we demonstrate here a telomere capping function of RPA that appears to be specific for ALT tumor cells.
MATERIALS AND METHODS
Cell lines
The cell lines GM-847 (SV40 transformed), U2OS (osteosarcoma), HeLa (cerevical cancer), HaCaT (immortalized keratinocytes), SK-LU-1 (non-small cell lung cancer), GM-637 (SV40 transformed) and G292 (osteosarcoma) were cultured in Iscove's modified Dulbecco's medium (IMDM) (BioWhittaker, Verviers, Belgium) containing 10% fetal calf serum (FCS) (BioWhittaker). HEK293 (adenovirus transformed kidney cells) and U87 (glioblastoma) cells were grown in Dulbecco's modified essential medium (DMEM) (BioWhittaker) containing 10% FCS. WI-38-VA13 (SV40 transformed fibroblasts) was grown in Eagle minimal essential medium (EMEM) (BioWhittaker) containing 10% FCS, and SA-OS-2 (Osteosarcoma) and MCF-7 (breast cancer) were grown in RPMI 1640 (BioWhittaker) containing 10% FCS.
Plasmids
Myc epitope-tagged RPA70 was generated by inserting a PCR-generated KpnI–NotI fragment containing the RPA70 coding region into pEF6/myc-His C (Invitrogen, Carlsbad, California). Myc epitope-tagged RPA32 was generated by inserting a PCR-generated KpnI–BstEII fragment containing the RPA32 coding region into pEF6/myc-His C (Invitrogen). All plasmid transfections were performed using the FUGENE6 reagent (Invitrogen).
siRNAs
Cells were transfected using the Oligofectamine transfection reagent (Invitrogen) as described previously (26). Transfected cells were analyzed on Day 3 following transfection. Target sequences were
RPA32#1: GGCTCCAACCAACATTGTT (Ambion)
RPA32#2: GCCTGGTAGCCTTTAAGAT (Ambion)
RPA70#1: GCACTATCATTGCGAATCC (Ambion)
RPA70#2: CACTCTATCCTCTTTCATG (Ambion)
Luciferase (GL2): CTTACGCTGAGTACTTCGA (Eurogentec, Seraing, Belgium)
The RPA32#1 and RPA70#1 targets were used throughout the article. The additional targets (RPA32#2 and RPA70#2) were used in Supplementary Figure S1.
BrdU labeling, immunofluorescence (IF) and laser scanning cytometry (LSC)
IF labeling of cells was performed as previously described (27). To detect ssDNA within PML-NBs, cells grown on coverslips were treated with 10 μM BrdU for 24 h (from Day 2 to Day 3 after transfection with siRNAs) prior to fixation and IF labeling. Cells were prepared for LSC analysis by staining the DNA with TO-PRO-3 (Invitrogen) as previously described (26). Rabbit polyclonal antibodies used were anti-PML (H238; Santa Cruz Biotechnology, Santa Cruz, California). Mouse monoclonal antibodies used were anti-PML (PG-M3; Santa Cruz Biotechnology), anti-RPA32 (Ab-3; Calbiochem, San Diego, California), anti-RPA70 [2H10 (28)], anti-BrdU (Amersham Buckinghamshire, UK), anti-myc (Invitrogen) and anti-TRF2 (Upstate, Waltham, Massachusetts).
Fluorescence in situ hybridization (FISH)-IF
Combined FISH and IF was performed as described previously (27) with some modifications. Cells grown on coverslips were first fixed in 1.5% paraformaldehyde (PFA) in PBS for 5 min and permeabilized in 0.5% Triton X-100 in PBS for 4 min. Subsequently, the cells were stained using rabbit anti-PML (H238; Santa Cruz) or anti-RPA32 (Ab-3; Calbiochem) and fixed again in 4% PFA to cross-link bound antibodies. Cells were then treated with 0.2 mg/ml RNase (Sigma, St Louis, MO) in 2× SSC at 37°C for 15 min and washed three times in PBS.
The telomere-specific probe used in combination with anti-RPA32 was a FITC-conjugated C-rich peptide nucleic acid (PNA) probe (OO[CCCTAA]3) from Applied Biosystems (Foster City, California). The probes used in combination with anti-PML were digoxigenin (DIG) labeled (TTAGGG)3 or (CCCTAA)3. Probes were ethanol precipitated with 20× Cot-1 DNA and diluted in hybridization mix (50% deionized formamide, 10% dextran sulfate, 2× SSC) to a final concentration of 0.6 µg/ml. The probe was denatured for 5 min at 70°C and cooled to 37°C before use. Cells on coverslips were immersed in hybridization mix, and the cellular DNA was denatured for 5 min at 80°C before hybridization at 37°C in a humid environment overnight. For hybridization at native conditions, the denaturing step at 80°C was omitted. Following hybridization, the coverslips were washed three times for 10 min in 0.1× SSC. The DIG-labeled probes were detected by IF as described above using a monoclonal anti-DIG antibody (Roche, Basel, Switzerland).
For preparation of metaphase spreads cells grown in 6 cm dishes were transfected with siRNAs. On Day 3 following transfection, cells were treated with 50 ng/ml colcemid for 1 h and subsequently subjected to hypotonic swelling in 75 mM KCl at room temperature for 15 min. Cells were then fixed and stored in methanol/acetic acid according to standard procedures (29). Telomere-FISH was then performed using the FITC-conjugated PNA probe from Applied Biosystems essentially as described previously (30).
Protein extraction and western blotting
Following transfection in 6 cm culture dishes, cells were detached from the plates by trypsin treatment, washed twice in PBS and subsequently suspended in lysis buffer (20 mM Tris, pH 7.5, 0.42 M KCl, 25% glycerol, 0.1 mM EDTA, 5 mM MgCl, 0.5% Triton X-100, 1 mM DTT, 0.5 mM PMSF, 1 µg/ml leupeptin, 1 µg/ml aprotinin). Following incubation on ice for 30 min, samples were centrifuged at 12 000xg for 10 min and the supernatant was collected. SDS–PAGE and western blots were performed using NuPAGE 4–12% gradient Bis-Tris gels (Invitrogen). Antibodies used were mouse monoclonal anti-RPA32 (Ab-3; Calbiochem), mouse monoclonal anti-RPA70 (2H10) and goat anti-β-actin (I-19; Santa Cruz Biotechnology). Quantitative analysis of western blots was achieved using the Multi Gauge software (Fugifilm).
Pulse-field gel electrophoresis and in-gel detection of telomeric DNA
Total DNA was isolated from transfected cells using the DNeasy tissue kit (Qiagen) and subsequently treated with the restriction endonucleases HinfI and RsaI (New England Biolabs). The digested DNA (3–5 μg) was fractionated on 1% Megabase agarose (Bio-Rad) using the CHEF-DRIII system (Bio-Rad). Electrophoresis was performed in 0.5× TBE, at 14°C for 11 h using 1–6 s switch time and a 120° angle. In-gel hybridization was then performed essentially as described (31). Briefly gels were dried on filter paper at 50°C until completely dry. The gels were rehydrated by soaking in deionized water. Gels were prehybridized in Church buffer (0.5 M Na2HPO4 pH 7.2, 1 mM EDTA, 7% SDS, 1% BSA) for 30 min at 50°C and hybridized at 50°C overnight in the same buffer containing a γ-32P-ATP-labeled (CCCTAA)3 or (TTAGGG)3 telomeric probe. Gels were washed three times for 30 min in 4× SSC at room temperature and once in 4× SSC at 55°C for 1 h and exposed to a PhosphorImager screen overnight. Following exposure, gels were denatured by incubating in 0.5 M NaOH, 1.5 M NaCl for 30 min and neutralized in buffer containing 0.5 M Tris–HCl pH 7.5, 1.5 M NaCl for 20 min. Gels were then rehybridized to the same probe overnight in Church buffer and subsequently washed as described above for the first hybridization.
Analysis of extrachromosomal telomeric DNA by neutral–neutral two-dimensional agarose gel electrophoresis
Extrachromosomal DNA was isolated from cells grown in 6 cm culture dishes as described previously using a modified Hirt-extraction procedure (27). Purified DNA (3 μg) was fractionated by two-dimensional gel electrophoresis essentially as described previously (32). In brief, fractionation in the first dimension was performed in 0.4% agarose in 1× TBE at 1 V/cm for 18 h. The gel was then incubated in 1× TBE buffer containing 0.3 μg/ml ethidium bromide and exposed to UV light in order to excise the lines of interest. The second dimension was run in 1% agarose in 1× TBE containing 0.3 μg/ml ethidium bromide at 5 V/cm for 3.5 h at 4°C. The gels were illuminated with UV light, photographed, and subsequently subjected to in-gel hybridization as described above.
Microscopy
Microscopic images were acquired using a Zeiss LSM 510 Meta laser confocal microscope connected (Jena, Germany) to a 63× oil immersion objective (Zeiss). Fluorescence intensity diagrams were generated using the Zeiss LSM image examiner software.
RESULTS
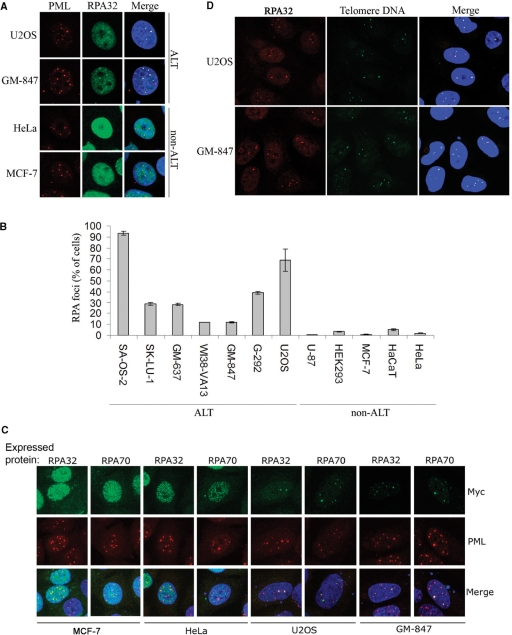
In the present study, we found that different cell lines exhibit a difference in the ability to sequester RPA32 within PML-NBs. For example, the ALT cell lines, GM-847 and U2OS, were found to contain PML-NB-associated RPA32 foci in 15–60% of the cells, whereas the telomerase-positive cell lines, HeLa and MCF-7, did not contain such foci and exhibited a dispersed nuclear RPA32 staining pattern (Figure 1A).
Figure 1.
RPA foci that colocalize with PML-NBs are formed in ALT cells, but not in telomerase-positive cells. (A) HeLa, MCF-7, GM-847 and U2OS cells were grown on coverslips, fixed, and fluorescently labeled using antibodies against PML (red) and RPA32 (green). DAPI is shown in blue in the merged images. (B) Quantification of cells with RPA32-containing foci in ALT and non-ALT cell lines. Cells that contained one or more distinct RPA32 focus that colocalized with a PML-NB were scored. For each sample, between 600 and 800 cells were examined. Data represent two independent experiments ± standard deviation (SD). (C) Visualization of ectopically expressed myc-tagged RPA32 and RPA70 in ALT and non-ALT cell lines. Myc-tagged proteins and PML-NBs were fluorescently labeled using anti-myc (green) and anti-PML (red) antibodies, respectively. (D) Asynchronous GM-847 and U2OS cells grown on coverslips were subjected to FISH-IF using a FITC-conjugated telomeric PNA probe (green) and anti-RPA32 (red).
To verify that localization of RPA to PML-NBs is a characteristic specifically associated with cells that use ALT, we examined the subnuclear distribution of RPA32 in a panel of cell lines with known telomere maintenance mechanisms. We found that cell cultures using ALT consistently contained a relatively high fraction (15–90%) of cells with RPA32-positive PML-NBs. In contrast, the fraction of cells containing such foci was considerably lower (below 5%) in all of the telomerase-positive cells tested (Figure 1B).
To further demonstrate a difference between ALT cells and telomerase-positive cells in the ability to sequester RPA within PML-NBs, we ectopically expressed myc-tagged RPA32 or RPA70 in ALT and non-ALT cell lines. Both of the expressed RPA subunits colocalized with PML-NBs in more than 20% of transfected U2OS or GM-847 cells, whereas the non-ALT cell lines, HeLa and MCF-7, generally exhibited a dispersed nuclear distribution of ectopically expressed RPA (Figure 1C).
The observation that RPA associates with PML-NBs in ALT cells, and not in telomerase-positive cells, is in accordance with a previous study showing that this protein represents one of the constituents of APBs (7). We therefore performed FISH on asynchronous growing U2OS and GM-847 cells to verify that the RPA-containing nuclear structures contained telomeric DNA. We found that nearly 100% of the RPA32 foci detected in these cells colocalized with telomere repeat sequences (Figure 1D). This result shows that the RPA-containing foci detected in ALT cell lines are equivalent to APBs and implicates a role of RPA in the ALT mechanism.
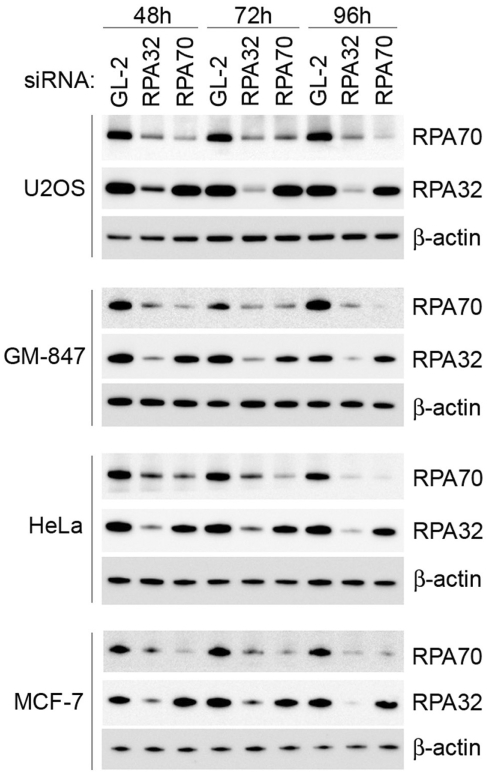
To study the function of RPA in ALT, we transfected four different cell lines, including two ALT cell lines (GM-847 and U2OS) and two telomerase-positive cell lines (HeLa and MCF-7), with a control siRNA against luciferase (GL-2) or siRNAs targeting either RPA32 or RPA70. All four cell lines showed reduced RPA expression at 48 h following transfection with RPA-specific siRNAs (Figure 2; see Supplementary Figure 1 for quantification of RPA expression). The siRNA against RPA32 caused decreased expression of both RPA32 and RPA70. This phenomenon was noted also in a previous study and may reflect a role of RPA32 in stabilizing the RPA complex (33).
Figure 2.
SiRNA depletion of RPA32 and RPA70 in ALT and telomerase-positive cell lines. Cells were transfected with the siRNAs indicated and subsequently harvested at 48, 72 and 96 h post-transfection. Western blots were probed with antibodies against RPA70 and RPA32. Blots were then stripped and subsequently re-probed with antibodies against β-actin. Luciferase siRNAs (GL-2) were used as a control. A quantitative representation of these data is presented in Supplementary Figure S1.
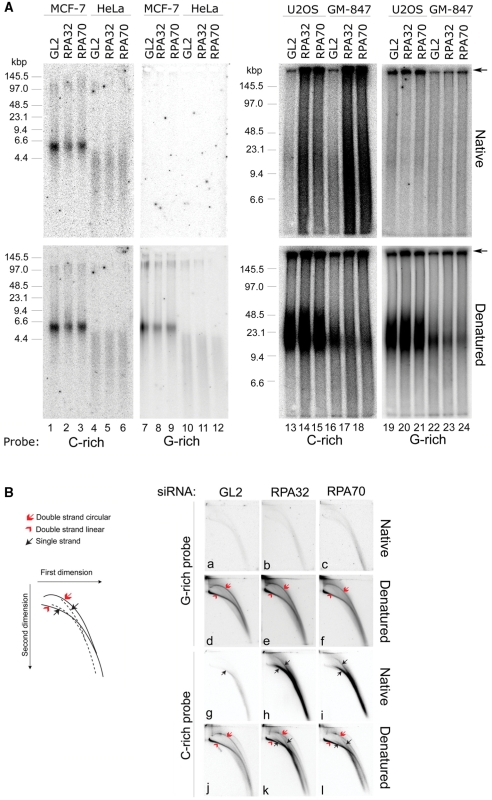
We first analyzed the telomeres in RPA-depleted and control-depleted cells using telomere restriction fractionation by pulse-field gel electrophoresis and subsequent in-gel hybridization under either native or denaturing conditions. Hybridization under native conditions revealed an increase in single-stranded G-rich telomeric DNA in GM-847 and U2OS cells following reduction of RPA32 or RPA70 expression (Figure 3A, upper panels, lanes 13–18). In contrast, we detected no changes in exposed G-rich telomeric ssDNA in response to RPA loss in the non-ALT cell lines HeLa or MCF-7 (Figure 3A, upper panels, lanes 1–6). In the ALT cells we also detected single-stranded C-rich telomeric DNA, but in this case we did not observe an increase following targeted depletion of RPA (Figure 3A, upper panels, lanes 19–24). In contrast, we did not detect single-stranded C-rich telomeric DNA in the non-ALT cell lines MCF-7 and HeLa (Figure 3A, upper panels, lanes 7–12). Interestingly, a large proportion of the single-stranded telomeric DNA detected in RPA-depleted ALT cells was retained near the well of the gel (Figure 3A, lanes 13–24). Such high molecular weight telomeric DNA species that do not enter the gel during electrophoresis are commonly detected in ALT cells by telomere restriction analysis and may represent complex recombination intermediates (7). This observation may indicate that RPA loss mainly affects DNA that is engaged in recombinational telomere elongation. Interestingly, hybridization at denatured conditions also revealed a net increase of these slow-migrating DNA species suggesting that RPA may prevent their formation (Figure 3A, lower panels, lanes 13–24). The increase in G-rich telomeric ssDNA detected in these experiments was not due to off-target effects caused by the siRNAs used, as the same result was obtained by using a second set of siRNAs targeting different regions of the RPA32 and RPA70 mRNA sequence (Supplementary Figure S2).
Figure 3.
Depletion of RPA32 or RPA70 in ALT cells leads to increased levels of single-stranded G-rich telomeric DNA. Luciferase siRNA (GL2) was used as a control. (A) Analysis of telomere restriction fragments by pulse-field gel electrophoresis and in-gel hybridization. The gels were first hybridized using native conditions (upper panels). The same gels were subsequently denatured and then rehybridized to the same probes (lower panels). A C-rich ([CCCTAA]3) or a G-rich ([TTAGGG]3) telomere-specific probe was used. Arrows indicate the position of the wells. (B) Analysis of Hirt-extracted extrachromosomal DNA from GM-847 cells by two-dimensional agarose gel electrophoresis and in-gel hybridization. Gels were first hybridized to telomere-specific probes using native conditions. The same gels were then denatured and rehybridized using the same probe. Triplicate panels were equally exposed.
To determine if RPA loss in ALT cells affected the integrity of extrachromosomal telomeric DNA, we analyzed low molecular weight DNA isolated from transfected GM-847 cells using the Hirt extraction procedure. For these experiments, the purified DNA species were fractionated by two-dimensional gel electrophoresis to allow separation of circular and linear as well as single-stranded and double-stranded DNA species (Figure 3B). Native in-gel hybridization using a C-rich telomeric probe revealed an increase in the level of single-stranded G-rich extrachromosomal telomere repeats following RPA loss (Figure 3B, panels g–i). Conversely, we detected no increase in single-stranded C-rich extrachromosomal DNA following RPA depletion (Figure 3B, panels a–c). Similar results were obtained by analyzing extrachromosomal DNA isolated from RPA- and control-depleted U2OS cells (Supplementary Figure S3). We did not detect any increase or decrease in the amount of circular double-stranded telomeric DNA in cells with reduced RPA expression (Figure 3B, Panels d–f and j–l). Although the Hirt extraction procedure is highly selective for low molecular extrachromosomal DNA, we cannot exclude the possibility that some of the telomeric DNA detected in these samples is of genomic origin as single-stranded DNA is expected to be particularly sensitive to shearing forces.
Due to the central role of RPA in DNA replication, reduced expression of this protein complex is expected to cause defects in DNA synthesis and, consequently, replication stress. To assess the possibility that the increase in single-stranded telomeric DNA is caused by frequent replication fork stalling or inhibition of DNA synthesis, we treated the ALT cell line U2OS with different concentrations of the DNA synthesis inhibitors hydroxyurea or aphidicolin and subsequently determined the level of single-stranded G-rich telomeric DNA. The DNA synthesis inhibitors did not cause a significant change in the levels of single-stranded G-rich telomeric DNA neither at low concentrations of the drugs that will cause frequent replication fork stalling and replication stress, nor at higher concentrations that is expected to cause severe inhibition of DNA synthesis (Supplementary Figure 4).
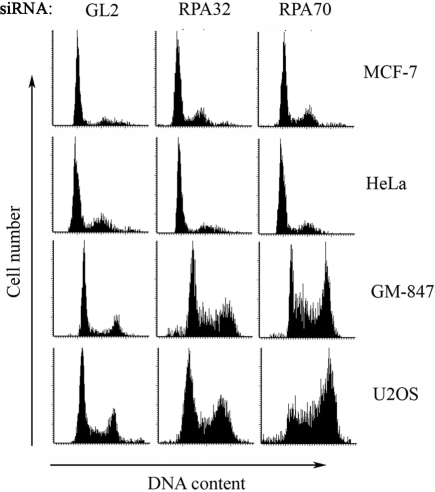
Figure 4.
RPA loss in ALT cells causes cell cycle arrest in G2/M phase. Cells on coverslips were transfected by the indicated siRNAs and subjected to laser scanning cytometry on Day 3 following transfection.
Previous studies have shown that loss of Cdc13 in S. cerevisiae or Pot1 in chicken causes an increase in single-stranded telomeric DNA and subsequent activation of cell cycle checkpoints (34,35). Furthermore, siRNA-mediated depletion of RPA70 in HeLa cells has been found to cause checkpoint activation and cell cycle arrest in G2/M phase at Day 4 following transfection (33,36). To determine the effect of RPA loss on cell cycle progression under the conditions used in the present study, we analyzed cell cycle distribution in control and RPA-depleted cells at Day 3 following siRNA transfection. Both ALT cell lines (GM-847 and U2OS) showed an increase of cells in late S and G2/M phase in response to RPA loss (Figure 4). The arrest was less pronounced in RPA32-depleted cells compared to RPA70-depleted cells, possibly due to a role of RPA32 (but not RPA70) in activating the G2/M checkpoint. In contrast to the ALT cells, we did not observe a dramatic alteration in the cell cycle distribution following transfection of the telomerase-positive cell lines, MCF-7 and HeLa, with siRNAs against RPA (Figure 4). Thus, ALT cells appear to arrest more readily than the telomerase-positive cell lines in G2/M phase following RPA loss.
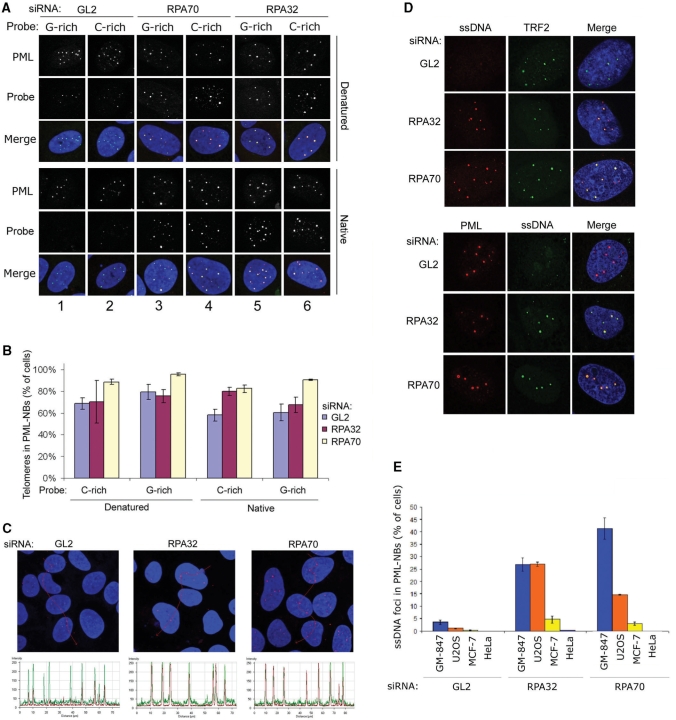
We next analyzed the effect of RPA depletion on the nuclear localization of telomeric DNA using FISH. Also for these experiments samples were hybridized at both native and denatured conditions and by using probes specific for both telomeric strands (Figure 5A). Loss of RPA70 or RPA32 did not cause a significant increase in the number of cells containing detectable telomeric DNA within PML-NBs (Figure 5B). However, the telomeric hybridization signal within individual PML-NBs was considerably intensified following RPA depletion (as indicated by the increase in telomere hybridization (red) relative to the PML staining (green) in Figure 5C and Supplementary Figure S5). This effect was evident using both C- and G-rich telomeric probes and for hybridization of both native and denatured samples (Figure 5C and Supplementary Figure S5).
Figure 5.
APBs accumulate ssDNA in response to RPA loss. (A) FISH analysis showing colocalization of telomeres and PML-NBs in U2OS cells transfected with control siRNAs (GL2) and siRNAs against RPA32 and RPA70. Cells were hybridized under native and denaturing conditions, with either a G-rich ([TTAGGG]3) or a C-rich ([CCCTAA]3) telomeric probe. (B) Quantification of U2OS cells containing detectable telomeric signals that colocalize with PML-NBs. Cells containing one or more telomeric signal that colocalize with a PML-NB were scored. For each sample between 70 and 80 cells were examined. Data represent two independent experiments ± SD. (C) Fluorescence intensity diagram of telomeric DNA and PML in siRNA-transfected cells. Red (telomeric DNA), green (PML) and blue (DAPI) is overlaid in each of the images. The intensity diagrams shown in the lower panels are plotted against the red line shown in the upper panels. Telomere signal is represented by red curves and PML signal is represented by green curves. The example shows U2OS cells hybridized using a C-rich probe at native conditions. The complete figure showing all of the hybridization conditions is shown in Supplementary Figure S5. (D) Visualization of single-stranded DNA in control-depleted or RPA-depleted U2OS cells using anti-BrdU antibodies in combination with anti-TRF2 (upper panels) or anti-PML (lower panels). (E) Quantification of ssDNA foci that colocalize with PML-NBs in control-transfected cells (GL2) and cells transfected with RPA32- and RPA70-specific siRNAs. Cells containing one or more distinct ssDNA foci within a PML-NB were scored. For each sample 600 to 800 cells were examined. Data represent two independent experiments ± SD.
We next assessed the effect of RPA depletion on the distribution of the general pool of ssDNA. For this experiment we employed a previously described ssDNA detection assay, which is based on metabolic labeling of cells with bromodeoxyuridine (BrdU) and subsequent detection of BrdU-containing ssDNA by fluorescence labeling under non-denaturing conditions (26). We found that siRNA-mediated knockdown of RPA32 or RPA70 in the ALT cell lines GM-847 and U2OS caused a considerable increase in the number of cells containing detectable ssDNA within PML-NBs (Figure 5D and E). These ssDNA foci were found to colocalize with the telomere-binding protein TRF2 in more than 90% of the cells examined, indicating that the ssDNA detected was telomeric (Figure 5D, upper panels). We observed a modest increase in cells containing ssDNA within PML-NBs following transfection of the non-ALT cell line MCF-7 with RPA32- and RPA70-specific siRNAs (Figure 5E). However, in these cases we detected only one or two ssDNA-positive PML-NB per nucleus, and they were never found to colocalize with TRF2 (data not shown). HeLa cells were virtually devoid of ssDNA foci both before and after depletion of RPA32 or RPA70 (Figure 5E).
We concluded from these experiments that RPA loss in ALT cells leads to an increase of telomere DNA repeats within PML-NBs. Furthermore, since these telomeric DNA species are detected under both native and denaturing hybridization conditions, and because of the general increase of ssDNA within this nuclear compartment following RPA loss, a proportion of the telomeric DNA residing in PML-NBs subsequent to RPA depletion is likely to be single stranded.
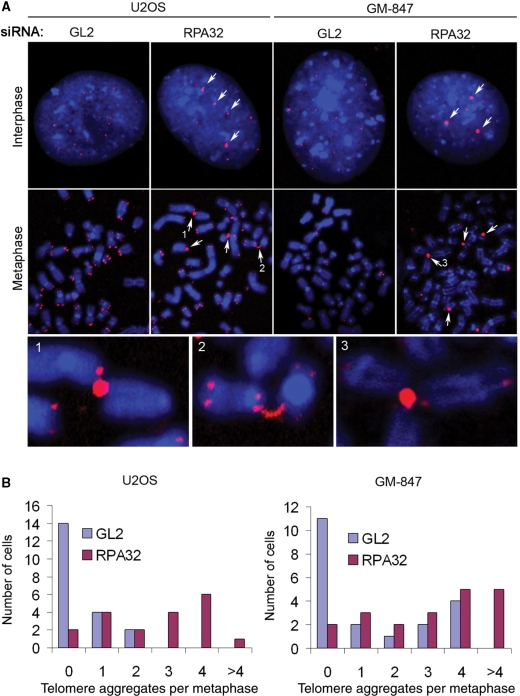
We then analyzed the telomeres in metaphase spreads prepared from control and RPA32-depleted cells using FISH. We made attempts to analyze spreads prepared from RPA70-depleted cells, but this proved difficult due to a low frequency of metaphases. In the RPA32-depleted U2OS and GM-847 cells, we detected telomere signals in metaphases that had the same shape and fluorescence intensity as those detected in interphase cells (Figure 6A, arrows). In most cases these abnormally strong telomere signals were found to associate with chromosomal ends, indicating they may represent very large telomeres. Furthermore, ∼30% of these telomeric aggregates appeared to have connections with more than one chromosomal end, suggesting that they may be involved in telomeric recombination events (Figure 6A, bottom panels). To quantify the occurrence of these large telomere aggregates, we analyzed 20 metaphases from each of the transfected samples. We found that reduced RPA32 expression in GM-847 and U2OS cells caused a considerable increase in the number of the abnormally intense telomeric signals, suggesting that RPA prevented their formation (Figure 6B). Large telomere aggregates at chromosomal ends were not detected in the non-ALT cells HeLa or MCF-7 (data not shown).
Figure 6.
Loss of RPA causes formation of telomere aggregates at chromosomal ends. U2OS and GM-847 cells were transfected with control (GL2) siRNAs or siRNAs specific for RPA32. On Day 3 following transfection metaphase spreads were prepared and telomeres were visualized using a C-rich ([CCCTAA]3) telomeric PNA probe. (A) Visualization of telomeres in metaphase and interphase of control-depleted (GL2) and RPA32-depleted U2OS and GM-847 cells. Arrows point to abnormally strong telomere signals. Selected telomere fusions (numbered 1 to 3) are shown at high magnification in the lower panels. (B) Quantification of abnormally strong telomere signals in metaphase cells. For each sample 20 metaphase spreads were scored.
We also noted other chromosomal aberrations in metaphases prepared from RPA32-depleted cells, including chromosomal breakage and fragmentation. However, these aberrations were detected with approximate equal frequency in both the ALT and non-ALT cell lines suggesting that they may be caused due to a general defect in DNA repair or DNA replication (data not shown).
DISCUSSION
In the present study, we have shown that loss of RPA in human ALT tumor cells causes an increase in single-stranded G-rich telomeric DNA, cell cycle arrest in G2/M phase, increased accumulation of telomeric DNA within APBs and generation of large telomeric aggregates that associate with the ends of metaphase chromosomes. The mechanism by which RPA interacts with telomeres in order to prevent the onset of these phenotypes may be similar to that of other single-stranded telomere-binding proteins. This is suggested by the structural conservation between RPA and members of the single-strand telomere-binding family of proteins (11,12), and by studies also showing that S. cerevisiae Cdc13 and chicken Pot1 prevent exposure of single-stranded G-rich telomere DNA and subsequent G2/M phase arrest (34,37).
Loss of RPA in U2OS and GM-847 cells appears to enhance several of the ALT-specific phenotypic features. First, RPA depletion induces the formation of telomeric DNA that has poor electrophoretic mobility in agarose gels following treatment with restriction enzymes. Such DNA fragments that are retained near the well of the gel are normally detected in ALT cells and may represent recombining DNA (7). Interestingly, a large proportion of the single-stranded telomeric DNA that formed in response to RPA loss was also found to associate with these putative recombination intermediates. This observation may suggest that formation of the single-stranded telomeric DNA in cells with reduced RPA expression is coupled to recombinational telomere elongation.
The second ALT-specific characteristic that becomes enhanced by reduced RPA expression is the presence of telomeric DNA repeats within APBs. RPA depletion did not lead to a significant increase in the number of cells containing detectable telomeric DNA within PML-NBs, suggesting that RPA loss did not lead to a net increase in the number of APBs. Rather, reduced expression of this protein complex seemed to cause an increase in the amount of telomeric DNA present within pre-existing APBs. RPA knockdown also caused accumulation of single-stranded DNA within APBs as determined by native IF labeling of metabolically incorporated BrdU. However, despite the clear increase of single-stranded G-rich versus C-rich telomeric strands that was revealed by in-gel hybridization, we were not able to demonstrate a specific increase of single-stranded telomeric G-strands within APBs using a native telomere FISH procedure (hybridization without prior heat denaturing). This result may be due to the presence of inherent denatured regions of duplex telomeric DNA during the FISH procedure. Alternatively, the increase in both G and C-strand signal in APBs under these conditions may reflect a recruitment mechanism that sequesters telomeric DNA within APBs, and consequently intensifies the signals from both strands.
Finally, analysis of metaphase spreads revealed that loss of RPA in the ALT cell lines GM-847 and U2OS caused an increase in large telomeric occlusions that are associated with one or multiple chromosomal ends. It is possible that these large telomeric structures consist of extremely long telomeres that have been generated at chromosomal ends. Alternatively, they may represent aggregates of extrachromosomal telomeric repeats that are contacting the ends of the chromosomes. The number and size of these metaphase structures were similar to the telomere aggregates detected in interphase cells within APBs, suggesting that they may represent the same structures. These observations are consistent with the notion that APBs are in contact with one or several chromosomal ends and that these nuclear compartments represent sites of recombinational telomere elongation.
One possible mechanism for recombinational telomere elongation in ALT cells that has received strong experimental support is telomere synthesis by rolling-circle replication (3–5,38–40). According to this model, relatively large tracts of telomeric DNA can be synthesized essentially in one step by using extrachromosomal circular telomeres as a template. This mechanism is predicted to cause an abrupt elongation of the telomere 3′ overhang and may therefore account for the increase in single-stranded telomeric G-rich DNA that is seen in ALT cells with reduced RPA expression. RPA may function to inhibit initiation of such template-directed DNA synthesis simply by preventing annealing of the single-stranded 3′ telomeric overhang to partially single-stranded telomere circles. Alternatively, RPA could occlude the single-stranded telomeric DNA in a way that prevents Rad51-mediated strand invasion of the 3′ end into a duplex telomere sequence. This latter possibility is consistent with a previous study showing that RPA has the ability to suppress direct-repeat recombination in S. cerevisiae (41).
The single-stranded G-rich DNA may also be formed in RPA-depleted cells due to exposure of the C-rich telomeric DNA to one or several 5′ exonucleases. Such protection of telomeres from C-strand resection has been proposed to be an important function of S. cerevisiae Cdc13, which together with Stn1 and Ten1 has been suggested to form an RPA-like protein complex (12). However, a role of RPA in protection from end resection seems unlikely to be the full explanation for the increase of telomeric DNA within PML-NBs and at chromosomal ends, which can be observed in RPA-depleted ALT cells. Nor does it explain why RPA loss affects the level of single-stranded G-rich telomeric DNA in ALT cells and not in telomerase-positive cells.
The rolling-circle mechanism of telomere extension also predicts the involvement of a helicase that unwinds the duplex DNA ahead of the elongating strand. Interestingly, a previous study has demonstrated that overexpression of the Bloom syndrome helicase BLM in the ALT cell line WI38-VA13 causes increased recruitment of telomeric DNA to PML-NBs in a manner similar to that observed in RPA-depleted ALT cells (42). Thus, RPA and BLM may cooperate in the process of telomere elongation in ALT cells. It is possible, for example, that RPA has an inhibitory effect on BLM at telomeric ends.
Until now, telomerase-mediated telomere elongation is the only telomere maintenance strategy being actively pursued as a target in cancer therapy. While such therapies are expected to prevent growth of telomerase-positive tumors, they are not likely to be effective against telomerase-negative cancer cells that use ALT. RPA represents the first example of a protein that is required for telomere protection in cells employing the ALT mechanism of telomere elongation. The present study suggests that the development of potent therapeutic strategies that specifically target tumor cells using ALT may involve the exploitation of RPA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The cost of this work was supported by the Cancer Gene Therapy Program funded by the Norwegian Health Department and Helse-Vest. Support from the Norwegian Cancer Society and the Norwegian Research Council is also acknowledged. Funding to pay the Open Access publication charges for this article was provided by the Norwegian Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 2.Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- 3.Cesare AJ, Griffith JD. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol. Cell. Biol. 2004;24:9948–9957. doi: 10.1128/MCB.24.22.9948-9957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Natarajan S, McEachern MJ. Recombinational telomere elongation promoted by DNA circles. Mol. Cell. Biol. 2002;22:4512–4521. doi: 10.1128/MCB.22.13.4512-4521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Compton SA, Choi JH, Cesare AJ, Ozgur S, Griffith JD. Xrcc3 and Nbs1 are required for the production of extrachromosomal telomeric circles in human alternative lengthening of telomere cells. Cancer Res. 2007;67:1513–1519. doi: 10.1158/0008-5472.CAN-06-3672. [DOI] [PubMed] [Google Scholar]
- 7.Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- 8.Fasching CL, Neumann AA, Muntoni A, Yeager TR, Reddel RR. DNA damage induces alternative lengthening of telomeres (ALT) associated promyelocytic leukemia bodies that preferentially associate with linear telomeric DNA. Cancer Res. 2007;67:7072–7077. doi: 10.1158/0008-5472.CAN-07-1556. [DOI] [PubMed] [Google Scholar]
- 9.Binz SK, Sheehan AM, Wold MS. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst.) 2004;3:1015–1024. doi: 10.1016/j.dnarep.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 10.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 11.Croy JE, Wuttke DS. Themes in ssDNA recognition by telomere-end protection proteins. Trends Biochem. Sci. 2006;31:516–525. doi: 10.1016/j.tibs.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 13.Salas TR, Petruseva I, Lavrik O, Bourdoncle A, Mergny JL, Favre A, Saintome C. Human replication protein A unfolds telomeric G-quadruplexes. Nucleic Acids Res. 2006;34:4857–4865. doi: 10.1093/nar/gkl564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl Acad. Sci. USA. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schramke V, Luciano P, Brevet V, Guillot S, Corda Y, Longhese MP, Gilson E, Geli V. RPA regulates telomerase action by providing Est1p access to chromosome ends. Nat. Genet. 2004;36:46–54. doi: 10.1038/ng1284. [DOI] [PubMed] [Google Scholar]
- 16.Chandra A, Hughes TR, Nugent CI, Lundblad V. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 2001;15:404–414. doi: 10.1101/gad.861001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colgin LM, Baran K, Baumann P, Cech TR, Reddel RR. Human POT1 facilitates telomere elongation by telomerase. Curr. Biol. 2003;13:942–946. doi: 10.1016/s0960-9822(03)00339-7. [DOI] [PubMed] [Google Scholar]
- 18.Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 19.Froelich-Ammon SJ, Dickinson BA, Bevilacqua JM, Schultz SC, Cech TR. Modulation of telomerase activity by telomere DNA-binding proteins in Oxytricha. Genes Dev. 1998;12:1504–1514. doi: 10.1101/gad.12.10.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 21.Kibe T, Ono Y, Sato K, Ueno M. Fission yeast Taz1 and RPA are synergistically required to prevent rapid telomere loss. Mol. Biol. Cell. 2007;18:2378–2387. doi: 10.1091/mbc.E06-12-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith J, Zou H, Rothstein R. Characterization of genetic interactions with RFA1: the role of RPA in DNA replication and telomere maintenance. Biochimie. 2000;82:71–78. doi: 10.1016/s0300-9084(00)00183-8. [DOI] [PubMed] [Google Scholar]
- 23.Larrivee M, Wellinger RJ. Telomerase- and capping-independent yeast survivors with alternate telomere states. Nat. Cell Biol. 2006;8:741–747. doi: 10.1038/ncb1429. [DOI] [PubMed] [Google Scholar]
- 24.Petreaca RC, Chiu HC, Eckelhoefer HA, Chuang C, Xu L, Nugent CI. Chromosome end protection plasticity revealed by Stn1p and Ten1p bypass of Cdc13p. Nat. Cell Biol. 2006;8:748–755. doi: 10.1038/ncb1430. [DOI] [PubMed] [Google Scholar]
- 25.Zubko MK, Lydall D. Linear chromosome maintenance in the absence of essential telomere-capping proteins. Nat. Cell Biol. 2006;8:734–740. doi: 10.1038/ncb1428. [DOI] [PubMed] [Google Scholar]
- 26.Boe SO, Haave M, Jul-Larsen A, Grudic A, Bjerkvig R, Lonning PE. Promyelocytic leukemia nuclear bodies are predetermined processing sites for damaged DNA. J. Cell Sci. 2006;119:3284–3295. doi: 10.1242/jcs.03068. [DOI] [PubMed] [Google Scholar]
- 27.Jul-Larsen A, Visted T, Karlsen BO, Rinaldo CH, Bjerkvig R, Lonning PE, Boe SO. PML-nuclear bodies accumulate DNA in response to polyomavirus BK and simian virus 40 replication. Exp. Cell Res. 2004;298:58–73. doi: 10.1016/j.yexcr.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 28.Kenny MK, Schlegel U, Furneaux H, Hurwitz J. The role of human single-stranded DNA binding protein and its individual subunits in simian virus 40 DNA replication. J. Biol. Chem. 1990;265:7693–7700. [PubMed] [Google Scholar]
- 29.Lee JJ, Warburton D, Robertson EJ. Cytogenetic methods for the mouse: preparation of chromosomes, karyotyping, and in situ hybridization. Anal. Biochem. 1990;189:1–17. doi: 10.1016/0003-2697(90)90036-9. [DOI] [PubMed] [Google Scholar]
- 30.Zijlmans JM, Martens UM, Poon SS, Raap AK, Tanke HJ, Ward RK, Lansdorp PM. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc. Natl Acad. Sci. USA. 1997;94:7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karlseder J, Smogorzewska A, de Lange T. Senescence induced by altered telomere state, not telomere loss. Science. 2002;295:2446–2449. doi: 10.1126/science.1069523. [DOI] [PubMed] [Google Scholar]
- 32.Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 33.Dodson GE, Shi Y, Tibbetts RS. DNA replication defects, spontaneous DNA damage, and ATM-dependent checkpoint activation in replication protein A-deficient cells. J. Biol. Chem. 2004;279:34010–34014. doi: 10.1074/jbc.C400242200. [DOI] [PubMed] [Google Scholar]
- 34.Churikov D, Wei C, Price CM. Vertebrate POT1 restricts G-overhang length and prevents activation of a telomeric DNA damage checkpoint but is dispensable for overhang protection. Mol. Cell. Biol. 2006;26:6971–6982. doi: 10.1128/MCB.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Araya R, Hirai I, Meyerkord CL, Wang HG. Loss of RPA1 induces Chk2 phosphorylation through a caffeine-sensitive pathway. FEBS Lett. 2005;579:157–161. doi: 10.1016/j.febslet.2004.11.066. [DOI] [PubMed] [Google Scholar]
- 37.Weinert TA, Hartwell LH. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993;134:63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin CY, Chang HH, Wu KJ, Tseng SF, Lin CC, Lin CP, Teng SC. Extrachromosomal telomeric circles contribute to Rad52-, Rad50-, and polymerase delta-mediated telomere-telomere recombination in Saccharomyces cerevisiae. Eukaryot. Cell. 2005;4:327–336. doi: 10.1128/EC.4.2.327-336.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomaska L, McEachern MJ, Nosek J. Alternatives to telomerase: keeping linear chromosomes via telomeric circles. FEBS Lett. 2004;567:142–146. doi: 10.1016/j.febslet.2004.04.058. [DOI] [PubMed] [Google Scholar]
- 40.Tomaska L, Nosek J, Makhov AM, Pastorakova A, Griffith JD. Extragenomic double-stranded DNA circles in yeast with linear mitochondrial genomes: potential involvement in telomere maintenance. Nucleic Acids Res. 2000;28:4479–4487. doi: 10.1093/nar/28.22.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith J, Rothstein R. A mutation in the gene encoding the Saccharomyces cerevisiae single-stranded DNA-binding protein Rfa1 stimulates a RAD52-independent pathway for direct-repeat recombination. Mol. Cell. Biol. 1995;15:1632–1641. doi: 10.1128/mcb.15.3.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stavropoulos DJ, Bradshaw PS, Li X, Pasic I, Truong K, Ikura M, Ungrin M, Meyn MS. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum. Mol. Genet. 2002;11:3135–3144. doi: 10.1093/hmg/11.25.3135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.