Abstract
To add a tag-peptide for degradation to the nascent polypeptide in a stalled ribosome, an unusual translation called trans-translation is facilitated by transfer-messenger RNA (tmRNA) having an upper half of the tRNA structure and the sequence encoding the tag-peptide except the first alanine. During this event, tmRNA enters the vacant A-site of the stalled ribosome without a codon–anticodon interaction, but with a protein factor SmpB. Here, we studied the sites and modes of binding of SmpB to the ribosome by directed hydroxyl radical probing from Fe(II) tethered to SmpB variants. It revealed two SmpB-binding sites, A-site and P-site, on the ribosome. Each SmpB can be superimposed on the lower half of tRNA behaving in translation. The sites of cleavages from Fe(II) tethered to the C-terminal residues of A-site SmpB are aligned along the mRNA path towards the downstream tunnel, while those of P-site SmpB are found almost exclusively around the region of the codon–anticodon interaction in the P-site. We propose a new model of trans-translation in that the C-terminal tail of SmpB initially recognizes the decoding region and the mRNA path free of mRNA by mimicking mRNA.
INTRODUCTION
Transfer-messenger RNA (tmRNA, also known as 10Sa RNA or SsrA RNA) is widely distributed among eubacteria and has also been found in some chloroplasts and mitochondria (1,2). It is a unique molecule in that it has both tRNA and mRNA properties. The upper half of this molecule structurally mimics the amino acid acceptor stem and the T-arm of tRNA (tRNA-like domain; TLD) (3–5), and its 3′-terminal is actually aminoacylated with alanine by alanyl-tRNA synthetase (3,4). In the lower half of tmRNA, the mRNA domain encoding the tag-peptide except the first alanine residue is surrounded by four pseudoknot structures (6,7). The molecular interplay between these two functions of this molecule facilitates an unusual translation reaction, trans-translation, in which a ribosome can switch from the translation of a truncated mRNA to the mRNA domain of tmRNA (8,9). This relieves the stalled translation from mRNAs lacking a stop codon or possessing a cluster of rare codons with the addition of a tag-peptide as a degradation signal to the truncated C-terminal of polypeptides decoded. These processes may promote recycling of ribosomes and degradation of truncated mRNAs and may prevent accumulation of abortively synthesized polypeptides (10,11). Thus, the trans-translation system is involved in normal cell growth (12) and other biological events (13–16).
The mechanism by which tmRNA resumes translation from the first GCA codon for a tag-peptide is yet mysterious, since no apparent codon–anticodon interaction is involved in binding to the ribosome. Some crucial interaction of the upstream of the tag-encoding region of tmRNA with anywhere on the ribosome has been assumed as a substitute for a codon–anticodon interaction (17,18). However, whether it occurs directly or via a trans-acting factor and how it is dynamically changed during the course of the trans-translation processes have yet to be clarified. Several tmRNA-binding proteins, EF-Tu, SmpB and S1, have been identified (19–22). Among them, SmpB is a sole factor shown to be essential for trans-translation, but it is dispensable for canonical translation (20,23–25). SmpB binds the TLD of tmRNA (23,26–28) to recruit tmRNA to the stalled ribosome (20,23), enhance the alanylation of tmRNA (23,24,29) and protect tmRNA from degradation (20,30). According to NMR studies, SmpB is comprised of an antiparallel beta barrel core with three alpha helices and C-terminal basic residues that are disordered in solution (31,32). In the crystal structure of SmpB in complex with a TLD fragment from Aquifex aeolicus (26), SmpB binds the elbow of TLD, and SmpB can be superimposed on the anticodon stem and loop of tRNA, if TLD is fixed on the amino acid acceptor stem and the T-arm. This raises the possibility that SmpB functions around the decoding region during trans-translation. A recent cryo-EM reconstitution of a complex of ribosome, alanyl-tmRNA, EF-Tu with GDP and kirromycin from Thermus thermophilus also supports this view (28). Although with no information about the structure and location of the C-terminal tail in the ribosome, its truncation leads to a loss of the ribosomal processes of trans-translation (33,34).
It has recently been reported that SmpB can bind to the ribosome in the absence of tmRNA (35), which has enforced us to reconsider the pathway of trans-translation. In the present study, we studied the sites and modes of binding of Escherichia coli SmpB to the ribosome by directed hydroxyl radical probing with Fe(II)-BABE. Fe(II)-BABE is a specific modifier of the cysteine residue of a protein, which generates hydroxyl radicals to cleave the RNA chain, allowing mapping amino acid residues of a binding protein on an RNA-based macromolecule such as the ribosome (36,37). It revealed two SmpB-binding sites on the ribosome. SmpB apparently mimics not only the anticodon stem and loop of tRNA but also mRNA during trans-translation.
MATERIALS AND METHODS
Preparation of factors for trans-translation
Mutations were introduced to a plasmid for overproduction of His-tagged SmpB from E. coli (23). SmpB and tmRNA were prepared as described previously (23,38). 70S ribosomes were prepared from E. coli W3110 ΔssrAΔSmpB (9).
Conjugation of Fe(II)-BABE to SmpB
1.5 nmol of each SmpB derivative was incubated with 20 nmol Fe(II)-BABE in 100 µl solution containing 50 mM MOPS (pH 8.2), 100 mM NaCl, 1 mM EDTA and 5% glycerol at 37°C for 1 h. Free Fe(II)-BABE was excluded by gel filtration.
Formation of the complex of SmpB and ribosome
70S ribosomes (20 pmol) were incubated with or without a synthetic mRNA with the sequence of 5′-AAGGAGGUAAAAAUG-3′ (50 pmol) and tRNAfMet (100 pmol) at 37°C for 10 min. They were subsequently incubated with Fe(II)-tethered SmpB (150 pmol) in 50 µl of 50 mM MOPS (pH 8.0), 120 mM NaCl, 0.1 mM EDTA, 10 mM MgCl2 and 10% glycerol at 37°C for 10 min.
Directed hydroxyl radical probing
Ribosome or tmRNA in complex with Fe(II)-tethered SmpB was probed by initiating hydroxyl radical formation with 6 µl of 250 mM ascorbic acid and 6 µl of 1.25% H2O2. The reaction mixtures were incubated on ice for 10 min and quenched with 100 mM thiourea. RNA was prepared by phenol extraction and ethanol precipitation (39). Reverse transcriptase reaction was carried out in a 12 µl reaction mixture containing 50 mM Tris–HCl (pH 8.3), 75 mM potassium chloride, 3 mM magnesium chloride, 10 mM dithiothreitol, 0.5 mM each of dNTP, 1 pmol of rRNA or tmRNA, 2 pmol of 5′ Texas Red labeled DNA primer complementary to a portion of the rRNA or tmRNA sequence, 3 units of ribonuclease inhibitor from human placenta (Takara) and 18 U of reverse transcriptase from Molony Murine Leukemia Virus (RNase H−, Takara) (40). After the addition of 3 µl of a stop solution containing 7 M urea and 0.5% bromophenol blue, the positions of cleavages were analyzed using a fluorescence DNA sequencer (Hitachi SQ-5500).
RESULTS
Directed hydroxyl radical probing of the ribosome by Fe(II)-tethered SmpB variants
To construct mutants of E. coli SmpB each having a single cysteine residue for attaching it to a Fe(II)-BABE probe, an SmpB mutant free of a cysteine residue, designating SmpB(-Cys), was prepared by replacing two naturally occurring cysteine residues at positions 82 and 123 with alanines. We confirmed that SmpB(-Cys) has activity of in vitro tag-peptide synthesis directed by poly (U) and tmRNA comparable to that for wild-type SmpB. Based on this background, we constructed 80 SmpB mutants, each having only one cysteine residue anywhere on the surface of SmpB. The mutant SmpB proteins were overexpressed, purified by anion exchange and affinity column chromatographies. SmpB variants having an activity of in vitro tag-peptide synthesis comparable to that of wild-type SmpB were selected for directed hydroxyl radical probing (Supplementary Figure 1). Then Fe(II) was tethered to the cysteine residue of each SmpB.
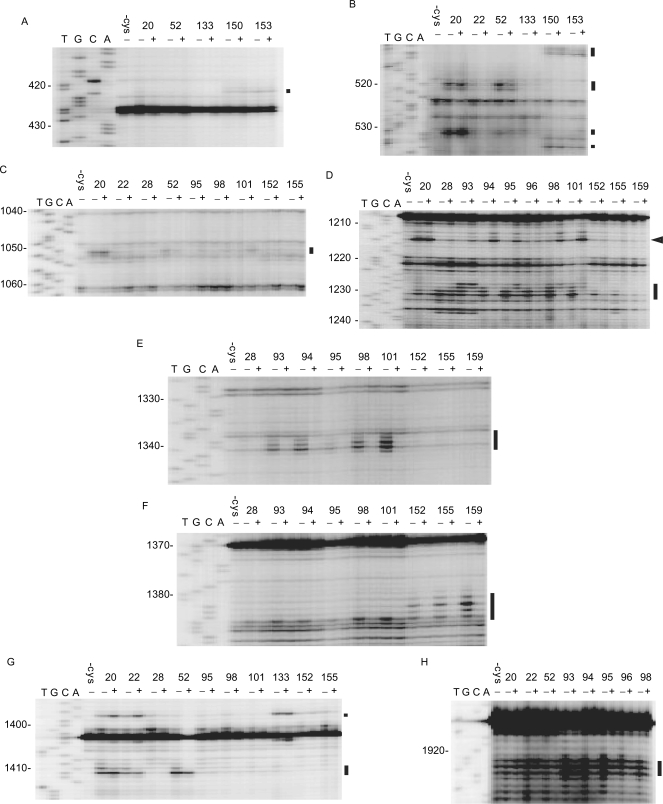
Figure 1.
Directed hydroxyl radical probing of 16S and 23S rRNAs. The sites of cleavage in 16S rRNA (A–G) and 23S rRNA (H) were detected by primer extension. 70S ribosome was probed with a bound SmpB variant in the presence (+) or absence (−) of the set of mRNA and tRNAfMet. The bands of interest are designated by bars. An arrowhead shows the band of cleavage from Fe(II) tethered to residue 94, 95, 98 or 101 only in the presence of the set of mRNA and tRNAfMet. The lanes of DNA sequencing with the same primer are designated by T, G, C and A. Some non-specific bands, which may represent the interruption of reverse transcription depending on the structure or modification of template rRNA, were detected in each panel.
Fe(II)-tethered SmpB was combined with the ribosome to form a complex, and cleavage of 16S rRNA or 23S rRNA by hydroxyl radicals generated from Fe(II) was detected by primer extension. Sixteen Fe(II)-SmpB variants cleaved 16S rRNA and eight variants cleaved 23S rRNA (Figure 1 and Table 1). The remaining 64 of 80 variants cleaved neither 16S rRNA nor 23S rRNA (Table 1). The sites of cleavage from Fe(II) tethered to the main body of SmpB (positions 1–132) were localized around the A-site and P-site predominantly in 16S rRNA (Figure 2A). Fe(II) tethered to the residues 20 and 22 in α1, 52 in loop 1 and 94, 95, 98 and 101 in α3 cleaved the A-site of the 16S rRNA, while Fe(II) tethered to 20 in α1, 28 in β2 and 93–96, 98 and 101 in α3 cleaved the P-site of the 16S rRNA. Helix 69 of 23S rRNA corresponding to the P-site was cleaved by Fe(II) tethered to 20 and 22 in α1, 52 in loop 1 and 93–96 and 98 in α3.
Table 1.
Sites of cleavage by directed hydroxyl radical probing
| Site of cleavage | |||||
|---|---|---|---|---|---|
| 16S rRNA | 23S rRNA | tmRNA | |||
| Site of cysteine in SmpB | A-site | P-site | A-site | P-site | |
| 20 (α1)a | 519–521 | 1231 | 1922–1924 | ||
| 531–532 | |||||
| 1052 | |||||
| 1215 | |||||
| 1398 | |||||
| 1411 | |||||
| 22 (α1) | 1398 | 1922–1924 | |||
| 1411 | |||||
| 28 (β2) | 1230–1231 | 19 | |||
| 52 (Loop 1) | 519–521 | 1922–1924 | |||
| 531–532 | |||||
| 1411 | |||||
| 76 (Loop 2) | 22–24 | ||||
| 77 (Loop 2) | 22–24 | ||||
| 78 (Loop 2) | 22–24 | ||||
| 79 (Loop 2) | 22–24 | ||||
| 80 (Loop 2) | 22–24 | ||||
| 93 (α3) | 1228–1231 | 1922–1924 | 19 | ||
| 1339–1341 | 27–31 | ||||
| 94 (α3) | 1215b | 1922–1924 | |||
| 1230–1231 | |||||
| 1339–1341 | |||||
| 95 (α3) | 1215b | 1230–1231 | 1922–1924 | 19 | |
| 1339–1341 | 27–31 | ||||
| 96 (α3) | 1230–1231 | 1922–1924 | 19 | ||
| 31 | |||||
| 98 (α3) | 1215b | 1230–1231 | 1922–1924 | ||
| 1339–1341 | |||||
| 1385 | |||||
| 99 (α3) | 19 | ||||
| 31 | |||||
| 101 (α3) | 1215b | 1230–1231 | |||
| 1339–1341 | |||||
| 1385 | |||||
| 119 (β7) | 19 | ||||
| 133 (C-terminal) | 1398 | ||||
| 150 (C-terminal) | 422 | ||||
| 512–513 | |||||
| 535 | |||||
| 152 (C-terminal) | 1231 | ||||
| 1382–1385 | |||||
| 153 (C-terminal) | 422 | ||||
| 512-513 | |||||
| 535 | |||||
| 155 (C-terminal) | 1231 | ||||
| 1382–1385 | |||||
| 159 (C-terminal) | 1382–1385 | ||||
aParentheses indicate the secondary structure in which cysteine residue is included (30).
bCleavage at 1215 from Fe(II) tethered to residue 94, 95, 98 or 101 occurs only in the presence of the set of mRNA and tRNAfMet, probably indicating a conformational change of the A-site by the addition of mRNA and tRNAfMet.
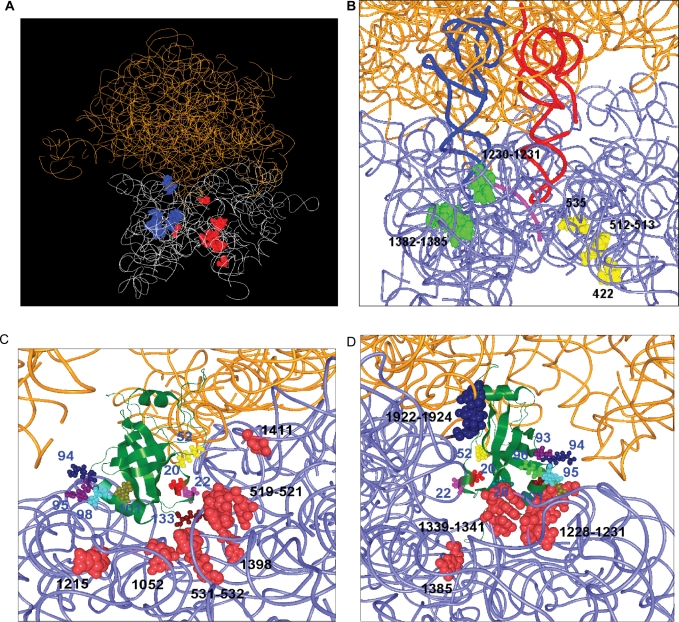
Figure 2.
A model of the binding of SmpB to the ribosome was built from the positions of cleavages. The tertiary structure models of SmpB from A. aeolicus (31) and 70S ribosome from E. coli (44) were used. 16S rRNA and 23S rRNAs are shown as white or light blue and orange wires, respectively. (A) All of the sites of cleavages by the SmpB variants used in the present study are designated as balls in the tertiary structure model of 70S ribosome from E. coli (44). The sites of cleavages in the presence of the set of mRNA and tRNAfMet are colored red. The sites of cleavages, which occurred in the absence but not in the presence of the set of mRNA and tRNAfMet, are shown as blue balls. (B) Sites in the 70S ribosome cleaved by Fe(II) tethered to residues in the C-terminal tail of SmpB. The sites of cleavages from A-site SmpB and P-site SmpB are colored yellow and green, respectively. A-site tRNA, P-site tRNA and mRNA are colored red, dark blue and pink, respectively. (C) A model of SmpB bound to the A-site. The residues in SmpB providing cleavages and the nucleotides of cleavage in rRNA are designated as small and large balls, respectively. (D) A model of SmpB bound to the P-site. The residues in SmpB providing cleavages are designated as small balls. The nucleotides of cleavage in 16S and 23S rRNA are designated as red and blue large balls, respectively.
Cleavage was also found along the mRNA path, when Fe(II) was introduced to the C-terminal region (positions 133–160) (Figure 2B and Table 1). Fe(II) tethered to residues 133, 150 and 153 cleaved the proximity of the A-site and its downstream region, while Fe(II) tethered to residues 152, 155 and 159 cleaved the proximity of the P-site.
Directed hydroxyl radical probing of the ribosome in complex with mRNA and P-site tRNA
Then synthetic mRNA encompassing the SD sequence to the P-site AUG codon together with tRNAfMet corresponding to the AUG codon was added to the ribosome fraction prior to the addition of Fe(II)-tethered SmpB. The signals of cleavage around the P-site disappeared, while those around the A-site remained (Figure 1). These results successfully distinguished the signals by the A-site SmpB from those by the P-site SmpB (Table 1), emphasizing the occurrence of two binding sites for SmpB in the ribosome.
We used the probing data as constraints to model position and orientation of SmpB in the 8.7 Å crystal structure of the 70S ribosome from E. coli in complex with tRNA and mRNA (41). For each rRNA nucleotide cleaved from a given SmpB probe, a sphere centered on the position of the target nucleotide was generated in the crystal structure of the 70S ribosome. It suggested two binding sites for SmpB in the ribosome; one is the A-site (Figure 2C) and the other is the P-site (Figure 2D).
Directed hydroxyl radical probing of tmRNA
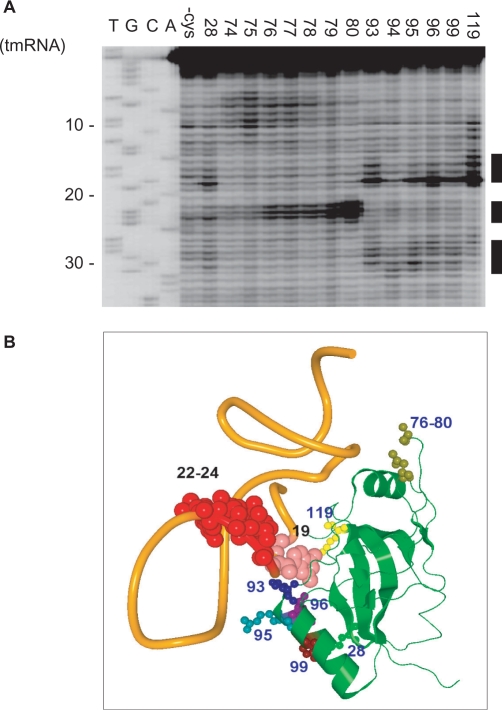
Directed hydroxyl radical probing was also performed towards a complex of Fe(II) tethered SmpB and tmRNA to constrain the orientation of SmpB with respect to tmRNA. Several cleavages were found at TLD. Fe(II) tethered to 28 in β2, 93, 95, 96 and 99 in α3 and 119 in β7 cleaved position 19 and Fe(II) tethered to 76–80 in loop 2 cleaved position 22–24 (Figure 3A), being consistent with a co-crystal structure of SmpB lacking the C-terminal tail and a TLD fragment from A. aeolicus (26) (Figure 3B). Then this structure as well as tRNA was superimposed on our model. It was found that the main body of SmpB and its bound TLD fit the lower and upper halves, respectively, of tRNA of each site (Supplementary Figure 2). Fe(II) tethered to 93, 95, 96 and 99 in α3 also cleaved the region 27–31 (Figure 3A), which is located in the long helix between PK1 and TLD. Several reports have suggested additional sites of SmpB binding outside the tRNA domain such as in PK1 and in the proximity of the translation resuming point (42–44). Thus, we searched the sites of cleavage in the 5′ half from the mRNA domain. However, no additional sites of cleavage were detected.
Figure 3.
Directed hydroxyl radical probing of tmRNA. (A) The sites of cleavage in tmRNA were detected by primer extension. All lanes from tmRNA probed with bound mock-derivatized SmpB(-cys) and those with mutants providing cleavages are aligned from the left. (B) The sites of cleavage in tmRNA are mapped on a co-crystal structure of SmpB (green) and a TLD fragment (orange) from A. aeolicus (26). The residues in SmpB providing cleavage and the site of cleavage in tmRNA are designated as small and large balls, respectively. Cleavages of positions 27–31 by Fe(II) tethered to several residues in α3 are not designated, because they are in the loop in the model TLD fragment but are actually in the middle of the long helix leading to PK1 in tmRNA.
DISCUSSION
In the present study, we demonstrated the position and orientation of SmpB in the 70S ribosome from directed hydroxyl radical probing. In agreement with an earlier study (35), SmpB binds the 70S ribosome without the help of tmRNA. From our data, two binding sites were identified in the 70S ribosome; one is the A-site and the other is the P-site. In either A-site or P-site, the main body of SmpB occupies the binding site of the anticodon arm of tRNA. The crystal structure of a complex containing SmpB and the TLD of tmRNA from A. aeolicus (26), which is consistent with the interaction between SmpB and tmRNA from E. coli or other species in solution revealed by the present directed hydroxyl radical probing and other biochemical studies (27,45), was applied to the present results. It revealed that two SmpB molecules are able to be accommodated at the A-site and P-site in the small ribosomal subunit so that TLD bound to each SmpB superimposes the amino acid acceptor stem of tRNA and its 3′-CCA end is oriented towards the peptidyl-transferase center in the large subunit (Supplementary Figure 2). It was also shown that SmpB, in the absence of tmRNA, binds to the vacant A-site of the ribosome in which the P-site was occupied by tRNA and a truncated mRNA.
The capacity of two SmpB molecules to bind a ribosome has been suggested by biochemical studies (35,46). A chemical footprinting study also suggests two SmpB-binding sites in a ribosome, one in the vicinity of the P-site and the other is below the L7/L12 stalk (47). Recent cryo-EM studies have shown that one molecule of SmpB from T. thermophilus is bound to the A-site, which is consistent with A-site SmpB in the present study (28,48). An additional SmpB has been observed around the GTPase-associated center in a pre-accommodated complex of the stalled ribosome with alanyl-tmRNA, EF-Tu, GDP and kirromycin (28). In the present study, no signals were found around the GTPase-associated center.
Although the C-terminal tail of SmpB has been suggested to have a crucial role in the ribosomal processes of trans-translation (33,34), its position, structure and behavior in the ribosome remain unknown. The present results reveal that the hydroxyl radicals generated from several residues in the C-terminal tail of A-site SmpB cleaved the 16S rRNA along with the mRNA path, encompassing the tunnel of the entrance of mRNA to the A-site, while those of P-site SmpB exclusively cleaved the region around the codon–anticodon interaction in the P-site. They indicate that the initial residue of the C-terminal tail at 133 of A-site SmpB locates near the decoding center, and the residues 150 and 153 of A-site SmpB are near helices 16 and 18. Thus the C-terminal tail of A-site SmpB may lie along the mRNA path towards the downstream tunnel. The appearance of signals at interval of three residues (150 and 153) implied an α helical structure of the C-terminal tail which has been predicted from the periodical occurrence of positively charged residues (34). Considering residues from 134–149 for A-site SmpB provided no apparent cleavages, the former half of the C-terminal tail might be out from the mRNA path, while the latter half might be extended along the mRNA path as the α helical structure. Fe(II) tethered to the residues 152 and 155 of P-site SmpB cleaved three portions of 16S rRNA, 1231 and 1382–1385, all around the site of the codon–anticodon interaction in the P-site. The latter portion was also cleaved by Fe(II) tethered to the penultimate residue 159 and residues 98 and 101 in α3 of P-site SmpB. Thus the C-terminal tail of P-site SmpB folds back to the P-site rather than extends along the mRNA path.
On the basis of the present results, we propose a model of trans-translation (Figure 4). In this model, SmpB molecules bound to the A-site and P-site shown in the present study reflect the pre- and post-translocation steps of trans-translation, respectively. Upon entrance of tmRNA to the stalled ribosome, whether it enters in complex with SmpB or independently after the SmpB binding to the ribosome is yet controversial (35). In either case, the C-terminal tail of SmpB may recognize the vacant A-site free of mRNA to trigger trans-translation. It has been reported that trans-translation can occur, but inefficiently, on an mRNA of a short, rather than long, 3′-extension from the A-site (49,50), indicating the competition on the A-site between translation of mRNA and trans-translation. In such a case, a transient detachment of the 3′-extension from the A-site may be required for trans-translation. This agrees with the present results showing the C-terminal tail of A-site SmpB overlaps mRNA in the downstream of the A-site. After peptidyl-transfer to Ala-tmRNA, translocation of peptidyl-Ala-tmRNA/SmpB from the A-site to the P-site may occur. During this event, the extended C-terminal tail folds around the region of the codon–anticodon interaction in the P-site, which drives out mRNA from the P-site, and consequently from the ribosome. Just after the movement of SmpB to the P-site, the A-site becomes free so that the resume codon of tmRNA can be accommodated.
Figure 4.
Schematic model of the early stage of trans-translation. Upon or before entrance of tmRNA to the stalled ribosome, the C-terminal tail of SmpB may recognize the vacant A-site free of mRNA to lie along the mRNA path towards the downstream tunnel. After peptidyl-transfer to tmRNA, translocation of peptidyl-tmRNA/SmpB from the A-site to the P-site, possibly with the help of EF-G, occurs to drive out mRNA from the ribosome. During this event, the extended C-terminal tail somehow folds to substitute for the codon–anticodon interaction in the P-site. Then the resume codon of tmRNA is accommodated in the decoding region. The regions that serve as mRNA are highlighted by black.
This model can provide several insights into the yet unidentified mechanism of trans-translation; how the stalled ribosome free of tRNA and mRNA is preferentially recognized, and what substitutes for a codon–anticodon interaction during trans-translation. Apparently, the main body of SmpB mimics the lower half of tRNA both in the A-site and P-site, while the upper half of tRNA is mimicked by TLD. The C-terminal tail of SmpB mimics mRNA and/or the codon–anticodon interaction both before and after translocation, during which it undergoes a drastic conformational change. Consequently, SmpB/TLD behaves as mRNA+tRNA so that SmpB/tmRNA might decode the first alanine encoded by the C-terminal tail of SmpB. It has been shown that the upstream of the tag-encoding sequence of tmRNA is crucial for trans-translation (17,18,51). How this upstream region interacts with SmpB/ribosome and how it rearranges during trans-translation should be highlighted in the future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank the staff of Gene Research Center of Hirosaki University for the use of the facility. We also thank the 21st COE program of Iwate University. This work was supported by a grant-in-aid for scientific research from the Ministry of Education, Science, Sports and Culture, Japan to A.M. and H.H. (No. 14035201), a grant-in-aid for scientific research from the Japan Society for the Promotion of Science to A.M. and H.H. (No. 17380061) and a Grant for Priority Research Designated by the President of Hirosaki University to H.H. Funding to pay the Open Access publication charges for this article was provided by the Japan Society for the Promotion of Science.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gueneau de Novoa P, Williams KP. The tmRNA website: reductive evolution of tmRNA in plastids and other endosymbionts. Nucleic Acids Res. 2004;32:D104–D108. doi: 10.1093/nar/gkh102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen ES, Rosenblad MA, Larsen N, Westergaard JC, Burks J, Wower IK, Wower J, Gorodkin J, Samuelsson T, et al. The tmRDB and SRPDB resources. Nucleic Acids Res. 2006;34:D163–D168. doi: 10.1093/nar/gkj142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ushida C, Himeno H, Watanabe T, Muto A. tRNA-like structures in 10Sa RNAs of Mycoplasma capricolum and Bacillus subtilis. Nucleic Acids Res. 1994;22:3392–3396. doi: 10.1093/nar/22.16.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komine Y, Kitabatake M, Yokogawa T, Nishikawa K, Inokuchi H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl Acad. Sci. USA. 1994;91:9223–9227. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felden B, Himeno H, Muto A, McCutcheon JP, Atkins JF, Gesteland RF. Probing the structure of the Escherichia coli 10Sa RNA (tmRNA) RNA. 1997;3:89–104. [PMC free article] [PubMed] [Google Scholar]
- 6.Nameki N, Felden B, Atkins JF, Gesteland RF, Himeno H, Muto A. Functional and structural analysis of a pseudoknot upstream of the tag-encoded sequence in E. coli tmRNA. J. Mol. Biol. 1999;286:733–744. doi: 10.1006/jmbi.1998.2487. [DOI] [PubMed] [Google Scholar]
- 7.Nameki N, Tadaki T, Himeno H, Muto A. Three of four pseudoknots in tmRNA are interchangeable and are substitutable with single-stranded RNAs. FEBS Lett. 2000;470:345–349. doi: 10.1016/s0014-5793(00)01349-1. [DOI] [PubMed] [Google Scholar]
- 8.Keiler KC, Waller PRH, Sauer RT. Role of a peptide tagging system in degradation of protein synthesized from messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 9.Himeno H, Sato M, Tadaki T, Fukushima M, Ushida C, Muto A. In vitro trans-translation mediated by alanine-charged 10Sa RNA. J. Mol. Biol. 1997;268:803–808. doi: 10.1006/jmbi.1997.1011. [DOI] [PubMed] [Google Scholar]
- 10.Muto A, Ushida C, Himeno H. A bacterial RNA that functions as both a tRNA and an mRNA. Trends Biochem. Sci. 1998;23:25–29. doi: 10.1016/s0968-0004(97)01159-6. [DOI] [PubMed] [Google Scholar]
- 11.Withey JH, Friedman DI. A salvage pathway for protein structures: tmRNA and trans-translation. Annu. Rev. Microbiol. 2003;57:101–123. doi: 10.1146/annurev.micro.57.030502.090945. [DOI] [PubMed] [Google Scholar]
- 12.Huang C, Wolfgang MC, Withey J, Koomey M, Friedman DI. Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. EMBO J. 2000;19:1098–1107. doi: 10.1093/emboj/19.5.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muto A, Fujihara A, Ito K, Matsuno J, Ushida C, Himeno H. Requirement of transfer-messenger RNA (tmRNA) for the growth of Bacillus subtilis under stresses. Genes Cells. 2000;5:627–636. doi: 10.1046/j.1365-2443.2000.00356.x. [DOI] [PubMed] [Google Scholar]
- 14.Abo T, Inada T, Ogawa K, Aiba H. SsrA-mediated tagging and proteolysis of LacI and its role in the regulation of lac operon. EMBO J. 2000;19:3762–3769. doi: 10.1093/emboj/19.14.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujihara A, Tomatsu H, Inagaki S, Tadaki T, Ushida C, Himeno H, Muto A. Detection of tmRNA-mediated trans-translation products in Bacillus subtilis. Genes Cells. 2002;7:343–350. doi: 10.1046/j.1365-2443.2002.00523.x. [DOI] [PubMed] [Google Scholar]
- 16.Keiler KC, Shapiro L. tmRNA in Caulobacter crescentus is cell cycle regulated by temporally controlled transcription and RNA degradation. J. Bacteriol. 2003;185:1825–1830. doi: 10.1128/JB.185.6.1825-1830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams KP, Martindale KA, Bartel DP. Resuming translation on tmRNA: a unique mode of determining a reading frame. EMBO J. 1999;18:5423–5433. doi: 10.1093/emboj/18.19.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S, Ishii M, Tadaki T, Muto A, Himeno H. Determinants on tmRNA for initiating efficient and precise trans-translation: some mutations upstream of the tag-encoding sequence of Escherichia coli tmRNA shift the initiation point of trans-translation in vitro. RNA. 2001;7:999–1012. doi: 10.1017/s1355838201010342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rudinger-Thirion J, Giegé R, Felden B. Aminoacylated tmRNA from Escherichia coli interacts with procaryotic elongation factor Tu. RNA. 1999;5:989–992. doi: 10.1017/s135583829999101x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karzai AW, Susskind MM, Sauer RT. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA) EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barends S, Wower J, Kraal B. Kinetic parameters for tmRNA binding to alanyl-tRNA synthetase and elongation factor Tu from Escherichia coli. Biochemistry. 2000;39:2652–2658. doi: 10.1021/bi992439d. [DOI] [PubMed] [Google Scholar]
- 22.Wower J, Zwieb C, Guven SA, Wower I. Binding and cross-linking of tmRNA to ribosomal protein S1, on and off the Escherichia coli ribosome. EMBO J. 2000;19:6612–6621. doi: 10.1093/emboj/19.23.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanawa-Suetsugu K, Takagi M, Inokuchi H, Himeno H, Muto A. SmpB functions in various steps of trans-translation. Nucleic Acids Res. 2002;30:1620–1629. doi: 10.1093/nar/30.7.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimizu Y, Ueda T. The role of SmpB protein in trans-translation. FEBS Lett. 2002;514:74–77. doi: 10.1016/s0014-5793(02)02333-5. [DOI] [PubMed] [Google Scholar]
- 25.Takada K, Takemoto C, Kawazoe M, Konno T, Hanawa-Suetsugu K, Lee S, Shirouzu M, Yokoyama S, Muto A, et al. In vitro trans-translation of Thermus thermophilus: ribosomal protein S1 is not required for the early stage of trans-translation. RNA. 2007;13:1–8. doi: 10.1261/rna.363207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutmann S, Haebel PW, Metzinger L, Sutter M, Felden B, Ban N. Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB. Nature. 2003;424:5503–5509. doi: 10.1038/nature01831. [DOI] [PubMed] [Google Scholar]
- 27.Nameki N, Someya T, Okano S, Suemasa R, Kimoto M, Hanawa-Suetsugu K, Terada T, Shirouzu M, Hirao I, et al. Interaction analysis between tmRNA and SmpB from Thermus thermophilus. J. Biochem. 2005;138:729–739. doi: 10.1093/jb/mvi180. [DOI] [PubMed] [Google Scholar]
- 28.Kaur S, Gillet R, Li W, Gursky R, Frank J. Cryo-EM visualization of transfer messenger RNA with two SmpBs in a stalled ribosome. Proc. Natl Acad. Sci. USA. 2006;103:16484–16489. doi: 10.1073/pnas.0607438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barends S, Karzai AW, Sauer RT, Wower J, Kraal B. Simultaneous and functional binding of SmpB and EF-Tu-GTP to the alanyl-acceptor arm of tmRNA. J. Mol. Biol. 2001;314:9–21. doi: 10.1006/jmbi.2001.5114. [DOI] [PubMed] [Google Scholar]
- 30.Hong SJ, Tran QA, Keiler K. Cell cycle-regulated degradation of tmRNA is controlled by RNase R and SmpB. Mol. Microbiol. 2005;57:565–575. doi: 10.1111/j.1365-2958.2005.04709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong G, Nowakowski J, Hoffman DW. Structure of small protein B: the protein component of the tmRNA-SmpB system for ribosome rescue. EMBO J. 2002;21:1845–1854. doi: 10.1093/emboj/21.7.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Someya T, Nameki N, Hosoi H, Suzuki S, Hatanaka H, Fujii M, Terada T, Shirouzu M, Inoue Y, et al. Solution structure of a tmRNA-binding protein, SmpB, from Thermus thermophilus. FEBS Lett. 2003;535:94–100. doi: 10.1016/s0014-5793(02)03880-2. [DOI] [PubMed] [Google Scholar]
- 33.Sundermeier TR, Dulebohn DP, Cho HJ, Karzai AW. A previously uncharacterized role for small protein B (SmpB) in transfer messenger RNA-mediated trans-translation. Proc. Natl Acad. Sci. USA. 2005;102:2316–2321. doi: 10.1073/pnas.0409694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacob Y, Sharkady SM, Bhardwaj K, Sanda AA, Williams KP. Function of the SmpB tail in transfer-messenger RNA translation revealed by a nucleus-encoded form. J. Biol. Chem. 2005;280:5503–5509. doi: 10.1074/jbc.M409277200. [DOI] [PubMed] [Google Scholar]
- 35.Hallier M, Ivanova N, Rametti A, Pavlov M, Ehrenberg M, Felden B. Pre-binding of small protein B to a stalled ribosome triggers trans-translation. J. Biol. Chem. 2004;279:25978–25985. doi: 10.1074/jbc.M314086200. [DOI] [PubMed] [Google Scholar]
- 36.Wilson KS, Ito K, Noller HF, Nakamura Y. Functional sites of interaction between release factor RF1 and the ribosome. Nat. Struct. Biol. 2000;7:866–870. doi: 10.1038/82818. [DOI] [PubMed] [Google Scholar]
- 37.Dallas A, Noller HF. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell. 2001;8:855–864. doi: 10.1016/s1097-2765(01)00356-2. [DOI] [PubMed] [Google Scholar]
- 38.Nameki N, Tadaki T, Muto A, Himeno H. Amino acid acceptor identity switch of Escherichia coli tmRNA from alanine to histidine in vitro. J. Mol. Biol. 1999;289:1–7. doi: 10.1006/jmbi.1999.2754. [DOI] [PubMed] [Google Scholar]
- 39.Lancaster L, Kiel MC, Kaji A, Noller HF. Orientation of ribosome recycling factor in the ribosome from directed hydroxyl radical probing. Cell. 2002;111:129–140. doi: 10.1016/s0092-8674(02)00938-8. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi T, Konno T, Muto A, Himeno H. Various effects of paromomycin on tmRNA-directed trans-translation. J. Biol. Chem. 2003;278:27672–27680. doi: 10.1074/jbc.M211724200. [DOI] [PubMed] [Google Scholar]
- 41.Vila-Sanjurjo A, Ridgeway WK, Seymaner V, Zhang W, Santoso S, Yu K, Cate JH. X-ray crystal structures of the WT and a hyper-accurate ribosome from Escherichia coli. Proc. Natl Acad. Sci. USA. 2003;100:8682–8687. doi: 10.1073/pnas.1133380100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wower J, Zwieb CW, Hoffman DW, Wower IK. SmpB: a protein that binds to double-stranded segments in tmRNA and tRNA. Biochemistry. 2002;41:8826–8836. doi: 10.1021/bi0201365. [DOI] [PubMed] [Google Scholar]
- 43.Metzinger L, Hallier M, Felden B. Independent binding sites of small protein B onto transfer-messenger RNA during trans-translation. Nucleic Acids Res. 2005;33:2384–2394. doi: 10.1093/nar/gki534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Konno T, Kurita D, Takada K, Muto A, Himeno H. A functional interaction of SmpB with tmRNA for determination of the resuming point of trans-translation. RNA. 2007 doi: 10.1261/rna.604907. doi:10.1261/rna.604907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dulebohn DP, Cho HJ, Karzai AW. Role of conserved surface amino acids in binding of SmpB protein to SsrA RNA. J. Biol. Chem. 2006;281:28536–28545. doi: 10.1074/jbc.M605137200. [DOI] [PubMed] [Google Scholar]
- 46.Hallier M, Desreac J, Felden B. Small protein B interacts with the large and the small subunits of a stalled ribosome during trans-translation. Nucleic Acids Res. 2006;34:1935–1943. doi: 10.1093/nar/gkl097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ivanova N, Pavlov MY, Bouakaz E, Ehrenberg M, Schiavone LH. Mapping the interaction of SmpB with ribosomes by footprinting of ribosomal RNA. Nucleic Acids Res. 2005;33:3529–3539. doi: 10.1093/nar/gki666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gillet R, Kaur S, Li W, Hallier M, Felden B, Frank J. Scaffolding as an organizing principle in trans-translation: the roles of small protein B and ribosomal protein S1. J. Biol. Chem. 2006;282:6356–6363. doi: 10.1074/jbc.M609658200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ivanova N, Pavlov MY, Felden B, Ehrenberg M. Ribosome rescue by tmRNA requires truncated mRNAs. J. Mol. Biol. 2004;338:33–41. doi: 10.1016/j.jmb.2004.02.043. [DOI] [PubMed] [Google Scholar]
- 50.Asano K, Kurita D, Takada K, Konno T, Muto A, Himeno H. Competition between trans-translation and termination or elongation of translation. Nucleic Acids Res. 2005;33:5544–5552. doi: 10.1093/nar/gki871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konno T, Takahashi T, Kurita D, Muto A, Himeno H. A minimum structure of aminoglycosides that causes an initiation shift of trans-translation. Nucleic Acids Res. 2004;32:4119–4126. doi: 10.1093/nar/gkh750. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.