Abstract
Rv3291c gene from Mycobacterium tuberculosis codes for a transcriptional regulator belonging to the (leucine responsive regulatory protein/regulator of asparigine synthase C gene product) Lrp/AsnC-family. We have identified a novel effector-binding site from crystal structures of the apo protein, complexes with a variety of amino acid effectors, X-ray based ligand screening and qualitative fluorescence spectroscopy experiments. The new effector site is in addition to the structural characterization of another distinct site in the protein conserved in the related AsnC-family of regulators. The structures reveal that the ligand-binding loops of two crystallographically independent subunits adopt different conformations to generate two distinct effector-binding sites. A change in the conformation of the binding site loop 100–106 in the B subunit is apparently necessary for octameric association and also allows the loop to interact with a bound ligand in the newly identified effector-binding site. There are four sites of each kind in the octamer and the protein preferentially binds to aromatic amino acids. While amino acids like Phe, Tyr and Trp exhibit binding to only one site, His exhibits binding to both sites. Binding of Phe is accompanied by a conformational change of 3.7 Å in the 75–83 loop, which is advantageously positioned to control formation of higher oligomers. Taken together, the present studies suggest an elegant control mechanism for global transcription regulation involving binding of ligands to the two sites, individually or collectively.
INTRODUCTION
Mycobacterium tuberculosis is a successful pathogen mainly because of its ability to persist in the human host for several years while evading the immune system (1). The rv3291c gene codes for an Lrp/AsnC (leucine responsive regulatory protein/regulator of asparigine synthase C gene product) type global transcriptional regulator (MtbLrp) (2) and belongs to the large family of DNA-binding transcriptional regulators. It has been suggested that Lrp/AsnC type proteins should more appropriately be called feast/famine regulatory proteins because of their involvement in regulation of metabolic pathways in response to the availability of amino acids and nitrogen bases in the external environment (3,4). These proteins are also involved in DNA bending, condensation of DNA into globular nucleo-protein structures, chromosome structure and organization, among other roles (3,5).
Individual subunits of these proteins have a molecular weight of ∼17 kDa and are divided into two domains namely DNA and effector-binding domains which occur at the N- and C-terminal regions, respectively. The DNA-binding domain contains the helix-turn-helix motif while the effector-binding domain is also involved in oligomeric interactions. Higher order assemblies usually exist as multiples of dimers and include formation of tetramers, octamers, dodecamers and chromatin-like cylinders (5–9). How these proteins translate the effector-binding event to the protein–DNA-binding site is unclear. Some global regulators like the Escherichia coli Lrp, which reportedly controls over 10% of all genes, are known to bind to a variety of amino acids including leucine, alanine, valine, proline and lysine, whose binding can effect a positive/negative regulation of the target genes (5,10). On the other hand, local regulation involving proteins like AsnC, is controlled by the binding of a specific effector molecule like asparigine (11,12). It has been suggested that binding of appropriate ligands, primarily amino acids, can effect functionally relevant tertiary structure conformational changes or to oligomeric association. Thus a small number of such regulators found in archaea and bacteria can regulate a large number of genes in response to the environmental changes (4).
Consistent with this, rv3291c has a molecular weight of ∼18 kDa and has been found to be up-regulated 15-fold in nutrition starved tuberculosis models designed to mimic the latent/persistent state (13). Our earlier sequence analysis and initial characterization (14) suggested that it belongs to the Lrp-family of regulators. It lies in the genomic region upstream of sigF involved in mycobacterial stress response (15) and its expression has also been observed on inhibition of septum formation and is inversely proportional to bacterial growth rate (16,17). More recently, in Mycobacterium fortuitum, the homologous gene was been found to be highly conserved and in vivo screening involving a murine infection model of persistence using this pathogen led to the identification of a mutant where this gene was disrupted. It was found that the mutation affected the ability of the pathogen to persist in the kidney in the model (personal communication, Dr R. Srivastava, Central Drug Research Institute, Lucknow, India). The above data suggest that the protein is important for maintenance/adaptation to long-term persistence although it is not essential for growth under normal conditions (18). Rv3291c therefore has the potential to be an important therapeutic target for developing strategies to counter persistence of the disease.
Structurally characterized examples of the Lrp/AsnC-family of transcription regulators includes the E. coli Lrp (19), E. coli AsnC complexed to asparigine (20), Neisseria Meningitides Lrp complexed to leucine and methionine, respectively (21), Bacillus subtilis LrpC and related archaeal regulators LrpA from Pyrococcus furiosus and P. sp OT3 FL11 (3,11,12,22–24). A comparison of the changes that occur upon effector binding could not be addressed in the AsnC structure due to the absence of the apo structure. The complexes with the N. meningitides regulator did not reveal significant changes to tertiary structure on amino acid binding. A sequence analysis of Lrp and AsnC type regulators show that the protein sub-families exhibit larger differences in the C-terminal/effector binding domain with an overall sequence identity of ∼25% (14,20) reflecting differences in effector binding between the protein sub-families.
Many aspects of structure and function-like variations in individual regulation mechanism are currently unclear; as also how ligand binding affects interactions with DNA and whether ligand binding effects significant changes to the tertiary structure. In the initial part of this work we report the X-ray structure of the apo MtbLrp solved through MIRAS methods. The protein associates as a tetramer of dimers to form an octamer. Fluorescence spectroscopy and X-ray co-crystal structures show that MtbLrp binds to a variety of amino acids. We used X-ray screening-based approaches to determine that the protein preferentially binds to aromatic amino acids and also were able to rank their order of preference. In the present structures we have identified a new small molecule-binding site at the dimer interface (called ‘type II’) in addition to the previously reported effector-binding site in the related E. coli AsnC and N. meningitides Lrp complexes (called ‘type I’). Both sites apparently have different preferences for ligand molecules. While amino acids like Phe, Leu and Trp bind to the site previously reported in the E. coli AsnC regulator, His binds to both sites in MtbLrp. The ligands occupy only 4 out of 8 possible sites of type I in contrast to full occupancy in the earlier reported complexes. There is a change in the binding site loop conformation in the unoccupied type I sites which lets the same loop interact with the bound histidine in type II binding sites while preventing ligand binding at site I. Taken together, the present studies suggest an elegant control mechanism for global transcription regulation involving binding of ligands to the two sites, individually or collectively.
MATERIALS AND METHODS
Overexpression and purification
Rv3291c was cloned, expressed and purified as reported earlier (14). Briefly the PCR-amplified construct was ligated into pET21d (M/s Novagen) vector between the NcoI and HindIII sites. His-tagged protein, with the tag at the C-terminal end, was purified by standard procedures after transforming the plasmid into BL21 (DE3) cells. The protein was concentrated to ∼20 mg/ml in 50 mM Tris–HCl pH 8.0, 50 mM NaCl and 5 mM EDTA after size exclusion chromatography using Superdex-200 HR 10/30 (M/s Amersham). Protein concentrations were determined as per the Bradford method (25).
Crystallization, data collection and structure determination
The protein was crystallized using the hanging drop technique by equilibrating protein drops against reservoirs containing 100 mM MES pH 6.0, 0.75 M lithium acetate at 22°C. Heavy atoms were screened by band shift assays on a native gel. Crystals were soaked in appropriate heavy atom solutions for varying times and concentrations. Data were collected using a Rigaku RU300 X-ray generator coupled with MAR345 detector or alternatively with MAR345-DTB detector on crystals mounted in quartz capillaries at room temperature. Data were processed using MOSFLM (26) implemented in the CCP4 package (27) and collection parameters are summarized in Supplementary Table 1. While crystals of the apo protein diffract to 2.5 Å resolutions, the crystals of the Phe complex diffract to 2.2 Å.
Extensive molecular replacement calculations using available archaeal and bacterial Lrp structures were inconclusive. Phases were therefore calculated by MIRAS methods using the SOLVE/RESOLVE package (28–30) using three derivatives; the first one being silver nitrate, (crystals soaked in 80 mM solution for 2 days) and the latter two data sets from potassium tetra-chloro-platinate (2.0 mM, overnight soak) and cadmium iodide (1.7 mM, 1 h soak) derivatives. The solution output by SOLVE had a figure of merit 0.28 before solvent flattening and this went up to 0.68 after density modification and phase extension using RESOLVE. The structure solution parameters are summarized in Supplementary Table 1a. RESOLVE could trace ∼15% of the residues. The map quality was sufficient to trace the remaining chains manually using TURBO–FRODO (31). Refinements were carried out using REFMAC (32) after setting aside ∼10% of the reflections for monitoring the free R-factor. Structures of the complexes were solved using difference Fourier techniques. Omit maps were examined to avoid bias. Quality of the structures was checked using PROCHECK (33) and the geometric parameters as also the Ramachandran plot (34) falls into acceptable ranges.
X-ray screening for ligands
Crystals were grown under above conditions, except that an amino acid mixture corresponding to 2 mM each of the following amino acids namely Phe, His, Leu, Trp, Met and Tyr, were added to the crystallization drop. The best binding amino acid was identified after each round of data collection and structure solution. The next rounds of crystallization were carried out serially using only the remaining compounds.
Qualitative fluorescence spectroscopy studies
A qualitative analysis of the amino acid binding was carried out using competitive displacement of ANS (1-Anilino-8-naphthalene-sulfonate) from the protein by the respective ligands (35). A Perkin-Elmer Life Sciences LS 50B fluorescence spectrophotometer was used in the experiments at 25°C. The excitation wavelength used was 380 nM and the emission was monitored between 400 and 600 nM, respectively. Excitation and emission slit widths were kept at 7 nM, respectively. All samples were incubated for 2 h under specified conditions before recording the spectra. Initially the protein (1 µM) was titrated against 0–50 µM ANS to determine its optimal concentration for subsequent experiments. In the displacement experiments 1 µM of protein incubated with 10 µM ANS was titrated with 0–25 mM of respective ligands. An inhibitor namely cyclohexylmethyl amine derivative, of Lysine ε-aminotransferase (36), a totally unrelated protein, was used as a control compound which should not exhibit any binding to the protein.
Analysis
Analysis of the inter-subunit contacts and ligand–protein interactions, accessible surface area, etc. were carried out using CONTACTS and other tools implemented in the CCP4 package (27). Structural superposition was carried out using ALIGN (37) and PROFIT (http://www.bioinf.org.uk/software/profit). Figures were generated using Pymol (38), CCP4MG (39) and InsightII (http://www.accelrys.com/products/insight).
RESULTS
Structure description
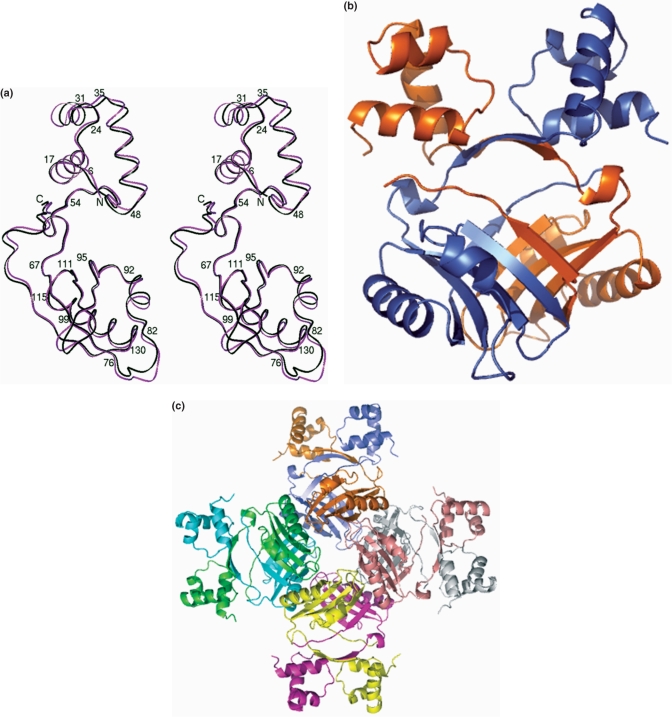
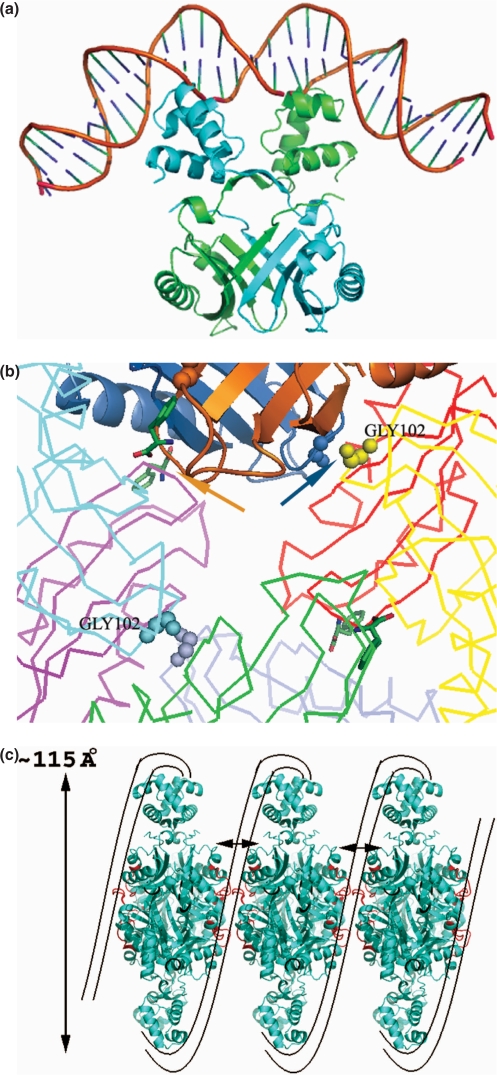
The protein was purified as reported earlier (14). The size of the protein was consistent with a stable octameric association in size exclusion chromatography experiments. Extensive molecular replacement calculations using the coordinates of previously reported archaeal Lrp structures were unsuccessful. We consequently used MIRAS approaches to solve the structure of the protein (Figure 1a). Structure solution and data collection statistics are given in Supplementary Table 1a. There are two independent subunits in the asymmetric unit, which form a dimer (Figure 1b). This dimer subsequently forms an octamer through crystal symmetry (Figure 1c). Each subunit of rv3291c (MtbLrp) folds into two distinct domains joined by a rather long linker as observed in other structural examples of this family (20,23,24). The N-terminal domain is the DNA-binding domain which initially forms a helix (residues 6–17) and subsequently folds into an helix-turn-helix motif (residues 24–48) observed earlier also in other structures from this protein family (4) (A structure-based sequence alignment is in Supplementary Figure 1). A rather long linker (residues 48–60) consisting of a β-strand and a helical stretch then brings the chain to the effector-binding domain. This latter domain is involved extensively in oligomeric interactions and contains four β-strands and two α-helices arranged in a βαββαβ topology (between residues 66 and 137). The chain ends in a β-strand (residues 139–143), which interacts with the β2 strand of the dimeric counterpart to form a five-stranded curved sheet in the dimer.
Figure 1.
(a) Crystal structure of MtbLrp. The two crystallographically independent subunits have been superposed and the Cα trace is depicted. Selected residues are labeled and the figure is shown in stereo. (b) The association of the subunits into a dimer. The two chains have been colored ‘blue’ and ‘golden’ respectively. (c) Octameric association in MtbLrp generated by crystal symmetry. The chains of the octamer have been colored distinctly for clarity.
Oligomeric association
The two independent subunits in the apo structure superpose onto each other with r.m.s.d. of 0.74 Å for all Cα atoms and reveals conformational differences in two loops; the first being the loop linking the initial β- and α-motifs (residues 75–83) of the effector-binding domain and the other forming part of the small molecule binding site (residues 100–105) (Figure 1a). The maximum displacements of 3.4 and 1.7 Å are centered at Ser79 and Glu103, respectively. When the subunits were superposed onto each other in the oligomer it became clear that the conformational changes are necessary for octamer association as otherwise it would lead to unacceptable steric contacts. The respective DNA-binding domains of each subunit of the octamer face the outside of the tightly packed oligomeric structure. This is in line with its observed role in formation of nucleo-protein structures and packaging of DNA (20,24). The domain exhibits a higher than average displacement of ∼1 Å after superposition for residues 24–48.
Structures of complexes with amino acids
We initially co-crystallized the protein with Se-Met to identify the effector-binding site unambiguously in MtbLrp. Clearly the binding sites for this molecule were spatially conserved with the E. coli AsnC and N. meningitides Lrp regulators where structures with amino acid complexes are available (20,21). We then co-crystallized the protein with a variety of amino acids including Leu, Arg, Gln, Thr, Phe, Tyr, Trp, Pro, Met and His. Unambiguous density for bound ligand was found in the structures with Phe, His, Tyr, Trp and Met (in addition to Se-Met which was used to identify the binding site) (Figure 2, Supplementary Figure 2). In the remaining structures including that with Leu, weaker density was observed. Data collection and refinement statistics of the respective complexes are in Supplementary Table 1b.
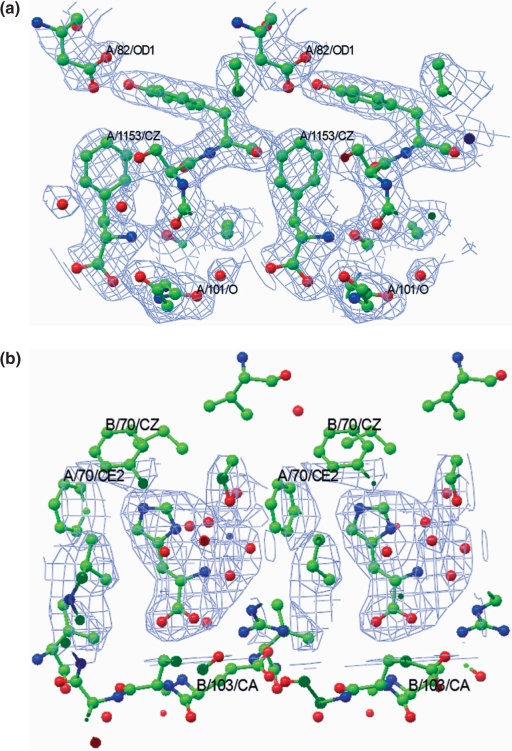
Figure 2.
Stereoview of the electron density maps at (a) binding site I in the MtbLrp–phenylalanine complex and (b) binding site II in the MtbLrp–histidine complex. The 2Fo−Fc maps have been contoured at 1σ level and depicted as a ‘blue’ mesh. Selected residues around the binding site have been shown and selected atoms labeled. Atoms are colored as per convention and water oxygens are shown as ‘red’ spheres. The figure was generated using CCP4MG (39).
Phe complex
Well-defined density for Phe at a resolution of 2.2 Å (Figure 2a), allows for a detailed analysis of the ligand–protein interactions. The ligand-binding site is partially formed by residues 100–106 of subunit A containing a conserved Gly (Gly 102 in MtbLrp) and also residues from symmetry related subunits in the octamer. This Gly is a conserved sequence feature in the respective binding sites of ACT/RAM domain containing proteins (20). Figure 3a shows a close-up of the interactions of Phe in its complex which is also tabulated (Supplementary Table 2a). The binding site is at an inter-dimer interface and comprises of residues from different subunits. In contrast to earlier complexes (20,21), only 4 sites out of 8 possible ligand-binding sites were occupied and binding to the other sites was precluded due to the conformational changes discussed above. The side chain of the ligand is stabilized mainly by ‘van der Waals’ interactions including those with Arg126, Tyr106, Asp82, Ser105 and Ser99. The main chain carboxylate atoms of Phe are stabilized by polar interactions involving Ser135 OG and N atoms. Thr135 OG also interacts with a main chain carboxylate atom of Phe. The main chain O and N atoms of Gly102, the conserved binding site residue mentioned above, interacts with the main chain N and carboxylate atoms of the ligand. Main chain N of the bound ligand also forms hydrogen bonds with the main chain O of Val100 and Glu104, respectively. The same ligand atom interacts with Ser105 OG and Glu103 N respectively through a water molecule. The same water molecule interacts with another binding site of water associated with symmetry-related Thr133 N and O atoms. Thus a network of polar interactions, which also involve symmetry-related atoms in the octamer stabilizes the bound ligand.
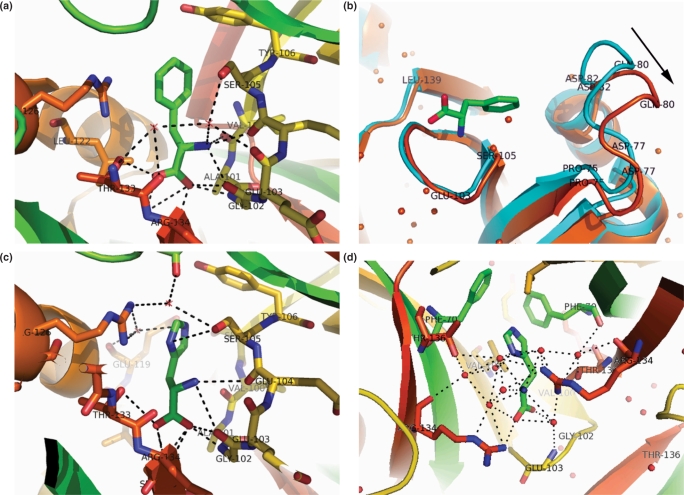
Figure 3.
(a) Interactions of phenylalanine in its complex with MtbLrp. Protein is shown in cartoon representation. Hydrogen bonds are indicated by black dotted lines. Selected binding-site residues are labeled and plotted in stick representation. Water molecules are indicated by red spheres. Atom coloring is as per convention except that carbon atoms of bound Phe (stick representation) are shown in ‘green’ while those of interacting subunits are shown in ‘orange’ and ‘yellow’ respectively. (b) Movement of loop 75–83 by 3.7 Å after binding of phenylalanine. The apo and complexed proteins are in ‘cyan’ and ‘orange’ respectively while the direction of the movement is indicated by an arrow. Selected residues are labeled. Bound ligand is depicted in ‘stick’ representation while structural water is depicted as ‘orange’ spheres. (c) Interactions of histidine with MtbLrp at site I. Color conventions and representations in this and the subsequent figure are as in (a) above. (d) Interactions of histidine at site II in MtbLrp.
The binding of Phe is accompanied by a change in the conformation of loop 75–83 of subunit A (Figure 3b). In fact the maximum main chain displacement of ∼3.7 Å occurs at the Cα atom of the Pro81 residue. On the other hand, the spatial disposition of loop 100–106 of the subunit as also that of the helix-turn-helix motif at the N-terminal domain are relatively unaffected by binding of the ligand.
His complex
The structure of the His complex resulted in the identification of a novel ligand-binding site in addition to the site occupied by Phe in its co-crystal structure. The effector has unambiguous density at both sites allowing for an analysis of the protein-His contacts in the complex (Figure 2b). The interactions of the main chain atoms of His with the protein at site I is similar to that of bound Phe with the main chain N and carboxylate atoms interacting with residues in the 100–106 loop and also with Thr133 and Ser135 of a neighboring subunit in the octamer (Figure 3c and Supplementary Table 2b). Additional interactions in this complex include those of ND1 atom of the side chain with Ser105 of the same subunit and with Arg126 of a neighboring subunit in the octamer. The nitrogen atoms of the side chain also interact with the latter arginine residue through a water molecule.
The type II binding site occurs at the intra-dimer interface. This site forms part of a hydrophobic core in the protein. In the apo structure as also in the other amino acid complexes two to three water molecules have a spatial disposition similar to that of the imidazole ring of His in this site. The main chain carboxylate atoms of bound His in this site interact with Glu B103 and B104 residues (Figure 3d). These latter residues form part of the binding site loop in the unoccupied type I sites in the octamer. The N atom of the main chain interacts with Arg A134. The ND1 and NE2 atoms of the side chain extensively interact with Thr B136 and Arg A134 through water bridges. The main chain atoms of the bound ligand interact with Arg134 from both subunits through water-mediated interactions as also with Thr B136 and Glu B103. Other residues within 4 Å of the ligand include Phe70 from both subunits and Val B100, which are spatially close to imidazole ring of the bound ligand. The protein–His interactions are tabulated in Supplementary Table 2b.
Tyr, Trp, Met, Leu and other complexes
The main chain interactions of Tyr in its complex are expectedly similar to those described earlier. Figure 4a contains a schematic of the interactions of the Tyr with the protein. The OH atom of its side chain exhibits polar interactions with Asp A82, Tyr B106* and Arg B126*. The latter two residues belong to the neighboring subunits in the octamer; incidentally the Tyr residue is part of the binding site loop in the unoccupied type I sites. On the other hand, Asp is part of the loop exhibiting maximum movement in the Phe complex.
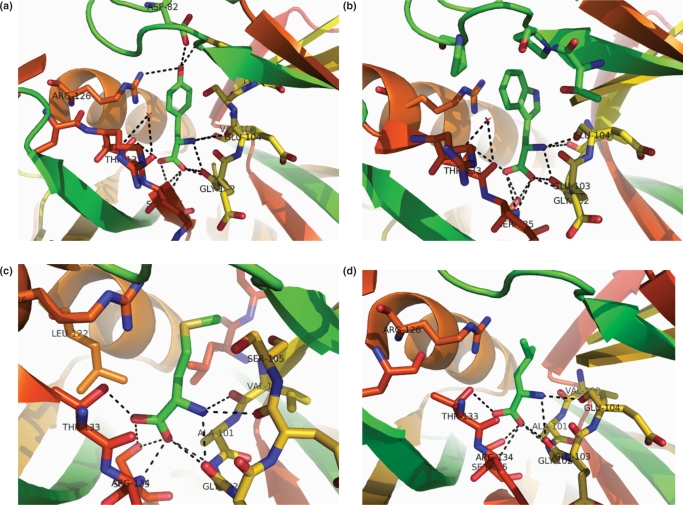
Figure 4.
Interactions of (a) Tyrosine, (b) Tryptophan, (c) Methionine and (d) Leucine in their respective site I complexes with MtbLrp. The coloring and representation of interacting atoms and ligand are as in the earlier figures.
In the Trp complex the indole side chain of the ligand mainly exhibits ‘van der Waals’ interactions with the protein (Figure 4b). Surprisingly, no polar interaction with the nitrogen atom in the side chain was observed in the structure. On the other hand the SD atom of the methionine side chain interacts with Arg 126 of a neighboring subunit in its complex (Figure 4c). In the latter structure also a change in the conformation of the loop 75–83, albeit to a lesser extent than in the Phe complex was observed. Interactions of the respective ligands are tabulated in Supplementary Table 2. The above three ligands also have clearly defined side chain electron density in the maps (Supplementary Figure 1).
The close-up of leucine in its complex with MtbLrp and interaction details is in Figure 4d and Supplementary Table 2f, respectively. Leu mainly interacts with the protein through its main chain atoms. The side chain is quite flexible as shown by the lack of clear density for it beyond the Cβ atom and exhibits only non-bonded interactions with the protein. A similar situation was observed in the complexes with Arg, Pro, Thr and Gln with which co-crystals could be obtained and the electron density maps reveal weak density at the position of the main chain atoms seen in the earlier structures (data not shown). It was therefore concluded that these amino acids exhibit weak binding to the protein.
X-ray based ligand screening
In an effort to identify the amino acids for which the protein has a natural affinity, we used X-ray based ligand screening as a tool where small molecules, which have the highest affinity for the protein are competitively selected from the crystallization solution. These screening techniques are sensitive in detecting and differentiating between even weakly binding small molecule fragments to a protein (40). We used this technique to characterize relative binding affinities of different amino acids to MtbLrp. Accordingly we crystallized the protein from a solution containing amino acids which showed well-defined density for at least the main chain and Cβ atoms in their respective co-crystal structures. The solution initially contained Leu, Phe, Tyr, Trp, Met and His. Crystals from the first round of experiments had unambiguous density for Phe, which was additionally confirmed by observing the displacement of the 75–83 loop observed in its complex. In the next round of screening, Phe was removed from the crystallization solution and this time His was identified from the electron density maps. This was aided by the fact that His is the only amino acid in this set that binds to type II sites also. Tyr and Trp formed co-crystals in subsequent screening experiments followed by Met. Identification of the respective ligands from the electron density maps was straightforward as we could also compare the densities with those obtained earlier from single co-crystallization experiments. Thus the relative binding affinities were identified to be Phe>His>Tyr>Trp> Met>(Leu, others). MtbLrp was therefore deduced as having a preference for aromatic amino acids.
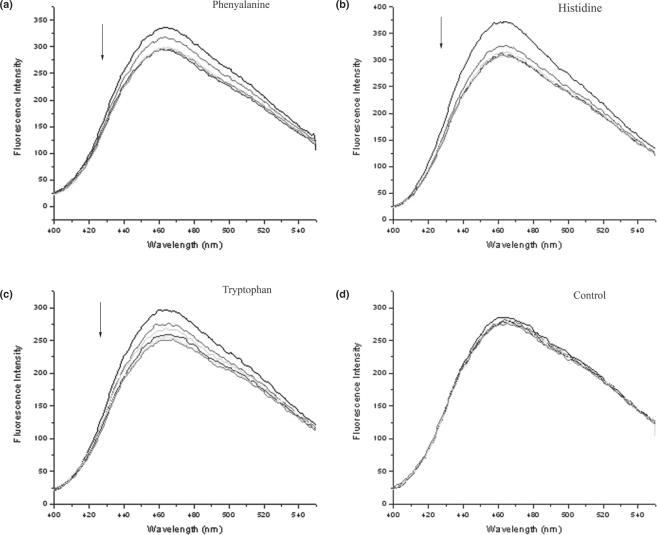
Qualitative fluorescence spectroscopy
We used competitive displacement of ANS by compounds being tested to qualitatively check the binding of the amino acids to MtbLrp (Figure 5). Binding of the amino acids led to reduction in the observed fluorescence of bound ANS. An inhibitor, namely a cyclohexylmethyl amine derivative, of a completely unrelated protein, lysine ε-amino transferase whose structure was reported by us recently (36) was used as a control compound, which should not bind to MtbLrp. As mentioned above, the binding exhibited by leucine can be rationalized by its co-crystal structure where only the main chain atoms are involved in binding to the protein. The spectroscopy results clearly support binding by a variety of amino acids. Taken in conjunction with the X-ray screening experiments, the results clearly demonstrate that MtbLrp preferentially binds aromatic amino acids.
Figure 5.
Competitive displacement of ANS by (a) Phenylalanine, (b) histidine, (c) tryptophan and (d) cyclohexylmethylamine derivative compound used as a non-binding control. Change in fluorescence was monitored by titrating the protein (1 µM)-ANS (10 µM) premix against 5, 10, 15, 20 and 25 mM concentrations of each compound in the respective experiments. Arrows indicate the change in fluorescence intensity upon titration. While the amino acids exhibit binding to the protein, the control compound did not exhibit any binding.
DNA binding and regulation
Earlier studies involving in silico modeling and solution studies with DNA (23), as also modeling with the current structure (Figure 6a) clearly suggests that DNA has to be bent for interactions with this family of regulators (41). The smallest functional unit of the protein is the dimer and we have modeled its mode of DNA interactions analogous to that reported earlier in the B. subtilis LrpC and E. coli AsnC structures (20). It can be conceptualized that multiple binding sites separated by non-binding regions in the DNA stretch can be threaded around the oligomer to form chromatin-like structures which have been observed earlier (42,43). The stable octameric association observed in the E. coli AsnC–asparigine complex and B. subtilis LrpC structures, differences in the diameters of the octamers of the bacterial and archaeal LrpA, along with an analysis of reported binding sites in the promoter region led the authors to suggest a model where adjacent Lrp octamers could wrap DNA into a solenoid-like structure (20). When we map the change in the conformation of the loop 75–83 in the presence of bound phenylalanine at site I, we find that the loop has an advantageous spatial disposition in the octamer to disrupt interactions with adjacent oligomers in the proposed model (Figure 6b and c). This role for bound ligands at site I agree with earlier co-crystal structures with E. coli AsnC and N. meningitides Lrp where binding of ligands to the site did not disrupt the octameric assembly. It also agrees with solution studies in E. coli Lrp, in which leucine-induced dissociation of hexadecamers into leucine-bound octamers by binding to a second distinct site (44). On the other hand, interactions of a ligand with the newly identified site II can lead to a breakdown in octamer symmetry as detailed in the ‘Discussion’ section.
Figure 6.
(a) Model of DNA binding to MtbLrp dimer. B-form DNA was taken from PDB 1CGP and adjusted using the program InsightII (M/s Accelrys) to fit the dimer. The DNA clearly has to be bent to interact with the protein. (b) Close-up of the arrangement of bound phenylalanine at site I (indicated by a ‘golden’ arrow) in its interactions with the protein octamer. Gly 102 in empty type I sites (indicated by a ‘blue’ arrow) are shown in ball-and-stick representation. Asp82 that forms part of the 75–83 loop, which moves by 3.7 Å on binding to the ligand is also indicated by ‘golden’ ball-and-sticks in the subunit with bound Phe. (c) Schematic of DNA wrapped around MtbLrp octamers placed side by side to form a solenoid-like arrangement. This class of proteins is known to form stable protein–DNA complexes like the nucleosome (43) and it is interesting that the diameter of the octamer is ∼115 Å which is close to the nucleosome diameter of 110 Å. The schematic is as per earlier experiments (43) and in silico modeling in B. subtilis LrpC (20). In this arrangement, the movement (indicated by double arrows) of the 75–83 loop (colored ‘red’) on ligand binding can control protein–DNA interactions and higher oligomeric association although the octameric structure is not disturbed.
DISCUSSION
In the present work, we have structurally characterized a global regulator from M. tuberculosis, which promises to be a novel target in developing therapies against persistence of the disease. The crystal structures of the complexes and X-ray based ligand-screening techniques, supported by the fluorescence spectroscopy results demonstrate that rv3291c has a strong preference for aromatic amino acids and suggests an important role in the metabolism of the latter. It is interesting that the MtbLrp preferentially binds aromatic amino acids while other characterized Lrp proteins prefer effectors like Val, Leu, Ala, etc. (5,10). Aromatic amino acids are known to be important for the viability of the pathogen (45). In fact the enzymes in the Shikimate pathway, which is important for aromatic amino acid metabolism, have been recognized as important targets for anti-bacterial therapies (46).
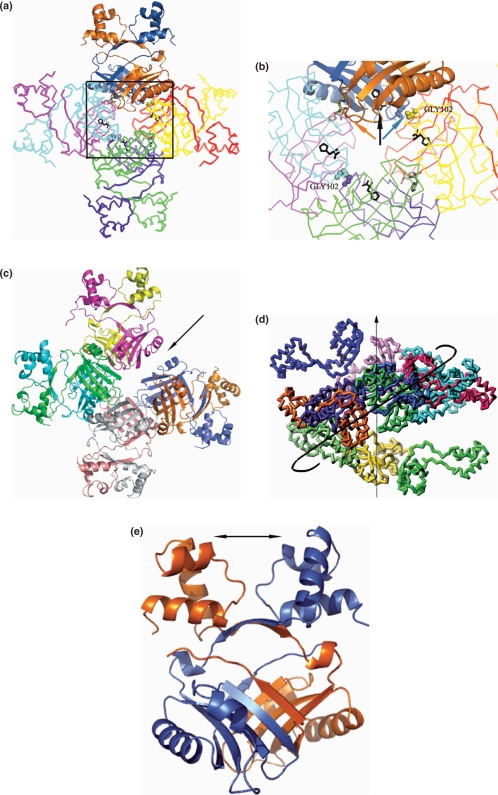
Importantly, the present structures have led to the identification of a novel small molecule-binding site II at the protein intra-dimer interface. There are four possible sites of this kind in the octamer (a tetramer of dimers, Figure 7a and b) and all sites are occupied in the structure with L-histidine. It is quite possible that other small molecules can also bind to the site. In fact histidine exhibits extensive water-mediated interactions with the protein and this structural water can be displaced to accommodate a bigger ligand also. It seems unlikely that effector molecules will disrupt the dimeric association; on the other hand a bound ligand at this site might effect small changes to the relative spatial disposition of the DNA-binding domains (Figure 7e). These changes will be amplified in the octamer and can lead to either strengthening/weakening of interaction with DNA. It may also be recalled that the binding site loop 100–106 of the B subunit interacts with the main chain atoms of His in its complex both directly and through water-mediated interactions. The conformation change observed in this loop is essential for octameric association in MtbLrp. It is therefore conceivable that a bound ligand can ‘push’ this loop and lead to a breakdown in octamer symmetry. Binding of a ligand to all four type II sites would then lead to disassociation of the octamer into lower oligomers (Figure 7). However, binding of a ligand to only two sites on adjacent dimers will break the octamer symmetry analogous to that observed in the recent E. coli Lrp-DNA co-crystals (19) where the octamer opens up in the presence of DNA into an open array (Figure 7c). Such a change in oligomeric symmetry could also form the starting point for the transition from octameric disks to a helical cylinder (Figure 7d) observed in the pyrococcus OT3 Lrp (24).
Figure 7.
Binding of ligands at site II in MtbLrp. (a) Octameric association in the MtbLrp–His complex. The region marked by a box is zoomed into (b) which shows details of histidine binding in its complex with the protein. Bound ligand in type I sites are depicted in ‘green’ sticks (indicated by a ‘golden’ arrow) while those bound to type II sites are in ‘black’ sticks (indicated by a ‘black’ arrow). A ‘blue’ arrow indicates the binding site loop, which can be ‘pushed’ out to distort the oligomeric association. Binding of ligand to only two sites can lead to the ‘open’ octamer arrangement seen in (c) E. coli Lrp structure (PDB 2GQQ) or also be the basis for a transformation into (d) a helical cylinder-like arrangement seen in P. furiosus sp OT3 Lrp (PDB 1RI7). Binding of ligands to this site are also advantageously situated to disturb (e) the spatial disposition of the DNA-binding domains.
The other effector-binding site (I) occurs at the inter-dimer interfaces in the octamer. There are therefore eight such possible binding sites (Figure 6b), out of which only four are accessible to a ligand. It is possible that these other sites may be occupied under different quaternary structural states observed in the class of proteins (19–21,23,24). In contrast, bound ligands were observed in all eight type I sites in the E. coli AsnC and N. meningitides Lrp structures. In fact in the latter structure, it was found that binding of leucine and methionine led to further stabilization of the octamer. In the present Phe complex a maximal conformational displacement of 3.7 Å was observed in the 75–83 loop and to a smaller extent in some of the other complexes. An important role for ligands bound at this site could therefore be in controlling higher oligomeric associations like the one suggested in Figure 6c.
It is interesting that residues of the binding site loop 100–106 of the unoccupied type I sites interact with bound His in type II sites in its complex. This strategy presumably allows for additional control of the oligomeric state of the protein through effector binding or fine-tuning of individual regulatory functions. Solution studies involving E. coli Lrp and a couple of its mutants (Asp113Glu & Leu135Arg), which prevent octamer–hexadecamer transition, also revealed two kinds of sites (44). Binding of leucine to a low affinity site induced dissociation of hexadecamers in the unmutated protein to octamers; on the other hand, the octameric form and the mutants retained high affinity for leucine. Structural mapping of the mutations onto the subsequently reported E. coli Lrp structure (19) suggests that the second weak-binding site is in a different inter-subunit interface compared to the type I and type II sites discussed in this work. The Asp113Glu mutation maps onto the 100–106 loop of MtbLrp and along with symmetry related E. coli Leu135 residue apparently generates a new site. On the other hand, it is likely that the high affinity site in E. coli Lrp corresponds to type I sites discussed here as binding of Leu to this site does not affect the octamer assembly. The above discussion suggests an unanticipated important role for the binding site loop (100–106 in MtbLrp) residues where they are involved in interacting with ligands in the three types of sites.
In conclusion, the present study has revealed a novel effector-binding site in the MtbLrp regulator. The large change in the disposition of the 75–83 loop by 3.7 Å is also the first seen in this regulator family upon ligand binding. The octameric association observed in MtbLrp is possible because of the change in the binding loop conformation in subunit B and this has probably evolved in order to interact with ligands in site II. We are only now beginning to understand the immense complexity of the regulation mechanisms employed by feast/famine regulatory proteins. Broad structural similarities and DNA recognition modes are apparently further fine-tuned by individual proteins by incorporating novel effector-binding sites, (the present study) and smaller changes in structure, as seen in the MtbLrp–Phe complex, to perform their regulatory roles.
ACCESSION NUMBER
Co-ordinates and structure factors of the apo protein have been deposited in the protein data bank with code 2IVM while those of the complexes have codes 2VBW, 2VBX, 2VBY, 2VBZ, 2VC0 and 2VC1, respectively.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
ACKNOWLEDGEMENTS
The work was funded by intramural grant MLP007 and Department of Biotechnology grant GAP0018. Computational work was supported by CSIR network grant CMM0017. T.S. is a CSIR senior research fellow. We thank Prof. T.P. Singh, AIIMS, New Delhi for collection of a couple of data sets of the protein complexes. This article bears CDRI communication number 7285. Funding to pay the Open Access publication charges for this article was provided by Council of Scientific & Industrial Research, India.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 2.Stewart GR, Robertson BD, Young DB. Tuberculosis: a problem with persistence. Nat. Rev. Microbiol. 2003;1:97–105. doi: 10.1038/nrmicro749. [DOI] [PubMed] [Google Scholar]
- 3.Calvo JM, Mathews RG. Leucine responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yokoyama K, Ishijima SA, Clowney L, Koike H, Aramaki H, Tanaka C, Makino K, Suzuki M. Feast/famine regulatory proteins (FFRPs): Escherichia coli Lrp, AsnC and related archaeal transcription factors. FEMS Microbiol. Rev. 2006;30:89–108. doi: 10.1111/j.1574-6976.2005.00005.x. [DOI] [PubMed] [Google Scholar]
- 5.Brinkman AB, Dahlke I, Tuininga JE, Lammers T, Dumay V, Heus E, Lebbink JHG, Thomm M, de Vos WM, et al. An Lrp-like transcription regulator from archaeon Pyrococcus furiosus is negatively autoregulated. J. Biol. Chem. 2000;275:38160–38169. doi: 10.1074/jbc.M005916200. [DOI] [PubMed] [Google Scholar]
- 6.Willins DA, Ryan CW, Platko JV, Calvo JM. Characterization of Lrp, an Escherichia coli regulatory protein that mediates a global response to leucine. J. Biol. Chem. 1991;266:10768–10774. [PubMed] [Google Scholar]
- 7.Madhusudhan KT, Huang N, Sokatch JR. Characterization of bkdR-DNA binding in the expression of the bkd operon of Pseudomonas putida. J. Bacteriol. 1995;177:636–641. doi: 10.1128/jb.177.3.636-641.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jafri S, Evoy S, Cho K, Craighead HG, Winans SC. An Lrp type transcriptional regulator from Agrobacterium tumefaciens condenses more than 100 nucleotides of DNA into globular nucleoprotein complexes. J. Mol. Biol. 1999;288:811–824. doi: 10.1006/jmbi.1999.2715. [DOI] [PubMed] [Google Scholar]
- 9.Chen S, Rosner MH, Calvo JM. Leucine-regulated self-association of leucine responsive regulatory protein (Lrp) Escherichia coli. J. Mol. Biol. 2001;312:625–635. doi: 10.1006/jmbi.2001.4955. [DOI] [PubMed] [Google Scholar]
- 10.Tani TH, Khodursky A, Blumenthal RM, Brown PO, Matthews RG. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl Acad. Sci. USA. 2002;99:13471–13476. doi: 10.1073/pnas.212510999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolling R, Lother H. AsnC – an autogenously regulated activator of asparagines synthetase A transcription in E. coli. J. Bacteriol. 1985;164:310–315. doi: 10.1128/jb.164.1.310-315.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Wind N, de Jong M, Meijer M, Stuitje AR. Site directed mutagenesis of the Escherichia coli chromosome near oriC: identification and characterization of asnC, a regulatory element in E. coli asparagines metabolism. Nucleic Acids Res. 1985;13:8797–8811. doi: 10.1093/nar/13.24.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 14.Shrivastava T, Kumar S, Ramachandran R. Cloning, expression, purification and crystallization of a transcriptional regulatory protein (Rv3291c) from Mycobacterium tuberculosis H37Rv. Acta Crystallogr. 2004;D60:1874–1876. doi: 10.1107/S0907444904016397. [DOI] [PubMed] [Google Scholar]
- 15.Wu QL, Kong DQ, Lam K, Husson RN. A mycobacterial extracytoplasmic function sigma factor involved in survival following stress. J. Bacteriol. 1997;179:2922–2929. doi: 10.1128/jb.179.9.2922-2929.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slayden RA, Knudson DL, Belisle JT. Identification of cell cycle regulators in Mycobacterium tuberculosis by inhibition of septum formation and global transcriptional analysis. Microbiology. 2006;152:1789–1797. doi: 10.1099/mic.0.28762-0. [DOI] [PubMed] [Google Scholar]
- 17.Landgraf JR, Wu J, Calvo JM. Effects of nutrition and growth rate on Lrp levels in Escherichia coli. J. Bacteriol. 1996;178:6930–6936. doi: 10.1128/jb.178.23.6930-6936.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 19.de Los Rios S, Perona JJ. Structure of the Escherichia coli Leucine-responsive regulatory protein Lrp reveals a novel octameric assembly. J. Mol. Biol. 2007;366:1589–1602. doi: 10.1016/j.jmb.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thaw P, Sedelnikova SE, Muranova T, Wiese S, Ayora S, Alonso JC, Brinkman AB, Akerboom J, van der Oost J, et al. Structural insight into gene transcriptional regulation and effector binding by Lrp/AsnC family. Nucleic Acids Res. 2006;34:1439–1449. doi: 10.1093/nar/gkl009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren J, Sainsbury S, Combs SE, Capper RG, Jordan PW, Berrow NS, Stammers DK, Saunders NJ, Owens RJ. The structure and transcriptional analysis of a global regulator from Neisseria meningitides. J. Biol. Chem. 2007;282:14655–14664. doi: 10.1074/jbc.M701082200. [DOI] [PubMed] [Google Scholar]
- 22.Beloin C, Jeusset J, Revet B, Mirambeau G, Le Hegarat F, Le Cam E. Characterization of lrp-like (LrpC) gene from Bacillus subtilis. Mol. Gen. Genet. 1997;256:63–71. doi: 10.1007/s004380050546. [DOI] [PubMed] [Google Scholar]
- 23.Leonard PM, Smits HJ, Sedelnikova SE, Brinkman AB, de Vos WM, van der Oost J, Rice DW, Rafferty JB. Crystal structure of the Lrp-like transcriptional regulator from the arachaeon Pyrococcus furiosus. EMBO J. 2001;20:990–997. doi: 10.1093/emboj/20.5.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koike H, Ishijima SA, Clowney L, Suzuki M. The archaeal feast/famine regulatory protein: potential roles of its assembly forms for regulating transcription. Proc. Natl Acad. Sci. USA. 2004;101:2840–2845. doi: 10.1073/pnas.0400109101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 26.Leslie AGW. 1992. Joint CCP4 + ESF-EAMCB Newsletter on protein crystallography, No. 26. [Google Scholar]
- 27.Collaborative Computing Project No. 4. The CCP4 suite: program for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 28.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. 1999;D55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terwilliger TC. Maximum likelihood density modification. Acta Crystallogr. 2000;D56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. 2003;D59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones TA. A graphic model building and refinement system for macromolecules. J. Appl. Crystallogr. 1989;11:268–272. [Google Scholar]
- 32.Murshudov GN, VaginA A, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–245. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 33.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereo-chemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 34.Ramachandran GN, Sasisekharan V. Conformation of polypeptides and proteins. Adv. Protein Chem. 1968;23:283–438. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- 35.Stryer L. The interaction of a naphthalene dye with apomyoglobin and apohemoglobin. A fluorescent probe of non-polar binding sites. J. Mol. Biol. 1965;13:482–495. doi: 10.1016/s0022-2836(65)80111-5. [DOI] [PubMed] [Google Scholar]
- 36.Tripathi SM, Ramachandran R. Direct evidence for a glutamate switch necessary for substrate recognition: crystal structure of lysine ε-aminotransferase (Rv3290c) from Mycobacterium tuberculosis. J. Mol. Biol. 2006;362:877–886. doi: 10.1016/j.jmb.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 37.Cohen GE. Align: a program to superimpose protein coordinates, accounting for insertion and deletion. J. Appl. Crystallogr. 1997;30:1160–1161. [Google Scholar]
- 38.DeLano WL. The PyMol Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- 39.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, et al. Developments in the CCP4 molecular graphics projects. Acta Crystallogr. 2004;D60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 40.Nienaber VL, Richardson PL, Klighofer V, Bouska JJ, Giranda VL, Greer J. Discovering novel ligands for macromolecules using x-ray crystallographic screening. Nat. Biotechnol. 2000;28:1105–1106. doi: 10.1038/80319. [DOI] [PubMed] [Google Scholar]
- 41.Wang Q, Calvo JM. Lrp, a major regulatory protein in Escherichia coli, bends DNA and can organize the assembly of a higher-order nucleoprotein structure. EMBO J. 1993;12:2495–2501. doi: 10.1002/j.1460-2075.1993.tb05904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tapias A, Lopez G, Ayora S. Bacillus subtilis LrpC is a sequence-independent DNA-binding and DNA-bending protein which bridges DNA. Nucleic Acids Res. 2000;28:552–559. doi: 10.1093/nar/28.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beloin C, Jeusset J, Revet B, Mirambeau G, Le Hegarat F, Le Cam E. Contribution of DNA conformation and topology in right handed DNA wrapping by the Bacillus subtilis LrpC protein. J. Biol. Chem. 2003;278:5333–5342. doi: 10.1074/jbc.M207489200. [DOI] [PubMed] [Google Scholar]
- 44.Chen S, Calvo JM. Leucine-induced dissociation of Escherichia coli Lrp hexadecamers to octamers. J. Mol. Biol. 2002;318:1031–1042. doi: 10.1016/S0022-2836(02)00187-0. [DOI] [PubMed] [Google Scholar]
- 45.Parish T, Stoker NG. The common aromatic amino acid biosynthesis pathway is essential in Mycobacterium tuberculosis. Microbiology. 2002;148:3069–3077. doi: 10.1099/00221287-148-10-3069. [DOI] [PubMed] [Google Scholar]
- 46.Ducati RG, Basso LA, Santos DS. Mycobacterial shikimate pathway enzymes as targets for drug design. Curr. Drug Targets. 2007;8:423–435. doi: 10.2174/138945007780059004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.