Abstract
This paper tests the hypothesis that salivary proteins and their counterpart mRNAs co-exist in human whole saliva. Global profiling of human saliva proteomes and transcriptomes by mass spectrometry (MS) and expression microarray technologies, respectively, revealed many similarities between saliva proteins and mRNAs. Of the function-known proteins identified in saliva, from 61 to 70% were also found present as mRNA transcripts. For genes not detected at both protein and mRNA levels, we made further efforts to determine if the counterpart is present. Of 19 selected genes detected only at the protein level, the mRNAs of 13 (68%) genes were found in saliva by RT-PCR. In contrast, of many mRNAs detected only by microarrays, their protein products were found in saliva, as reported previously by other investigators. The saliva transcriptome may provide preliminary insights into the boundary of the saliva proteome.
Keywords: microarray, mass spectrometry, saliva proteome, saliva transcriptome, correlation analysis
INTRODUCTION
Human saliva contains informative components that can be used as diagnostic markers for human diseases. Our laboratory is using patient-based genome-wide and proteome-wide technologies to identify disease biomarkers from saliva. We have recently identified thousands of human mRNAs in saliva, using expression microarrays (Li et al., 2004a), and discovered salivary RNA markers for oral cancer detection. Four saliva mRNA biomarkers (OAZ-1, SAT, IL1b, and IL8) collectively exhibit a 91% sensitivity and specificity for human oral cancer (Li et al., 2004b).
We have recently conducted human saliva proteome analysis using mass spectrometry (Hu et al., 2004, 2005). Cataloguing of the total proteins in human saliva will form a solid foundation for disease biomarker discovery study. In this study, we hypothesized that proteins and their counterpart mRNAs co-exist in human saliva. We tested the hypothesis by performing correlation analysis of saliva proteomes and transcriptomes from three healthy volunteers. The salivary transcriptome may provide preliminary insights into the boundary of the saliva proteome.
MATERIALS & METHODS
Saliva Sample Collection and Processing
Unstimulated saliva was collected between 9 a.m. and 10 a.m. according to published protocols (Navazesh, 1993). Participants were asked to refrain from eating, drinking, smoking, or oral hygiene procedures, and a water mouthrinse was required prior to sample collection. The study population was composed of six males and four females, with an average age of 42 yrs (range, 32–55 yrs). The following inclusion criteria were used: age ≥ 30 yrs, and no history of malignancy, immunodeficiency, autoimmune disorders, hepatitis, HIV infection, or smoking. Each participant underwent a physical examination by a dentist, to ensure that no suspicious mucosal lesion or inflammation was present in the oral cavity. The oral mucosa appeared healthy, without erythema and epithelial desquamations. There was also no active decay observed. A male volunteer (age, 33 yrs) was used for comparative saliva proteome and transcriptome analysis. All participants signed the institutional-review-board-approved consent form. RNase inhibitor (Superase-In, Ambion Inc., Austin, TX, USA) and protease inhibitor cocktail (Sigma, St. Louis, MO, USA) were immediately added to the saliva. The samples were centrifuged at 2600 g for 15 min (4°C), and the supernatant was aliquoted and stored at −80°C.
Saliva Proteome Analysis
Both “shotgun” and 2-D electrophoresis/MS (2-DE/MS) approaches were utilized for saliva proteome analysis of a single participant (Hu et al., 2005). For the “shotgun” approach, saliva samples (1 mL) were pre-fractionated with the use of Millipore ultracentrifuge filters (Millipore Corp., Billerica, MA, USA). Individual fractions were treated with 10 mM dithreitol for 30 min and then 50 mM iodoacetamide for 30 min. Afterward, each fraction was digested overnight with 100 ng trypsin at 37°C. The resulting peptide digests were then analyzed with LC-MS/MS (QSTAR® Pulsar XL QqTOF-MS; LC Packings and Applied Biosystems, Sunnyvale, CA, USA) on an LC Packings PepMap C18 pre-column and C18 analytical column. For 2-DE/MS experiments, saliva proteins were sequentially separated by isoelectric focusing and SDS-PAGE, and were visualized with Sypro-Ruby (Molecular Probes, Eugene, OR, USA). Gel spots were then excised and digested for MALDI-MS analysis (Micromass-Waters, Beverly, MA, USA), with α-cyano-4-hydroxycinnamic acid as the matrix. Database searching was performed with Mascot software (Matrix Science, London, UK).
RNA Isolation, Amplification, and HG-U133A Microarray Analysis
RNA was isolated from saliva supernatant with the use of a QIAamp Viral RNA kit (Qiagen, Valencia, CA, USA) as previously reported (Li et al., 2004a). Aliquots of RNA were treated with RNase-free DNase (DNase I-DNA-free, Ambion Inc., Austin, TX, USA) and then subjected to linear amplification in a RiboAmp™ RNA Amplification kit (Arcturus, Mountain View, CA, USA). We then used the Human Genome U133A Array (Affymetrix, Santa Clara, CA, USA) to perform gene expression analysis. The arrays were scanned, and the fluorescence intensities were measured by Microarray Suite 5.0 software (Affymetrix). The data were imported into DNA-Chip Analyzer software (Affymetrix) for normalization and model-based analysis (Li and Wong, 2001). A detection p-value was obtained for each probe set, and any probe sets with p < 0.04 were assigned as “present”, indicating that the matching gene transcript was reliably detected (Affymetrix, 2001). The raw data were then exported to Microsoft Excel software for data-sorting and -mining.
RT-PCR
We performed RT-PCR to validate a subset of selected transcripts that were assigned as “absent” on all 10 microarrays. Total RNA was reverse-transcribed in 40 μL of reaction mixture containing 2.5 U of Moloney murine leukemia virus reverse transcriptase and 50 pmol of random hexanucleotides (Applied Biosystems Inc.) at 42°C for 45 min. Oligonucleotide primers for PCR were obtained from Fisher Scientific (Tustin, CA, USA) (Table). Amplification of the complementary DNA (cDNA) was carried out by 50 cycles at 95°C for 20 sec, customized annealing Tm for 30 sec, and at 72°C for 30 sec, followed by a final extension cycle at 72°C for 7 min. Specificity of the PCR products was verified by the predicted size and by restriction digestion. To establish the specificity of the responses, we used negative controls in which either input RNA or the reverse transcriptase was omitted. As a positive control, mRNA was extracted from total salivary gland RNA (Clontech, Palo Alto, CA, USA).
Table.
RT-PCR Validation of Human Saliva mRNAs Undetectable by Microarray
| Gene Name (symbol) | Accession | Primer Sequence (5′ to 3′) |
|---|---|---|
| Human serum albumin (ALB) | M12523 | F: TGTTGCCCATTGTCCTGTTC
R: CCCAGAGTATTCCACTGCTGAG |
| Heat-shock 60-kDa protein 1 (HSPD1) | NM_002156 | F: TGGAATGGGAGGTGGTATG
R: GGTGAGGAACACTGCCTTG |
| Defensin, alpha-3 (DEFA3) | NM_005217 | F: CATCCTTGCTGCCATTCTC
R: AGGGAAACAACCACTTCTGG |
| Neutrophil defensin 3 (HNP-3) | M23281 | F: TCCTTGCTGCCATTCTCCTG
R: CACTTCTGGGATGTCCGCTG |
| Integrin, beta 6 (ITGB6) | NM_000888 | F: GGATGGTTCTGTTTCCTGCTC
R: GAATGTTTGGAGGCTTCGG |
| Kallikrein 1 (KLK1) | NM_002257 | F: CTGTGAAGGTCGTGGAGTTG
R: TAGGCAGGATTTTGAGGTCC |
| Ribosomal protein S11 (RPS11) | NM_001015 | F: CCCTTCACTGGTAATGTGTCC
R: AGATAGTCTCGGCGGATGAC |
| Transferrin (TF) | NM_001063 | F: GGAGGAGTATGCGAACTGCC
R: CCAAATAGGTGCTGCTGTTGAC |
| Zinc-alpha-2-glycoprotein precursor (LOC340333) | XM_294344 | F: CCGTGCCTTCTTCCACTACAAC
R: TGCTTCTCCCAGTCCTCTACTCC |
| Cystatin SN (CYST1) | NM_001898 | F: ACAAGGCCACCAAAGATGAC
R: GGGCTGGGACTTGGTACATA |
| Cystatin SA (CYST2) | NM_001322 | F: GGCCGAACCATATGTACCAA
R: TTGACACCTGGAATTCACCA |
| Carbonic anhydrase IV (CA4) | NM_000717 | F: ATACCAGGCCAAACAGTTGC
R: ATTCCTCGATGTCCCCTTCT |
| Transcobalamin I (TCN1) | NM_001062 | F: CATGCAGGCCCTCTTTGTAT
R: TTGCTGAATGCTCCTTGAGA |
| Hepatocellular carcinoma-associated protein TB6 (PIGR) | AF272149 | F: ATTTGGCATCTCCTGTCCTG
R: TGGGAAAGATCTCCCTCCTT |
| Myeloperoxidase (MPO) | NM_000250 | F: TCGGTACCCAGTTCAGGAAG
R: GTGGTGATGCCTGTGTTGTC |
| Hemopexin (HPX) | BC005395 | F: TTGGCCCTAACTCATGTTCC
R: CCCAGGAGACTGGTCACATT |
| Alpha-1-antitrypsin (SERPINA1) | K01396 | F: CACCCACGATATCATCACCA
R: CCCCATTGCTGAAGACCTTA |
RESULTS
Analysis of Salivary Proteomes and Transcriptomes
Using proteomics profiling, we have identified 309 saliva proteins from a healthy participant (Participant 1), including 220 proteins with known biological functions (Hu et al., 2004, 2005). A 2-D virtual display (pI vs. Mr) illustrates the isoelectric point and molecular-weight distributions of all identified saliva proteins (pI 4.4–12.1; Mr 2.9–590 kDa) (Fig. 1A). Such a virtual gel can allow for the location of saliva proteins of interest on a real 2-D electrophoresis gel. Recently, we have conducted the proteome analysis of saliva samples from two additional healthy volunteers. In total, we have identified 282 (213 function-known) and 297 (229 function-known) distinct saliva proteins from Participants 2 and 3, respectively.
Figure 1.
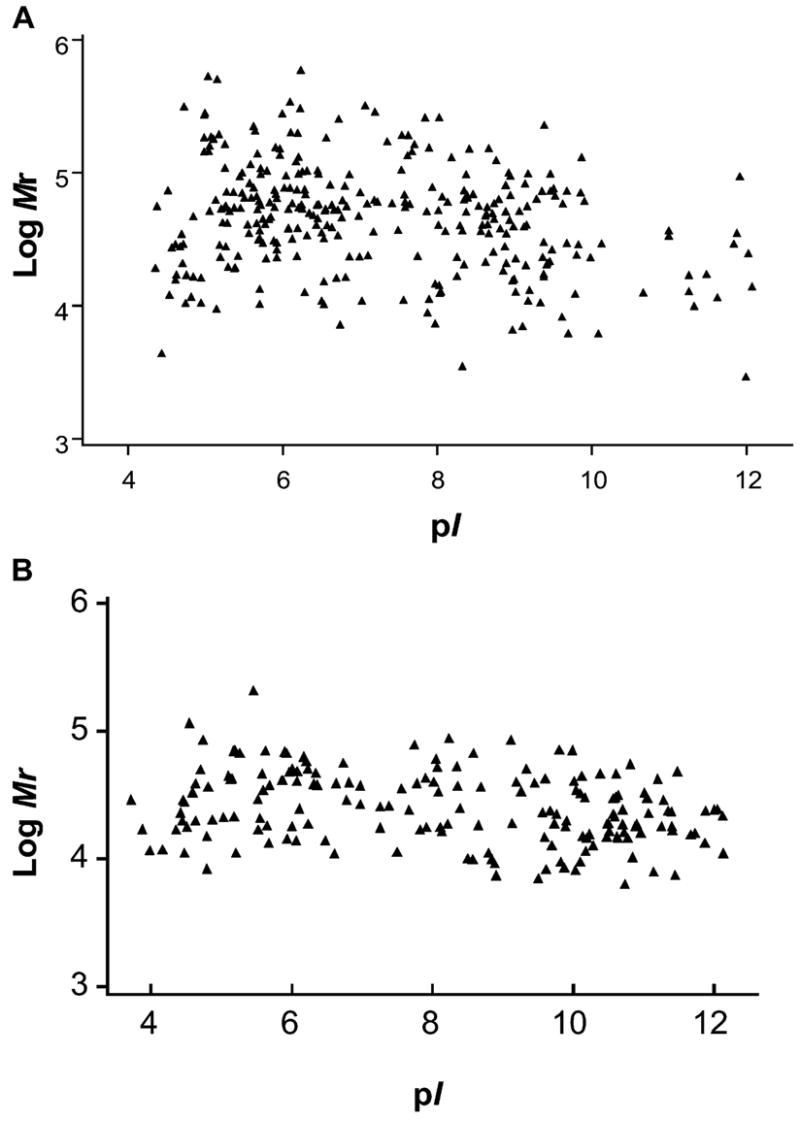
2-D virtual display (pI vs. log Mr) of all identified proteins in human saliva (A) (n = 1), as well as the proteins complementary to the Normal Saliva Core Transcriptome list (B) (n = 10).
Saliva mRNA profiles of the same three participants were obtained with the use of HG U133A expression arrays, which contain 22,215 human gene cDNA probe sets, representing ~ 19,000 genes. For the three participants, 3160, 2698, and 3638 probe sets (p < 0.04) on the array were assigned as present, indicating that 2940, 2521, and 3363 saliva mRNAs, respectively, were reliably detected.
Normal Saliva Core Transcriptome (NSCT)
We have profiled the saliva mRNA of 10 normal individuals using the U133A arrays. On average, 3143 ± 665 probe sets (p < 0.04, n = 10) were assigned as present (Li et al., 2004a). Two hundred seven probe sets, representing 185 genes, were detected among all 10 participants (detection p < 0.01). Using the same criterion, we found 570 genes present in 8/10 (80%) arrays, and 417 genes in 9/10 (90%) arrays. Among these, 49 and 37 salivary mRNAs, respectively, were found to be present in saliva at protein levels as determined by proteomic approaches. The genes present among all 10 arrays (100%) are referred to as “Normal Saliva Core Transcriptome (NSCT)” (APPENDIX). Their nucleotide sequences were virtually translated into amino acid sequences, and then the corresponding pI and molecular weights were displayed in a virtual 2-D format (Fig. 1B). These core genes were evenly distributed across a wide pI range (pI 3.7–12.1), but their Mr fell into a relatively narrow range, from around 10 to 100 kDa. Of the 185 core mRNAs, 43 (23%, highlighted in the (APPENDIX) were also found present at the protein level in the 3 saliva samples examined by proteomics approaches.
APPENDIX.
Normal Saliva Core Transcriptome (NSCT)
| Gene Name | NCBI ID |
|---|---|
| 6.2-kDa protein | NM_019059 |
| Acidic protein rich in leucines | NM_006401 |
| Actin-related protein 2/3 complex, subunit 2 (34 kDa) | NM_005731 |
| Actin, beta | NM_001101 |
| Actin, gamma 1 | NM_001614 |
| Activating transcription factor 4 (tax-responsive enhancer element B67) | NM_001675 |
| Adenylyl cyclase-associated protein | AA806142 |
| Annexin A1 | NM_000700 |
| Annexin A2 | NM_004039 |
| Beta-2-microglobulin | NM_004048 |
| Brain-abundant, membrane-attached signal protein 1 | NM_006317 |
| BTG family, member 2 | NM_006763 |
| Calmodulin 1 (phosphorylase kinase, delta) | NM_006888 |
| Calmodulin 2 (phosphorylase kinase, delta) | NM_001743 |
| CD24 antigen (small cell lung carcinoma cluster 4 antigen) | AA761181 |
| CD47 antigen (Rh-related antigen, integrin-associated signal transducer) | BG230614 |
| CGI-75 protein | NM_016020 |
| Chromobox homolog 3 (HP1 gamma homolog, Drosophila) | NM_016587 |
| Chromosome 1 open reading frame 10 | NM_016190 |
| Chromosome 1 open reading frame 8 | NM_004872 |
| Cofilin 1 (non-muscle) | NM_005507 |
| Cold-inducible RNA-binding protein | NM_001280 |
| Cold-shock domain protein A | AL556190 |
| Consensus includes gb:AA477655/FEA=EST/DB_XREF=gi:2206289/DB_XREF=est:zu37h09.s1/CLONE=IMAGE:740225/UG=Hs.181307 H3 histone, family 3A | AA477655 |
| Consensus includes gb:AI345238/FEA=EST/DB_XREF=gi:4082444/DB_XREF=est:tb81b07.×1/CLONE=IMAGE:2060725/UG=Hs.111334 ferritin, light polypeptide | AI345238 |
| Consensus includes gb:AI721229/FEA=EST/DB_XREF=gi:5038485/DB_XREF=est:as68c10.×1/CLONE=IMAGE:2333874/UG=Hs.326456 hypothetical protein FLJ20030 | AI721229 |
| Consensus includes gb:AI923984/FEA=EST/DB_XREF=gi:5659948/DB_XREF=est:wn49d12.×1/CLONE=IMAGE:2448791/UG=Hs.46320 Small proline-rich protein SPRK human, odontogenic keratocysts, mRNA Partial, 317 nt | AI923984 |
| Consensus includes gb:AL078596/DEF=Human DNA sequence from clone RP3-429G5 on chromosome 6q21-22.1. Contains the NR2E1 gene for nuclear receptor 2E1 (tailless, TLL, TLX, XTLL), the 3′ end of the SNX3 gene for sorting nexin 3, ESTs, STSs, GSSs and 4 pr | AL078596 |
| Consensus includes gb:AL121916/DEF=Human DNA sequence from clone RP1-189G13 on chromosome 20. Contains an RPL7A (60S ribosomal protein L7A) (SURF3) pseudogene, part of an RPS4 (40S ribosomal protein S4) pseudogene, ESTs, STSs and GSSs/FEA=CDS_2/DB_XRE | AL121916 |
| Consensus includes gb:AL356115/DEF=Human DNA sequence from clone RP11-486O22 on chromosome 10. Contains the 3′ part of a gene for KIAA1128 protein, a novel pseudogene, a gene for protein similar to RPS3A (ribosomal protein S3A), ESTs, STSs, GSSs and CpG is | AL356115 |
| Consensus includes gb:AW304232/FEA=EST/DB_XREF=gi:6713921/DB_XREF=est:xv82g01.×1/CLONE=IMAGE:2825040/UG=Hs.181357 laminin receptor 1 (67kD, ribosomal protein SA) | AW304232 |
| Consensus includes gb:BE869922/FEA=EST/DB_XREF=gi:10318698/DB_XREF=est:601446568F1/CLONE=IMAGE:3850432/UG=Hs.181307 H3 histone, family 3A | BE869922 |
| Consensus includes gb:BE963164/FEA=EST/DB_XREF=gi:11766582/DB_XREF=est:601656973R1/CLONE=IMAGE:3865650/UG=Hs.2186 eukaryotic translation elongation factor 1 gamma | BE963164 |
| Consensus includes gb:BG168283/FEA=EST/DB_XREF=gi:12674986/DB_XREF=est:602340822F1/CLONE=IMAGE:4448789/UG=Hs.82202 ribosomal protein L17 | BG168283 |
| Consensus includes gb:BG537190/FEA=EST/DB_XREF=gi:13528922/DB_XREF=est:602565589F1/CLONE=IMAGE:4690079/UG=Hs.111334 ferritin, light polypeptide | BG537190 |
| Consensus includes gb:L13283.1/DEF=Homo sapiens (clone MG2-5-12) mucin (MG2) mRNA, complete polyA site./FEA=mRNA/DB_XREF=gi:292518/UG=Hs.103944 Homo sapiens (clone MG2-5-12) mucin (MG2) mRNA, complete polyA site | L13283 |
| Cyclin I | AF135162 |
| Cystatin A (stefin A) | NM_005213 |
| Cystatin B (stefin B) | NM_000100 |
| Cytochrome c oxidase subunit IV isoform 1 | AA854966 |
| Cytochrome c oxidase subunit VIIa polypeptide 2 (liver) | NM_001865 |
| Cytochrome c oxidase subunit VIIa polypeptide 2 like | NM_004718 |
| Dual-specificity phosphatase 1 | NM_004417 |
| eIEF-associated protein HSPC021 | NM_016091 |
| Epithelial membrane protein 1 | NM_001423 |
| Eukaryotic translation elongation factor 1 alpha 1 | NM_001403 |
| Eukaryotic translation elongation factor 1 gamma | AF119850 |
| Eukaryotic translation initiation factor 3, subunit 3 (gamma, 40kD) | NM_003756 |
| Farnesyl-diphosphate farnesyltransferase 1 | BC003573 |
| Ferritin, heavy polypeptide 1 | NM_002032 |
| Ferritin, light polypeptide | BF312331 |
| Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox-derived); ribosomal protein S30 | NM_001997 |
| G-protein-coupled receptor kinase 7 | NM_017572 |
| gb:BC001865.1/DEF=Homo sapiens, Similar to cadherin 1, type 1, E-cadherin (epithelial), clone MGC:1151, mRNA, complete cds./FEA=mRNA/PROD=Similar to cadherin 1, type 1, E-cadherin (epithelial)/DB_XREF=gi:12804838/UG=Hs.194657 cadherin 1, type 1, E-c | BC001865 |
| gb:L08666.1/DEF=Homo sapiens porin (por) mRNA, complete cds and truncated cds./FEA=mRNA/GEN=por; por/PROD=porin; porin/DB_XREF=gi:190199/FL=gb:L08666.1 | L08666 |
| gb:NM_000970.1/DEF=Homo sapiens ribosomal protein L6 (RPL6), mRNA./FEA=mRNA/GEN=RPL6/PROD=ribosomal protein L6/DB_XREF=gi:4506656/UG=Hs.174131 ribosomal protein L6/FL=gb:BC004138.1 gb:D17554.1 gb:NM_000970.1 gb:AF261087.1 | NM_000970 |
| Glioma tumor suppressor candidate region gene 2 | NM_015710 |
| Glutamate-ammonia ligase (glutamine synthase) | AL161952 |
| Glyceraldehyde-3-phosphate dehydrogenase | AK026525 |
| GNAS complex locus | NM_000516 |
| Guanine nucleotide-binding protein (G protein), beta polypeptide 2-like 1 | NM_006098 |
| H3 histone, family 3A | AI955655 |
| Heat-shock 70-kDa protein 8 | AA704004 |
| Heat-shock 90-kDa protein 1, alpha | R01140 |
| Heterogeneous nuclear ribonucleoprotein A1 | NM_002136 |
| Histatin 1 | NM_002159 |
| Histidine triad nucleotide-binding protein | N32864 |
| HSPC019 protein | Z98200 |
| Hypothetical protein | BG167522 |
| Hypothetical protein FLJ20030 | NM_017627 |
| Hypothetical protein FLJ20897 | AI613383 |
| Hypothetical protein R33729_1 | Z78330 |
| Hypothetical protein SMAP31 | AB059408 |
| Interleukin 1 receptor antagonist | U65590 |
| Interleukin 1, beta | M15330 |
| Interleukin 8 | NM_000584 |
| Jumping translocation breakpoint | BC004239 |
| Keratin 13 | NM_002274 |
| Keratin 4 | X07695 |
| Keratin 6A | J00269 |
| Keratin 6B | AI831452 |
| KIAA1919 protein | AK000168 |
| Lactate dehydrogenase A | NM_005566 |
| LPS-induced TNF-alpha factor | AB034747 |
| Lysyl-tRNA synthetase | NM_00554 |
| Major histocompatibility complex, class I, C | AK024836 |
| Mal, T-cell differentiation protein | NM_002371 |
| Mitochondrial carrier homolog 1 | AF189289 |
| Mitogen-activated protein kinase kinase 3 | AA780381 |
| Myosin, light polypeptide 6, alkali, smooth muscle and non-muscle | BE734356 |
| Myosin, light polypeptide, regulatory, non-sarcomeric (20 kDa) | NM_006471 |
| NICE-1 protein | NM_019060 |
| Nuclear receptor co-activator 4 | AL162047 |
| Nuclease-sensitive element-binding protein 1 | BC002411 |
| Ornithine decarboxylase antizyme 1 | D87914 |
| Peptidylprolyl isomerase F (cyclophilin F) | BC005020 |
| Periplakin | NM_002705 |
| Phosphatase and tensin homolog (mutated in multiple advanced cancers 1), Pseudogene 1 | AF023139 |
| Phosphoglycerate mutase 1 (brain) | NM_002629 |
| Poly(A) binding protein, cytoplasmic 1 | AI734929 |
| Poly(A) binding protein, cytoplasmic 3 | NM_030979 |
| Pre-B-cell colony-enhancing factor | NM_005746 |
| Proteoglycan 1, secretory granule | NM_002727 |
| Prothymosin, alpha (gene sequence 28) | NM_002823 |
| Putative lymphocyte G0/G1 switch gene | NM_015714 |
| Putative Rab5-interacting protein | NM_018840 |
| Putative translation initiation factor | BF246436 |
| Putative translation initiation factor | W67644 |
| RAB11A, member RAS oncogene family | AI215102 |
| RAB13, member RAS oncogene family | NM_002870 |
| RAN binding protein 9 | AF306510 |
| Ras homolog enriched in brain 2 | D78132 |
| Ribosomal protein L12 | NM_000976 |
| Ribosomal protein L13 | BC004954 |
| Ribosomal protein L13a | NM_012423 |
| Ribosomal protein L14 | AA838274 |
| Ribosomal protein L15 | NM_002948 |
| Ribosomal protein L18 | AV738806 |
| Ribosomal protein L19 | NM_000981 |
| Ribosomal protein L21 | NM_000982 |
| Ribosomal protein L22 | BG152979 |
| Ribosomal protein L23a | NM_000984 |
| Ribosomal protein L24 | AI560573 |
| Ribosomal protein L27 | NM_000988 |
| Ribosomal protein L27a | NM_000990 |
| Ribosomal protein L3 | NM_000967 |
| Ribosomal protein L30 | L05095 |
| Ribosomal protein L31 | NM_000993 |
| Ribosomal protein L32 | NM_000994 |
| Ribosomal protein L34 | NM_000995 |
| Ribosomal protein L37 | BF216701 |
| Ribosomal protein L37a | NM_000998 |
| Ribosomal protein L39 | BC001019 |
| Ribosomal protein L4 | NM_000968 |
| Ribosomal protein L41 | NM_021104 |
| Ribosomal protein L5 | NM_000969 |
| Ribosomal protein L7 | NM_000971 |
| Ribosomal protein L7a | NM_000972 |
| Ribosomal protein S10 | AA320764 |
| Ribosomal protein S11 | NM_001015 |
| Ribosomal protein S14 | AF116710 |
| Ribosomal protein S15a | NM_001019 |
| Ribosomal protein S17 | NM_001021 |
| Ribosomal protein S18 | NM_022551 |
| Ribosomal protein S19 | NM_001022 |
| Ribosomal protein S2 | NM_002952 |
| Ribosomal protein S21 | NM_001024 |
| Ribosomal protein S23 | NM_001025 |
| Ribosomal protein S27 (metallopanstimulin 1) | NM_001030 |
| Ribosomal protein S28 | BC000354 |
| Ribosomal protein S3A | NM_001006 |
| Ribosomal protein S4, X-linked | AW132023 |
| Ribosomal protein S6 | NM_001010 |
| Ribosomal protein S7 | AI970731 |
| Ribosomal protein S8 | NM_001012 |
| Ribosomal protein S9 | BE348997 |
| Ribosomal protein, large P2 | NM_001004 |
| Ribosomal protein, large, P0 | NM_001002 |
| Ribosomal protein, large, P1 | NM_001003 |
| S100 calcium-binding protein A2 | NM_005978 |
| S100 calcium-binding protein A8 (calgranulin A) | NM_002964 |
| S100 calcium-binding protein A9 (calgranulin B) | NM_002965 |
| Salivary proline-rich protein | NM_006685 |
| Signal recognition particle 14 kDa (homologous Alu RNA-binding protein) | NM_003134 |
| Small proline-rich protein 1A | NM_005987 |
| Small proline-rich protein 1B (cornifin) | NM_003125 |
| Small proline-rich protein 3 | NM_005416 |
| Solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 | NM_002635 |
| Spermidine/spermine N1-acetyltransferase | NM_002970 |
| Stratifin | X57348 |
| Stress-associated endoplasmic reticulum protein 1 | NM_014445 |
| Superoxide dismutase 2, mitochondrial | W46388 |
| Tax1 (human T-cell leukemia virus type I) binding protein 1 | NM_006024 |
| Transaldolase 1 | NM_006755 |
| Transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) | BF976260 |
| tRNA isopentenylpyrophosphate transferase | BE964125 |
| Tropomyosin 4 | AI214061 |
| Tubulin, alpha 3 | AF141347 |
| Tumor protein, translationally controlled 1 | AL565449 |
| Ubiquitin B | NM_018955 |
| Ubiquitin C | AB009010 |
| Ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) | NM_003340 |
| Ubiquitin-conjugating enzyme E2L 6 | NM_004223 |
| Uncharacterized hypothalamus protein HT011 | BE565675 |
| Zinc finger protein 36, C3H type, homolog (mouse) | NM_003407 |
Co-existence of Saliva Protein and mRNA
The saliva protein and mRNA lists obtained from the three participants’ saliva were compared, and a co-existence phenomenon was observed. In Participant 1, 309 saliva proteins were identified, including 220 proteins with known biological functions and 89 hypothetical proteins without annotated function. Microarray profiling indicated that 154 (154/220, 70%) proteins with known functions were detected at the mRNA level, and the remaining 66 were not detectable. Similarly, 61% (130/213) and 65% (149/229) of function-known saliva proteins identified in Participants 2 and 3, respectively, were also found to “co-exist” with their saliva mRNAs.
Because microarray profiling may give false-negative results pertaining to the detected absence of saliva mRNA, we proceeded to test if the “undetectable” mRNAs were actually absent or present in saliva. We used RT-PCR to validate the presence of 9 mRNAs randomly selected from the “undetectable” 66 mRNAs from Participant 1. These mRNAs included serum albumin (ALB), heat-shock 60-kDa protein 1 (HSPD1), defensin, alpha-3 (DEFA3), neutrophil defensin 3 (HNP-3), integrin, beta 6 (ITGB6), kallikrein 1 (KLK1), ribosomal protein S11 (RPS11), transferrin (TF), and zinc-alpha-2-glycoprotein precursor (LOC340333) (Table). RT-PCR results indicated that mRNAs of HSPD1, DEFA3, ITGB6, KLK1, RPS11, TF, and HNP-3 were actually present in the saliva from Participant 1, and that LOC340333 and ALB mRNAs were undetectable (Fig. 2). These results suggested that the co-existence of salivary proteins and their mRNAs may reach a higher extent than the 70% (154/220) obtained by comparison of the proteomics and microarray outcomes. Of the remaining 66 (30%) salivary mRNAs that were undetectable by microarray, 7/9 (78%) could be independently detected by RT-PCR. This suggested that ~ 51 (78% of 66) additional saliva proteins may have a corresponding mRNA co-existing in saliva. Therefore, we may predict a co-existence rate of 93% (205/220) for saliva proteins and their counterpart saliva mRNAs. Similarly, we were able to validate 3 out of 5 randomly selected genes initially undetectable by microarray profiling in Participant 2 or Participant 3. RT-PCR analysis indicated that mRNAs of transcobalamin I (TCN1), myeloperoxidase (MPO), and hemopexin (HPX) were actually present in the saliva from Participant 2, whereas hepatocellular carcinoma-associated protein TB6 (PIGR) and alpha-1-antitrypsin (SERPINA1) mRNAs were undetectable. Meanwhile, in Participant 3, cystatin SA (CYST2), carbonic anhydrase IV (CA4), and HPX were detected, whereas cystatin SN (CYST1) and SERPINA1 were not (Figs. 2B, 2C).
Figure 2.
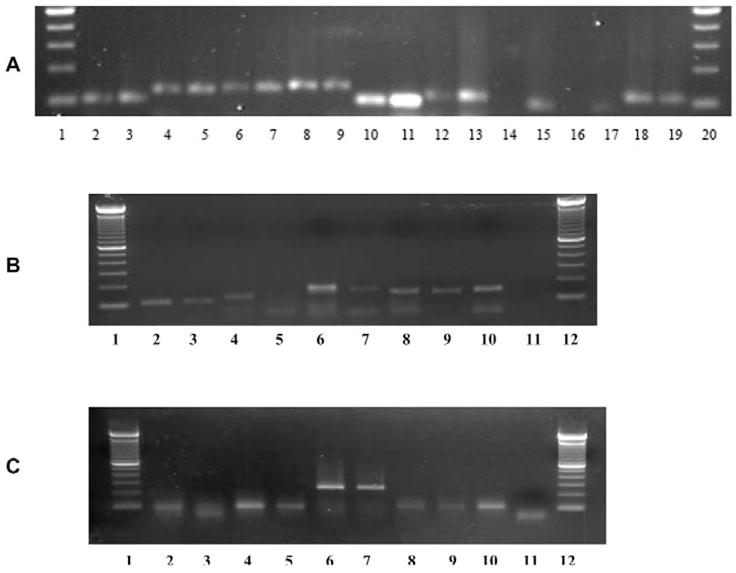
RT-PCR validation of randomly selected mRNAs in Participant 1 (A), Participant 2 (B), and Participant 3 (C). (A) Lanes 1 and 20 are DNA ladders. Saliva RNAs were detected in lanes 2 (HSPD1), 4 (DEFA3), 6 (ITGB6), 8 (KLK1), 10 (RPS11), 12 (TF), and 18 (HNP-3). LOC340333 (lane 14) and ALB (lane 16) were not detectable. Total salivary gland RNA (Clontech, Palo Alto, CA, USA) was used as a positive control, shown in lanes 3 (HSPD1), 5 (DEFA3), 7 (ITGB6), 9 (KLK1), 11 (RPS11), 13 (TF), 15 (LOC340333), 17 (ALB), and 19 (Defensin, HNP-3), respectively. Negative controls were used in which input RNA was omitted, or in which RNA was used but reverse-transcriptase was omitted (data not shown). (B) Lanes 1 and 12 are DNA ladders. Lane 2 (TCN1), lane 4, (PIGR), lane 6 (MPO), lane 8 (HPX), and lane 10 (SERPINA1) are PCR products from total salivary gland RNA, which served as positive controls. Corresponding mRNAs of TCN1 (lane 3), MPO (lane 7), and HPX (lane 9) were detected in the participant’s saliva, but PIGR (lane 5) and SERPINA1 (lane 11) were not detected. (C) Lane 2 (CYST1), lane 4 (CYST2), lane 6 (CA4), lane 8 (HPX), and lane 10 (SERPINA1) are positive controls. CYST2 (lane 5), CA4 (lane 7), and HPX (lane 9) were detected in saliva, but CYST1 (lane 3) and SERPINA1 (lane 11) were not detectable.
DISCUSSION
The correlative analysis of saliva proteome and transcriptome is meaningful only when the comparison is performed within the same individual. Within all three participants examined, certain concordance was observed between saliva proteomes and transcriptomes, as revealed by proteomic and microarray profiling. In Participant 1, 70% (154/220) of the function-known proteins identified in saliva were found present as mRNA transcripts. This may fluctuate somehow, due to incompatible gene names or accession numbers, but it indicates the co-existence phenomenon between saliva proteins and mRNAs. Complementarity was obvious for common saliva proteins (such as statherin, histatins, cystatins, and proline-rich proteins), saliva enzymes (such as lysozyme, amylase, lactate dehydrogenase, and cytochrome c oxidase), and structural proteins (such as actins, tubulins, and keratins). Both proteome and transcriptome profiling revealed a similar percentage of function-unknown genes (28.7% and 27.5%, respectively). Correlation of these function-unknown genes at protein and RNA levels is difficult, due to the lack of their annotation in the databases. Similarly, 61% (130/213) and 65% (149/229) of function-known proteins identified in saliva from Participants 2 and 3 were found present as mRNA transcripts, as revealed by microarray profiling. This might have been underestimated, because, as suggested by the RT-PCR study, microarray profiling may miscall the absence of those remaining mRNAs. While it is highly desirable to analyze the salivary proteome of additional healthy individuals (e.g., 10), the current limited throughput of proteomics technologies renders this goal impractical. Typically, it takes several months to complete the analysis of an individual’s saliva proteome, even with state-of-the-art proteomics technologies. However, through the comparative analyses of three pairs of saliva proteomes and transcriptomes, we achieved a consistent outcome that allowed us to estimate conclusively the correlation between saliva proteins and mRNAs.
Many genes were found only at either the protein or the mRNA level. For instance, proteomics revealed all salivary cystatin isoforms; however, microarray detected only 2 mRNAs from this gene family. This may be simply because there are no matching probe sets on the U133A array for other cystatins. Conversely, microarray revealed many ribosomal mRNAs, while proteomics discovered only some of the ribosomal proteins. It is likely that our preliminary MS analysis yielded only a subset of the entire saliva proteome. An additional pre-fractionation strategy can help expand the catalogue of saliva proteins. Currently, for a specific gene, validation at either the protein or the mRNA level is required for accurate RNA-protein correlation analysis.
In Participant 1, our analysis indicated that 70% (154/220) of the function-known saliva proteins had a corresponding saliva mRNA detectable by microarray. Of the remaining 66 (30%) genes, 9 candidates were selected for detection of the corresponding mRNA by RT-PCR (Table). These mRNA candidates were not found by microarray profiling, possibly because of the absence of matching probe sets on the U133A array. Seven of 9 genes were detectable by RT-PCR in saliva from Participant 1 (Fig. 2A), suggesting a higher correlation between saliva mRNAs and proteins than we obtained from proteomics and microarray profiling. Of the remaining 66 (30%) salivary mRNAs that were undetectable by microarray, 7/9 (78%) could be independently detected by RT-PCR. This suggests that ~ 51 (78% of 66) additional saliva proteins actually have a corresponding mRNA co-existing in saliva, and predicts a co-existence rate of 93% (205/220) for saliva proteins and their counterpart mRNAs. Similarly, further RT-PCR analysis suggested that there was a higher co-existence rate (85% for Participant 2 and 86% for Participant 3) than the one obtained from direct correlation of proteomics and microarray data (61% for Participant 2 and 65% for Participant 3).
The proteins discovered by proteomics represent only a partial list of all proteins in whole saliva. Many saliva proteins at lower concentrations remain to be identified. Therefore, only 7% of saliva mRNAs detected by microarray profiling were found to be present in the protein list. But this does not necessarily mean that the remaining (93%) mRNAs lack counterpart proteins. Several genes—including calmodulin, β-2-microglobulin, epidermal growth factor, ferritin, fibroblast growth factor, heat-shock protein 70, hepatocyte growth factor, histones, interleukin-2, interleukin-6, interleukin-8, superoxide dismutase, etc.—were detected by microarray, but not by proteomic profiling. Other investigators have previously found these proteins to be present in human saliva (Balekjian and Longton, 1973; Law and Henkin, 1986; Maddali et al., 1995; Trubnikov et al., 1998; Kagami et al., 2000; Streckfus et al., 2001; Nagler et al., 2002; Ohshima et al., 2002; Fabian et al., 2003; Eckley et al., 2004). In conclusion, comparative analysis of the saliva proteome and transcriptome from three healthy individuals allowed us to conclude that many salivary proteins and mRNA are concordantly present in human saliva. This is an important observation, since we, and others, are actively deciphering the human salivary proteome. One of the problems inherent in saliva proteome analysis is how to define the boundary of the proteome and decide the ‘finish line’ for the analysis. Considering the relatively high co-existence rate for saliva proteins and their counterpart mRNAs, the salivary transcriptome may serve as a good indicator of the diversity and range of the salivary proteome, and can be used as a reference guideline for human saliva proteome analysis. The specificity and coverage of RNA and protein profiling technologies need to be improved to yield more accurate and complete RNA-protein correlation analysis. In addition to high-throughput MS and microarray profiling, complementary validation methods, such as Western blots/ELISA or RT-PCR, will be required in the conduct of RNA-protein correlation studies.
Acknowledgments
This work was supported by NIH U01 DE015018 (to D.T. Wong) and NIH U01 U01DE016275 (to D.T. Wong and J.A. Loo), and by a faculty seed grant from the UCLA School of Dentistry (to S. Hu). We thank Drs. Jim Kerwin, Tianwei Yu, and Weihong Yan and Mr. Shawn Than for editing the manuscript and uploading the data to the UCLA human salivary proteome Web site (www.hspp.ucla.edu).
Footnotes
A supplemental appendix to this article is published electronically only at http://www.dentalresearch.org.
References
- Affymetrix (2001). Affymetrix technical note: new statistical algorithms for monitoring gene expression on GeneChip® probe arrays. Santa Clara, CA: Affymetrix, Inc., www.affymetrix.com
- Balekjian AY, Longton RW. Histones isolated from human parotid fluid. Biochem Biophys Res Commun. 1973;50:676–682. doi: 10.1016/0006-291x(73)91297-7. [DOI] [PubMed] [Google Scholar]
- Eckley CA, Michelsohn N, Rizzo LV, Tadakoro CE, Costa HO. Salivary epidermal growth factor concentration in adults with reflux laryngitis. Otolaryngol Head Neck Surg. 2004;131:401–406. doi: 10.1016/j.otohns.2004.01.020. [DOI] [PubMed] [Google Scholar]
- Fabian TK, Gaspar J, Fejerdy L, Kaan B, Balint M, Csermely P, Fejerdy P. Hsp70 is present in human saliva. Med Sci Monit. 2003;9:BR62–BR65. [PubMed] [Google Scholar]
- Hu S, Denny P, Denny P, Xie Y, Loo JA, Wolinsky LE, et al. Differentially expressed protein markers in human submandibular and sublingual secretions. Int J Oncol. 2004;25:1423–1430. [PubMed] [Google Scholar]
- Hu S, Xie Y, Ramachandran P, Loo RRO, Li Y, Loo JA, et al. Large-scale identification of proteins in human salivary proteome by liquid chromatography/mass spectrometry and two-dimensional gel electrophoresis-mass spectrometry. Proteomics. 2005;5:1714–1728. doi: 10.1002/pmic.200401037. [DOI] [PubMed] [Google Scholar]
- Kagami H, Hiramatsu Y, Hishida S, Okazaki Y, Horie K, Oda Y, et al. Salivary growth factors in health and disease. Adv Dent Res. 2000;14:99–102. doi: 10.1177/08959374000140011601. [DOI] [PubMed] [Google Scholar]
- Law JS, Henkin RI. Low parotid saliva calmodulin in patients with taste and smell dysfunction. Biochem Med Metab Biol. 1986;36:118–124. doi: 10.1016/0885-4505(86)90115-5. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhou X, St John MA, Wong DT. RNA profiling of cell-free saliva using microarray technology. J Dent Res. 2004a;83:199–203. doi: 10.1177/154405910408300303. [DOI] [PubMed] [Google Scholar]
- Li Y, St John MA, Zhou X, Kim Y, Sinha U, Jordan RC, et al. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res. 2004b;10:8442–8450. doi: 10.1158/1078-0432.CCR-04-1167. [DOI] [PubMed] [Google Scholar]
- Maddali Bongi S, Campana G, D’Agata A, Palermo C, Bianucci G. The diagnosis value of beta 2-microglobulin and immunoglobulins in primary Sjögren’s syndrome. Clin Rheumatol. 1995;14:151–156. doi: 10.1007/BF02214934. [DOI] [PubMed] [Google Scholar]
- Nagler RM, Klein I, Zarzhevsky N, Drigues N, Reznick AZ. Characterization of the differentiated antioxidant profile of human saliva. Free Radic Biol Med. 2002;32:268–277. doi: 10.1016/s0891-5849(01)00806-1. [DOI] [PubMed] [Google Scholar]
- Navazesh M. Methods for collecting saliva. Ann NY Acad Sci. 1993;694:72–77. doi: 10.1111/j.1749-6632.1993.tb18343.x. [DOI] [PubMed] [Google Scholar]
- Ohshima M, Fujikawa K, Akutagawa H, Kato T, Ito K, Otsuka K. Hepatocyte growth factor in saliva: a possible marker for periodontal disease status. J Oral Sci. 2002;44:35–39. doi: 10.2334/josnusd.44.35. [DOI] [PubMed] [Google Scholar]
- Streckfus C, Bigler L, Navazesh M, Al-Hashimi I. Cytokine concentrations in stimulated whole saliva among patients with primary Sjögren’s syndrome, secondary Sjögren’s syndrome, and patients with primary Sjögren’s syndrome receiving varying doses of interferon for symptomatic treatment of the condition: a preliminary study. Clin Oral Investig. 2001;5:133–135. doi: 10.1007/s007840100104. [DOI] [PubMed] [Google Scholar]
- Trubnikov GA, Sukharev AE, Uklistaia TA, Orlova EA. Clinico-diagnostic role of ferritin and lactoferrin assays in benign and malignant affections of lungs and pleura. Klin Med (Mosk) 1998;76:21–26. article in Russian. [PubMed] [Google Scholar]


