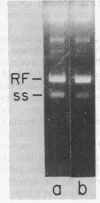
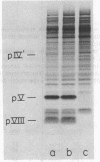
Abstract
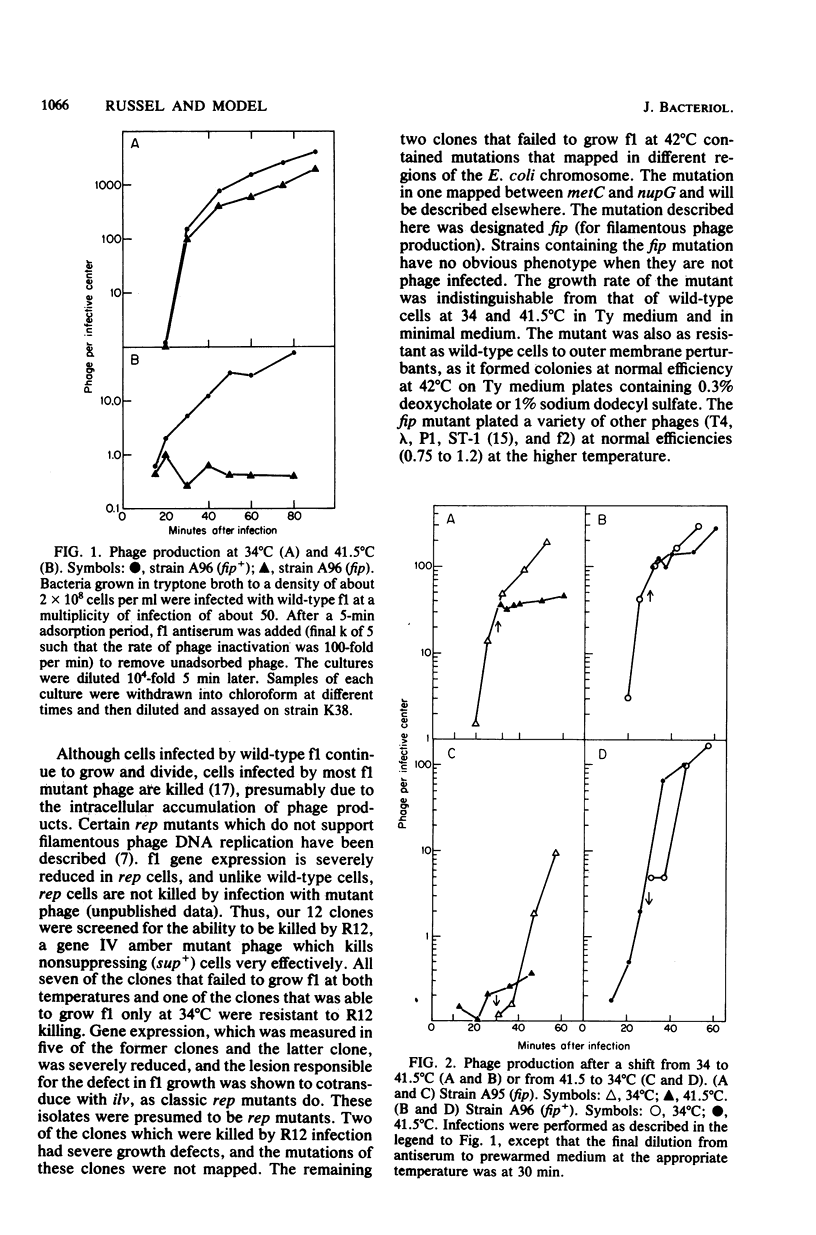
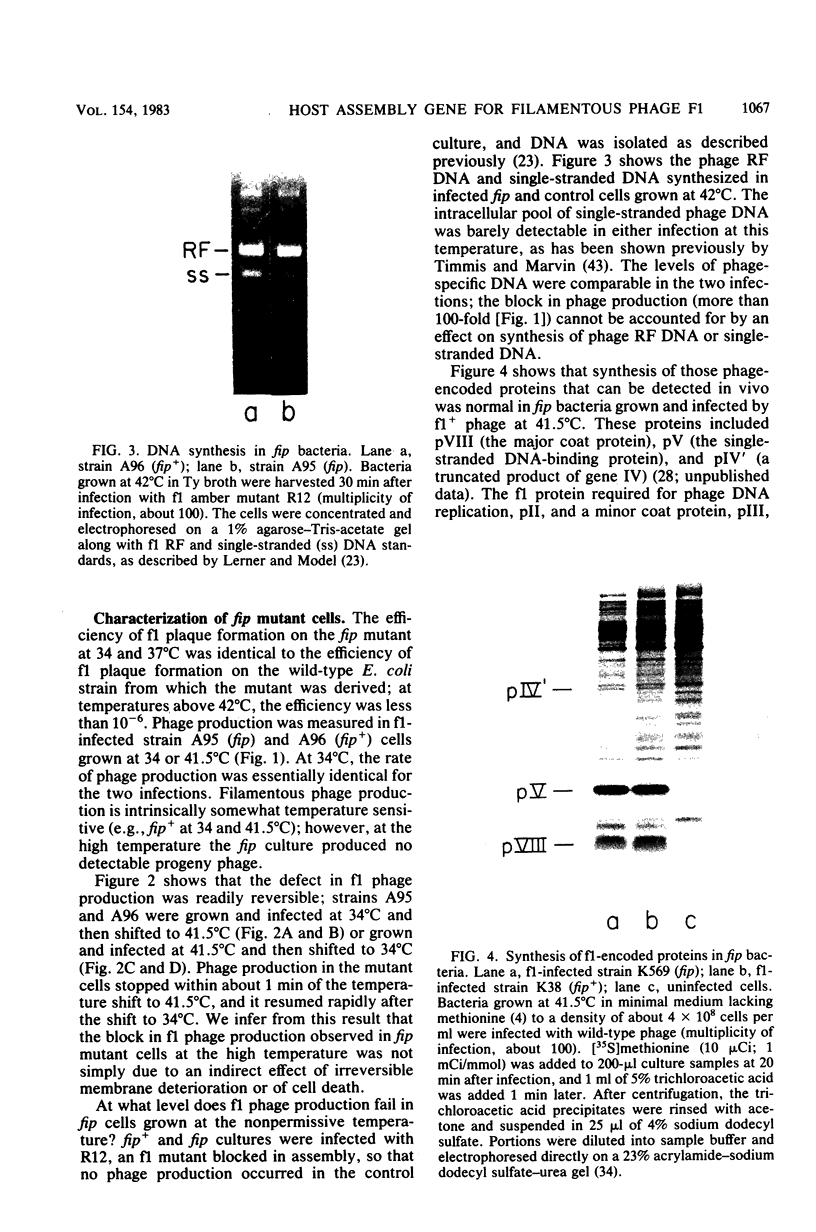
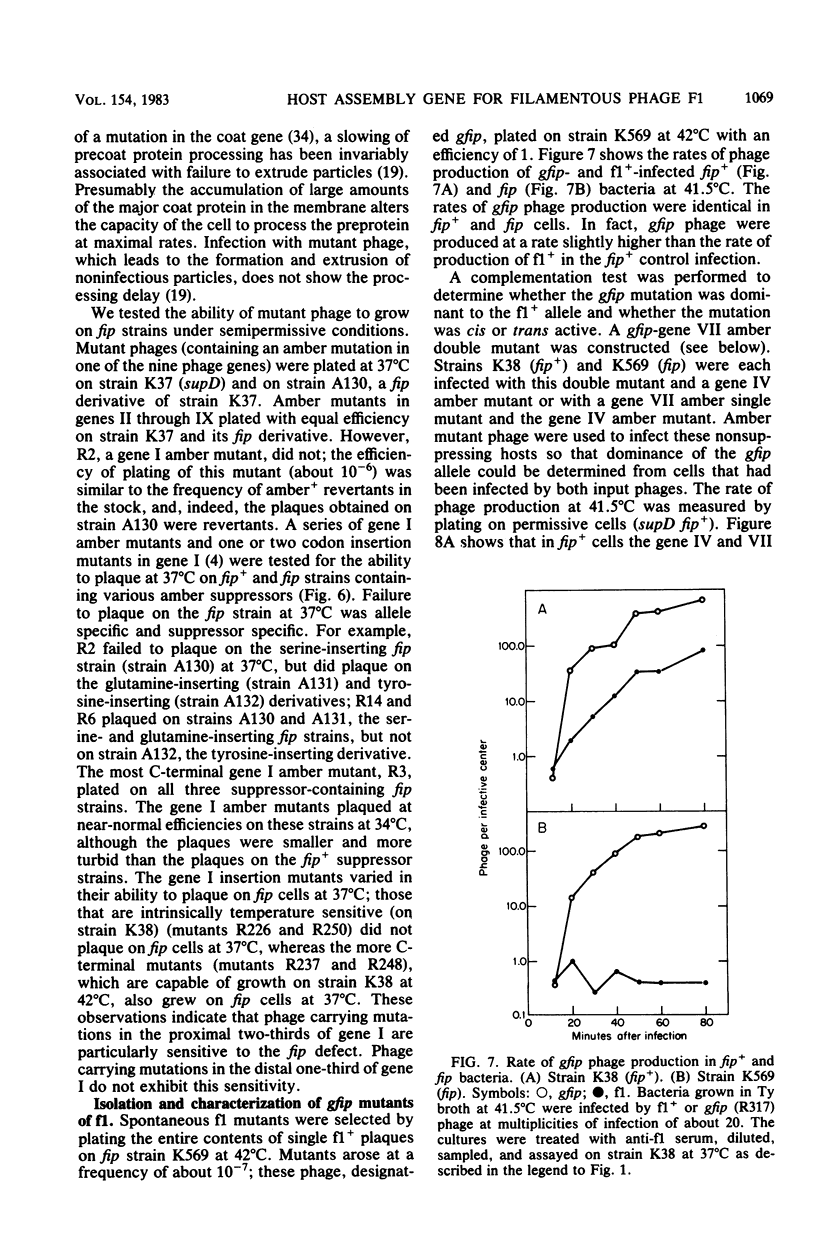
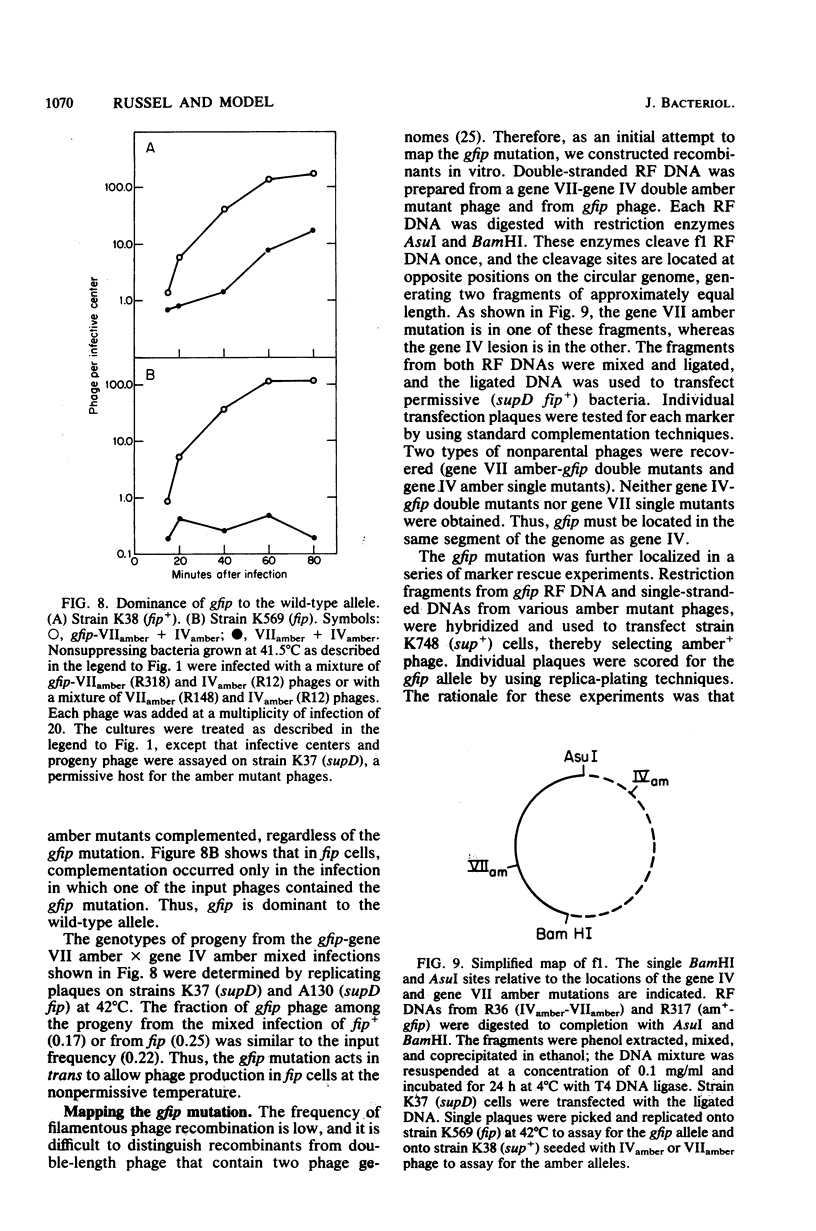
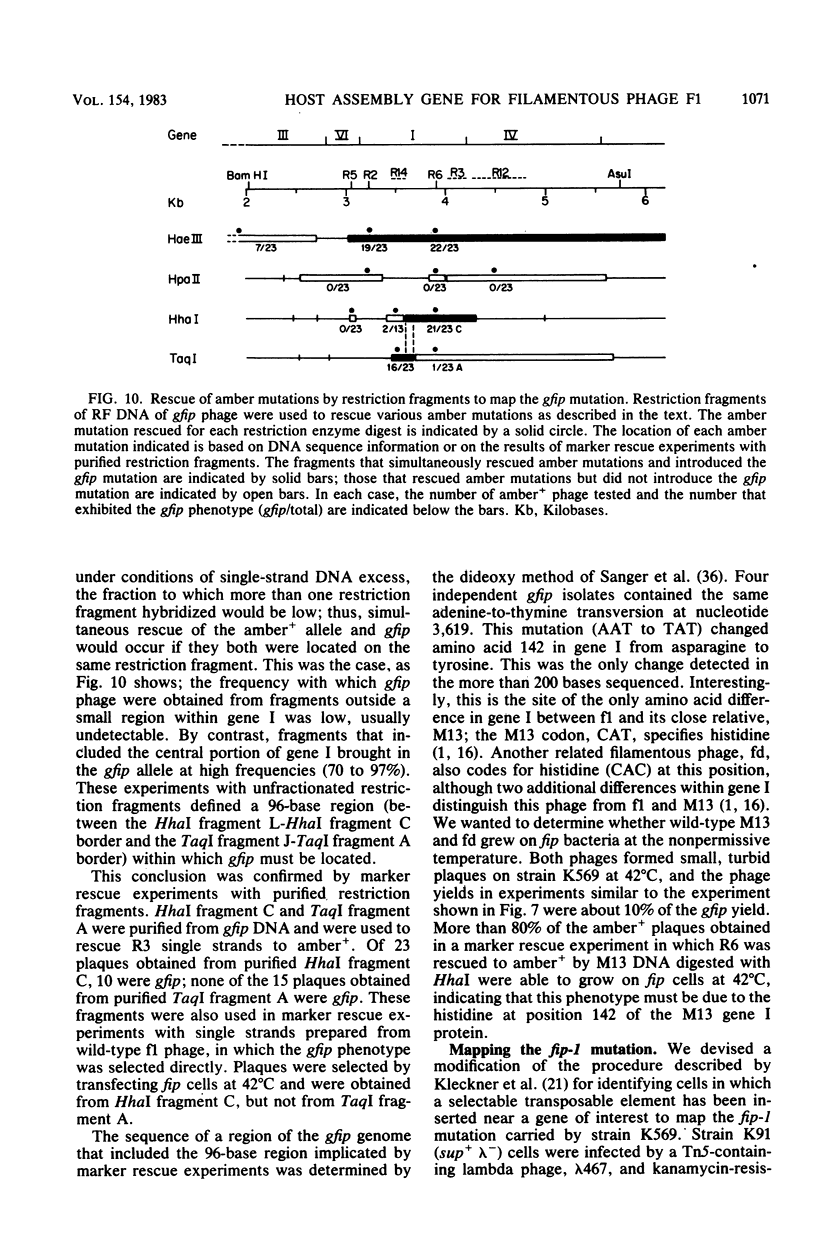
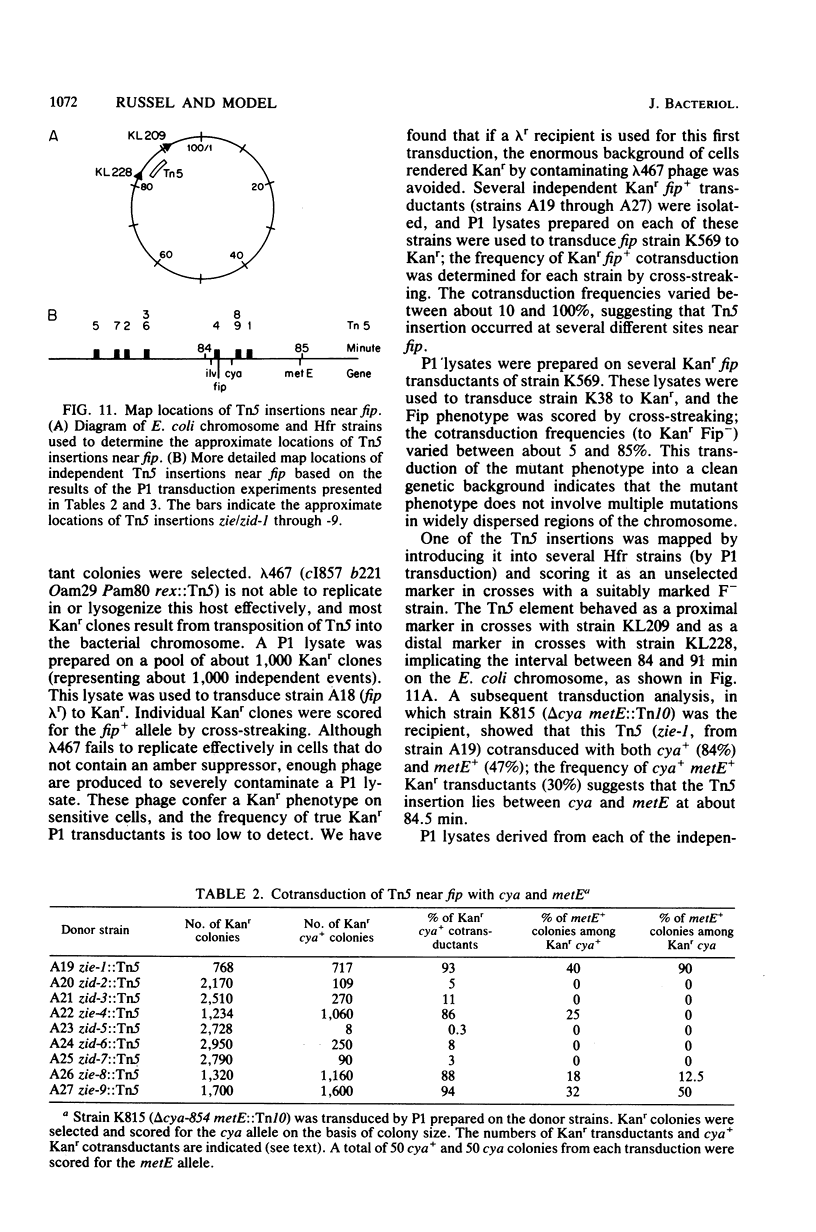
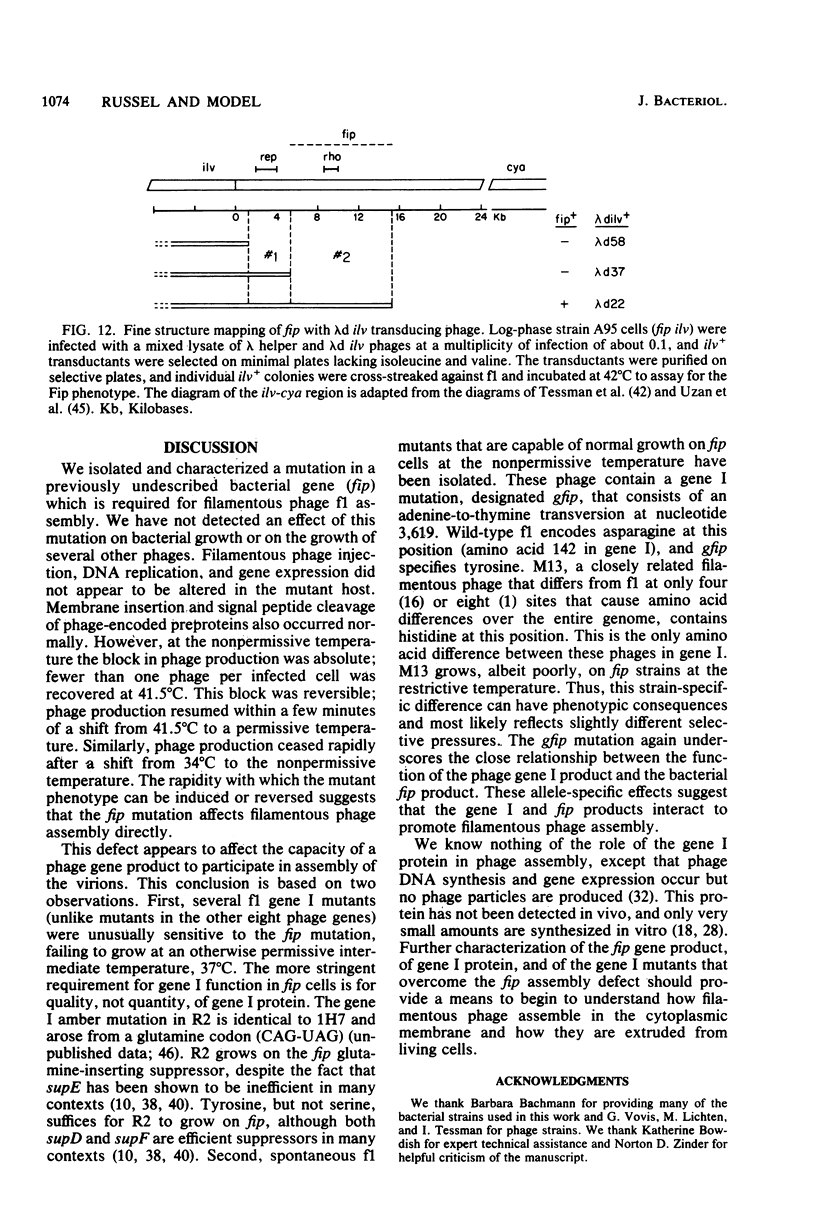
An Escherichia coli mutant which does not support the growth of filamentous bacteriophage fl allows phage fl DNA synthesis and gene expression in mutant cells, but progeny particles are not assembled. The mutant cells have no other obvious phenotype. On the basis of experiments with phage containing nonlethal gene I mutations and with mutant fl selected for the ability to grow on mutant bacteria, we propose an interaction between the morphogenetic function encoded by gene I of the phage and the bacterial function altered in this mutant. The bacterial mutation defines a new gene, fip (for filamentous phage production), located near 84.2 min on the E coli chromosome.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beck E., Zink B. Nucleotide sequence and genome organisation of filamentous bacteriophages fl and fd. Gene. 1981 Dec;16(1-3):35–58. doi: 10.1016/0378-1119(81)90059-7. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Model P. A prokaryotic membrane anchor sequence: carboxyl terminus of bacteriophage f1 gene III protein retains it in the membrane. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5200–5204. doi: 10.1073/pnas.79.17.5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D. One and two codon insertion mutants of bacteriophage f1. Mol Gen Genet. 1981;181(3):288–291. doi: 10.1007/BF00425599. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Russel M., Model P. Processing of filamentous phage pre-coat protein. Effect of sequence variations near the signal peptidase cleavage site. J Mol Biol. 1980 Dec 5;144(2):103–116. doi: 10.1016/0022-2836(80)90027-3. [DOI] [PubMed] [Google Scholar]
- Coppo A., Manzi A., Pulitzer J. F. Host mutant (tabD)-induced inhibition of bacteriophage T4 late transcription. II. Genetic characterization of mutants. J Mol Biol. 1975 Aug 25;96(4):601–624. doi: 10.1016/0022-2836(75)90141-2. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Thomas M., Cameron J., St John T. P., Scherer S., Padgett R. A. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65(1):404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T., Dressler D. H., Hathaway A. THE ABORTIVE REPLICATION OF PhiX174 DNA IN A RECOMBINATION-DEFICIENT MUTANT OF Escherichia coli. Proc Natl Acad Sci U S A. 1967 Mar;57(3):813–820. doi: 10.1073/pnas.57.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emr S. D., Hanley-Way S., Silhavy T. J. Suppressor mutations that restore export of a protein with a defective signal sequence. Cell. 1981 Jan;23(1):79–88. doi: 10.1016/0092-8674(81)90272-5. [DOI] [PubMed] [Google Scholar]
- Fujiki H., Palm P., Zillig W., Calendar R., Sunshine M. Identification of a mutation within the structural gene for the a subunit of DNA-dependent RNA polymerase of E. coli. Mol Gen Genet. 1976 Apr 23;145(1):19–22. doi: 10.1007/BF00331552. [DOI] [PubMed] [Google Scholar]
- Garen A., Garen S., Wilhelm R. C. Suppressor genes for nonsense mutations. I. The Su-1, Su-2 and Su-3 genes of Escherichia coli. J Mol Biol. 1965 Nov;14(1):167–178. doi: 10.1016/s0022-2836(65)80238-8. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C. P. Bacterial mutants in which the gene N function of bacteriophage lambda is blocked have an altered RNA polymerase. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2977–2981. doi: 10.1073/pnas.68.12.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos C. P., Hohn B. Identification of a host protein necessary for bacteriophage morphogenesis (the groE gene product). Proc Natl Acad Sci U S A. 1978 Jan;75(1):131–135. doi: 10.1073/pnas.75.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos C., Tilly K., Drahos D., Hendrix R. The B66.0 protein of Escherichia coli is the product of the dnaK+ gene. J Bacteriol. 1982 Mar;149(3):1175–1177. doi: 10.1128/jb.149.3.1175-1177.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D. F., Petersen G. B. Nucleotide sequence of bacteriophage f1 DNA. J Virol. 1982 Oct;44(1):32–46. doi: 10.1128/jvi.44.1.32-46.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn B., von Schütz H., Marvin D. A. Filamentous bacterial viruses. II. Killing of bacteria by abortive infection with fd. J Mol Biol. 1971 Feb 28;56(1):155–165. doi: 10.1016/0022-2836(71)90091-x. [DOI] [PubMed] [Google Scholar]
- Ito K., Mandel G., Wickner W. Soluble precursor of an integral membrane protein: synthesis of procoat protein in Escherichia coli infected with bacteriophage M13. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1199–1203. doi: 10.1073/pnas.76.3.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen P., Collins J., Fiil N., von Meyenbourg K. A ribosomal RNA gene, rrnC, of Escherichia coli, mapped by specialized transducing lambdadilv and lambda drbs phages. Mol Gen Genet. 1978 Jul 11;163(2):223–228. doi: 10.1007/BF00267413. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Chan R. K., Tye B. K., Botstein D. Mutagenesis by insertion of a drug-resistance element carrying an inverted repetition. J Mol Biol. 1975 Oct 5;97(4):561–575. doi: 10.1016/s0022-2836(75)80059-3. [DOI] [PubMed] [Google Scholar]
- LOEB T., ZINDER N. D. A bacteriophage containing RNA. Proc Natl Acad Sci U S A. 1961 Mar 15;47:282–289. doi: 10.1073/pnas.47.3.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lerner T. J., Model P. The "steady state" of coliphage f1: DNA synthesis late in infection. Virology. 1981 Dec;115(2):282–294. doi: 10.1016/0042-6822(81)90111-2. [DOI] [PubMed] [Google Scholar]
- Lyons L. B., Zinder N. D. The genetic map of the filamentous bacteriophage f1. Virology. 1972 Jul;49(1):45–60. doi: 10.1016/s0042-6822(72)80006-0. [DOI] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Model P., Zinder N. D. In vitro synthesis of bacteriophage f1 proteins. J Mol Biol. 1974 Feb 25;83(2):231–251. doi: 10.1016/0022-2836(74)90389-1. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Kurihara T., Saito H., Uchida H. Sigma subunit of Escherichia coli RNA polymerase affects the function of lambda N gene. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4593–4597. doi: 10.1073/pnas.76.9.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F. C., Phillips T. A., VanBogelen R. A., Smith M. W., Georgalis Y., Subramanian A. R. Identity of the B56.5 protein, the A-protein, and the groE gene product of Escherichia coli. J Bacteriol. 1981 Jan;145(1):513–520. doi: 10.1128/jb.145.1.513-520.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieczenik G., Model P., Robertson H. D. Sequence and symmetry in ribosome binding sites of bacteriophage f1 RNA. J Mol Biol. 1974 Dec 5;90(2):191–124. doi: 10.1016/0022-2836(74)90368-4. [DOI] [PubMed] [Google Scholar]
- Pratt D., Tzagoloff H., Erdahl W. S. Conditional lethal mutants of the small filamentous coliphage M13. I. Isolation, complementation, cell killing, time of cistron action. Virology. 1966 Nov;30(3):397–410. doi: 10.1016/0042-6822(66)90118-8. [DOI] [PubMed] [Google Scholar]
- Ravetch J., Horiuchi K., Model P. Mapping of bacteriophage f1 ribosome binding sites to their cognate genes. Virology. 1977 Sep;81(2):341–351. doi: 10.1016/0042-6822(77)90150-7. [DOI] [PubMed] [Google Scholar]
- Russel M., Model P. A mutation downstream from the signal peptidase cleavage site affects cleavage but not membrane insertion of phage coat protein. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1717–1721. doi: 10.1073/pnas.78.3.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel M., Model P. Filamentous phage pre-coat is an integral membrane protein: analysis by a new method of membrane preparation. Cell. 1982 Jan;28(1):177–184. doi: 10.1016/0092-8674(82)90387-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer E. R., Beckwith J. R., Brenner S. Mapping of suppressor loci in Escherichia coli. J Mol Biol. 1965 Nov;14(1):153–166. doi: 10.1016/s0022-2836(65)80237-6. [DOI] [PubMed] [Google Scholar]
- Smilowitz H., Carson J., Robbins P. W. Association of newly synthesized major f1 coat protein with infected host cell inner membrane. J Supramol Struct. 1972;1(1):8–18. doi: 10.1002/jss.400010103. [DOI] [PubMed] [Google Scholar]
- Soll L., Berg P. Recessive lethals: a new class of nonsense suppressors in Escherichia coli. Proc Natl Acad Sci U S A. 1969 Jun;63(2):392–399. doi: 10.1073/pnas.63.2.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Sugisaki H., Okamoto T., Takanami M. Studies on bacteriophage fd DNA. IV. The sequence of messenger RNA for the major coat protein gene. J Mol Biol. 1977 Apr 25;111(4):487–507. doi: 10.1016/s0022-2836(77)80065-x. [DOI] [PubMed] [Google Scholar]
- Tessman I., Fassler J. S., Bennett D. C. Relative map location of the rep and rho genes of Escherichia coli. J Bacteriol. 1982 Sep;151(3):1637–1640. doi: 10.1128/jb.151.3.1637-1640.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis K., Marvin D. A. Filamentous bacterial viruses. XVI. Inherent temperature sensitivity of gene 5 protein and its involvement in abortive infection. Virology. 1974 May;59(1):293–300. doi: 10.1016/0042-6822(74)90225-6. [DOI] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- Uzan M., Favre R., Gallay E., Caro L. Genetical and structural analysis of a group of lambda ilv and lambda rho transducing phages. Mol Gen Genet. 1981;182(3):462–470. doi: 10.1007/BF00293936. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Webster R. E., Rementer M. Replication of bacteriophage f1: a complex containing gene II protein in gene V mutant-infected bacteria. J Mol Biol. 1980 May 25;139(3):393–405. doi: 10.1016/0022-2836(80)90137-0. [DOI] [PubMed] [Google Scholar]
- Wechsler J. A., Gross J. D. Escherichia coli mutants temperature-sensitive for DNA synthesis. Mol Gen Genet. 1971;113(3):273–284. doi: 10.1007/BF00339547. [DOI] [PubMed] [Google Scholar]
- van Wezenbeek P. M., Hulsebos T. J., Schoenmakers J. G. Nucleotide sequence of the filamentous bacteriophage M13 DNA genome: comparison with phage fd. Gene. 1980 Oct;11(1-2):129–148. doi: 10.1016/0378-1119(80)90093-1. [DOI] [PubMed] [Google Scholar]