Abstract
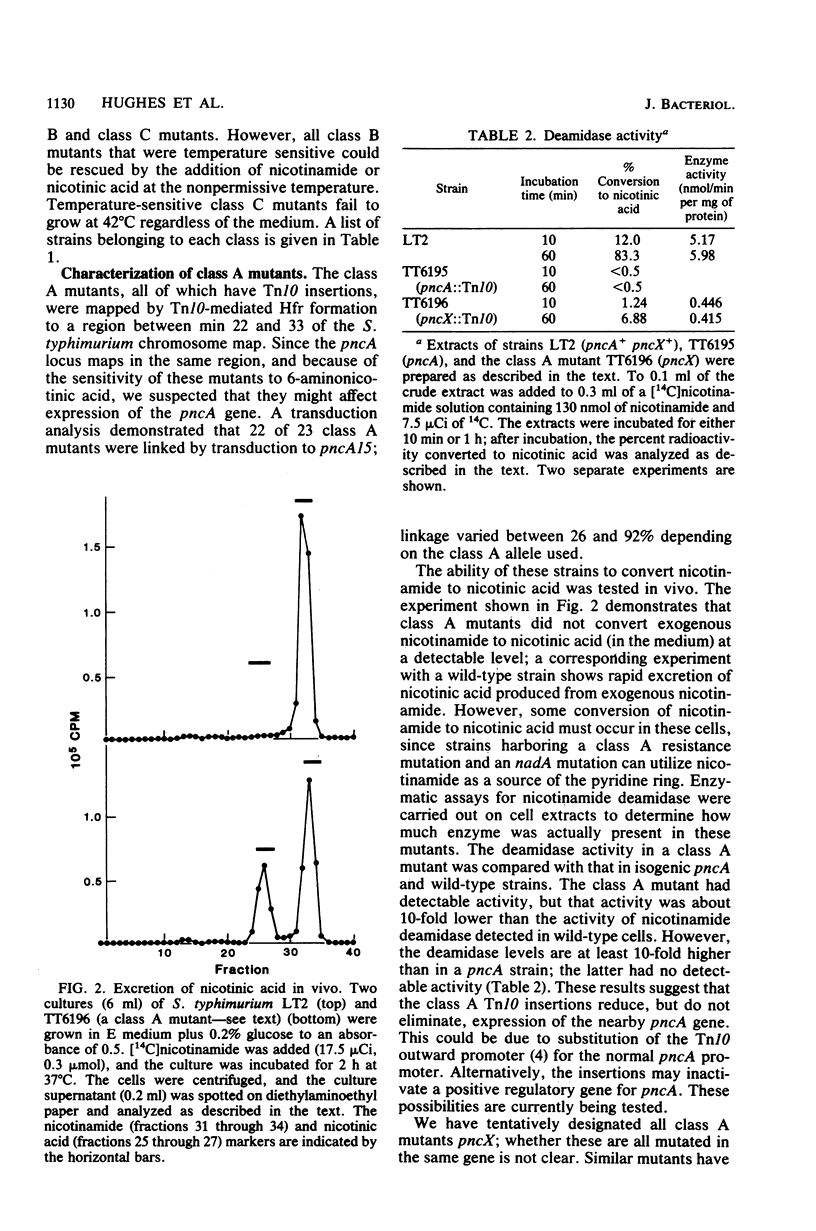
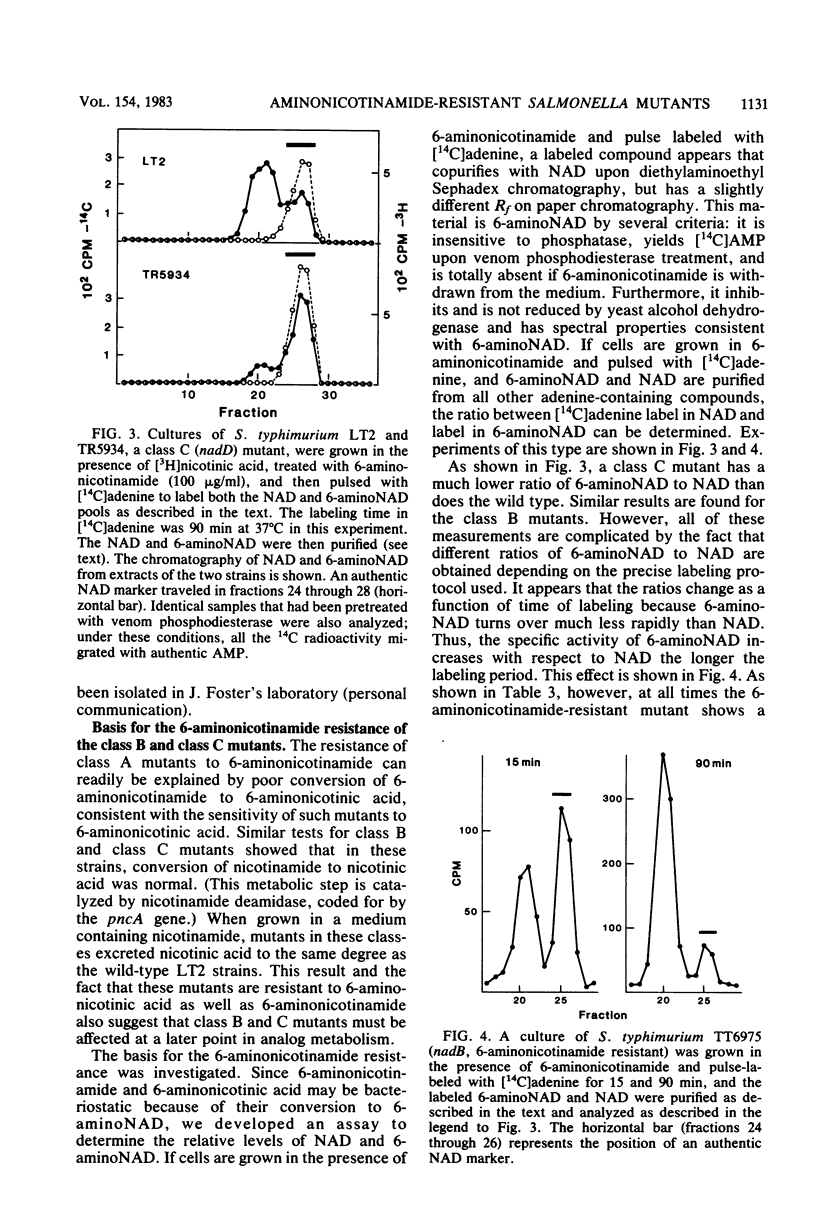
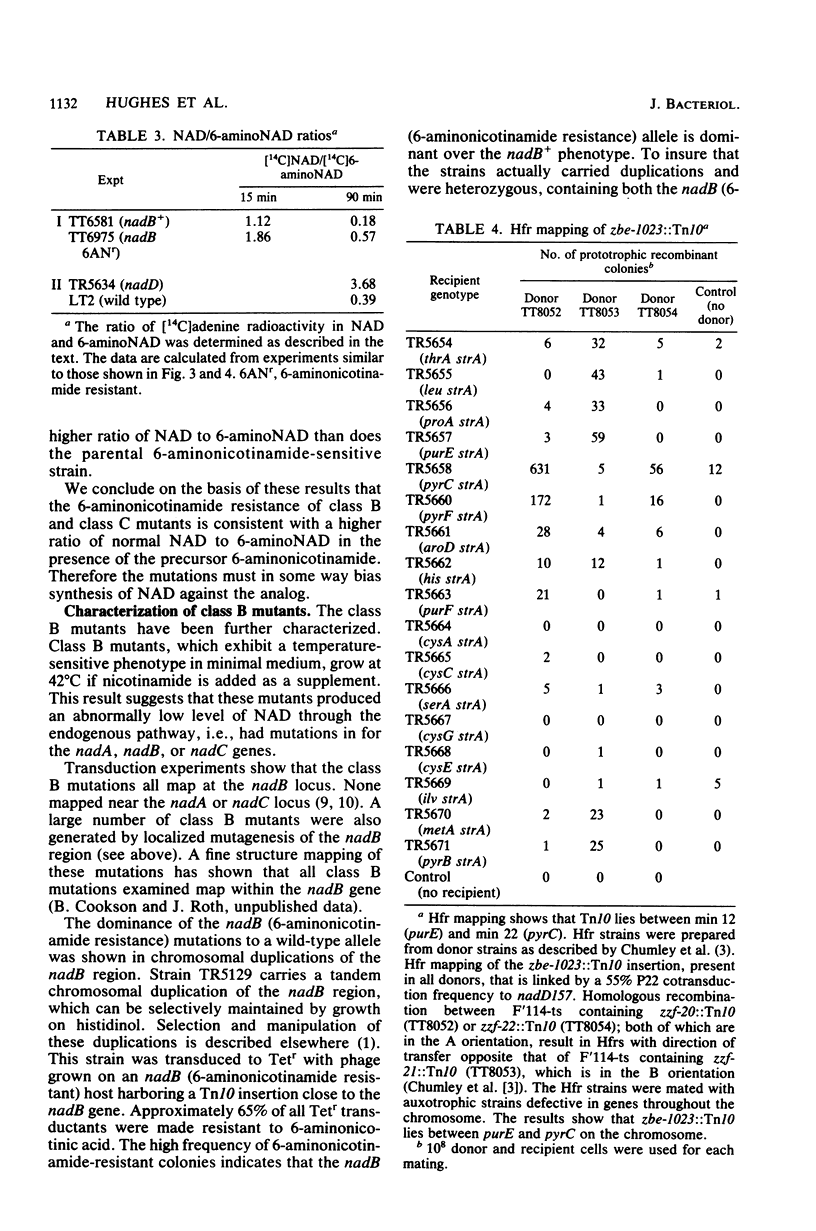
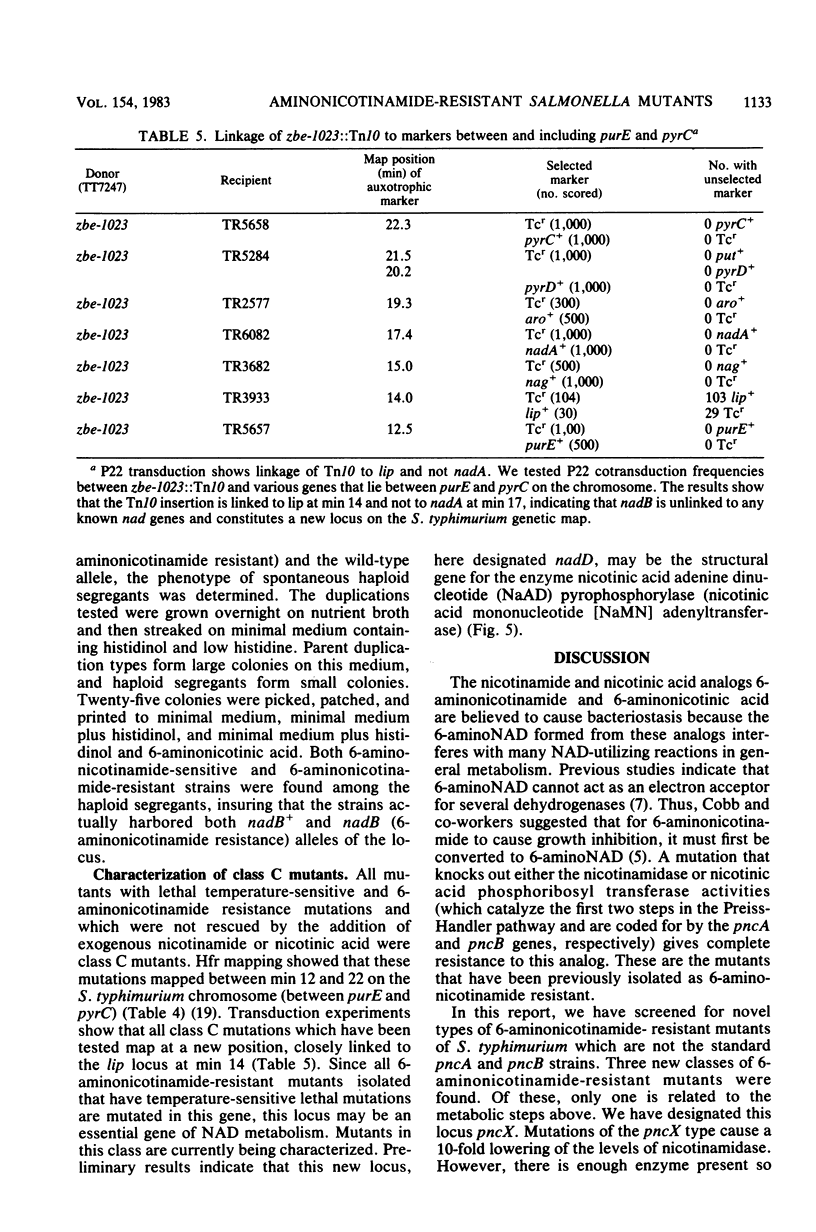
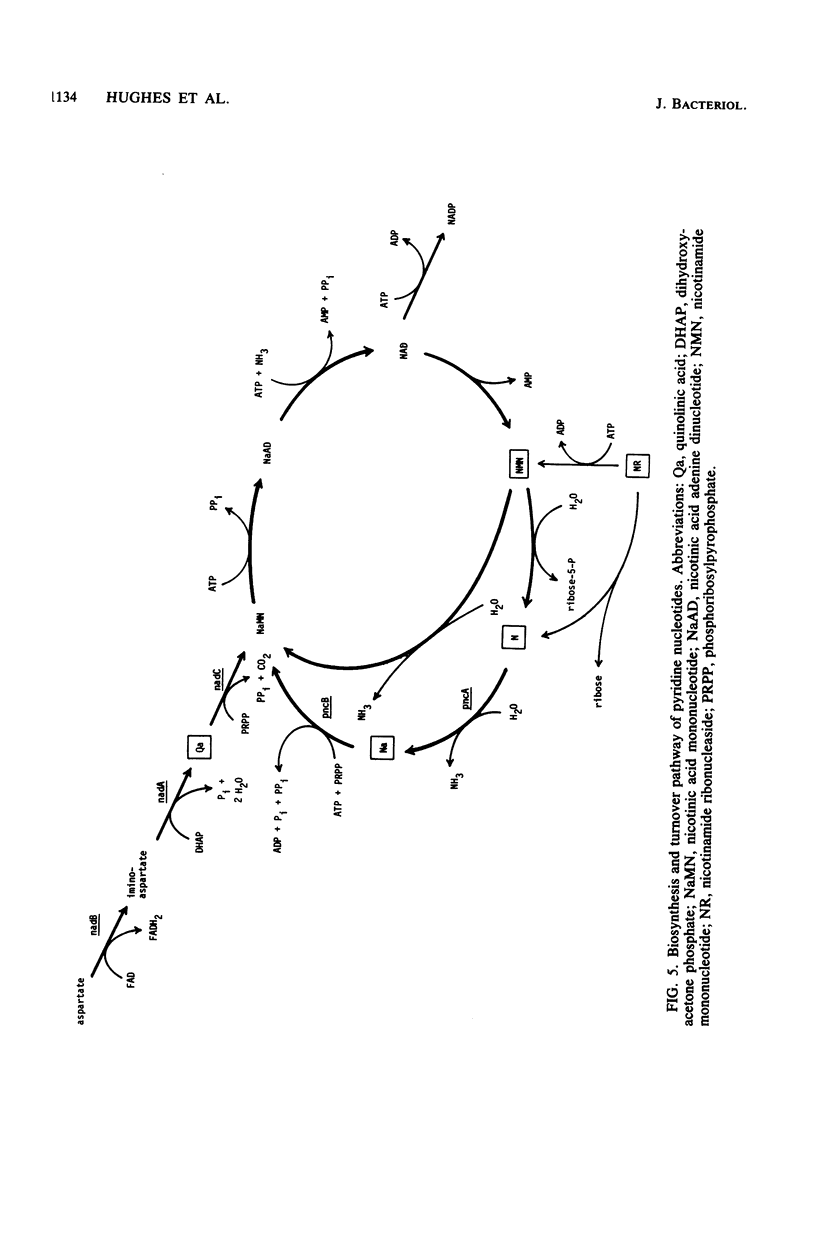
Resistance to the nicotinamide analog 6-aminonicotinamide has been used to identify the following three new classes of mutants in pyridine nucleotide metabolism. (i) pncX mutants have Tn10 insertion mutations near the pncA locus which reduce but do not eliminate the pncA product, nicotinamide deamidase. (ii) nadB (6-aminonicotinamide-resistant) mutants have dominant alleles of the nadB gene, which we propose are altered in feedback inhibition of the nadB enzyme, L-aspartate oxidase. Many of these mutants also exhibit a temperature-sensitive nicotinamide requirement phenotype. (iii) nadD mutants have mutations that affect a new gene involved in pyridine nucleotide metabolism. Since a high proportion of nadD mutations are temperature-sensitive lethal mutations, this appears to be an essential gene for NAD and NADP biosynthesis. In vivo labeling experiments indicate that in all the above cases, resistance is gained by increasing the ratio of NAD to 6-aminonicotinamide adenine dinucleotide. 6-Aminonicotinamide adenine dinucleotide turns over significantly more slowly in vivo than does normal NAD.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson R. P., Roth J. R. Tandem chromosomal duplications in Salmonella typhimurium: fusion of histidine genes to novel promoters. J Mol Biol. 1978 Feb 15;119(1):147–166. doi: 10.1016/0022-2836(78)90274-7. [DOI] [PubMed] [Google Scholar]
- Chumley F. G., Menzel R., Roth J. R. Hfr formation directed by tn10. Genetics. 1979 Apr;91(4):639–655. doi: 10.1093/genetics/91.4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampi M. S., Schmid M. B., Roth J. R. Transposon Tn10 provides a promoter for transcription of adjacent sequences. Proc Natl Acad Sci U S A. 1982 Aug;79(16):5016–5020. doi: 10.1073/pnas.79.16.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb J. R., Pearcy S. C., Gholson R. K. Metabolism of 6-aminonicotinic acid in Escherichia coli. J Bacteriol. 1977 Sep;131(3):789–794. doi: 10.1128/jb.131.3.789-794.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIETRICH L. S., FRIEDLAND I. M., KAPLAN L. A. Pyridine nucleotide metabolism: mechanism of action of the niacin antagonist, 6-aminonicotinamide. J Biol Chem. 1958 Oct;233(4):964–968. [PubMed] [Google Scholar]
- Foster J. W., Kinney D. M., Moat A. G. Pyridine nucleotide cycle of Salmonella typhimurium: isolation and characterization of pncA, pncB, and pncC mutants and utilization of exogenous nicotinamide adenine dinucleotide. J Bacteriol. 1979 Mar;137(3):1165–1175. doi: 10.1128/jb.137.3.1165-1175.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J. W., Moat A. G. Mapping and characterization of the nad genes in Salmonella typhimurium LT-2. J Bacteriol. 1978 Feb;133(2):775–779. doi: 10.1128/jb.133.2.775-779.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J. W., Moat A. G. Nicotinamide adenine dinucleotide biosynthesis and pyridine nucleotide cycle metabolism in microbial systems. Microbiol Rev. 1980 Mar;44(1):83–105. doi: 10.1128/mr.44.1.83-105.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith G. R., Chandler J. L., Gholson R. K. Studies on the de novo biosynthesis of NAD in Escherichia coli. The separation of the nadB gene product from the nadA gene product and its purification. Eur J Biochem. 1975 May;54(1):239–245. doi: 10.1111/j.1432-1033.1975.tb04133.x. [DOI] [PubMed] [Google Scholar]
- Hong J. S., Ames B. N. Localized mutagenesis of any specific small region of the bacterial chromosome. Proc Natl Acad Sci U S A. 1971 Dec;68(12):3158–3162. doi: 10.1073/pnas.68.12.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- Nasu S., Wicks F. D., Gholson R. K. L-Aspartate oxidase, a newly discovered enzyme of Escherichia coli, is the B protein of quinolinate synthetase. J Biol Chem. 1982 Jan 25;257(2):626–632. [PubMed] [Google Scholar]
- Olivera B. M., Lehman I. R. Diphosphopyridine nucleotide: a cofactor for the polynucleotide-joining enzyme from Escherichia coli. Proc Natl Acad Sci U S A. 1967 Jun;57(6):1700–1704. doi: 10.1073/pnas.57.6.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PREISS J., HANDLER P. Biosynthesis of diphosphopyridine nucleotide. I. Identification of intermediates. J Biol Chem. 1958 Aug;233(2):488–492. [PubMed] [Google Scholar]
- Ratzkin B., Roth J. Cluster of genes controlling proline degradation in Salmonella typhimurium. J Bacteriol. 1978 Feb;133(2):744–754. doi: 10.1128/jb.133.2.744-754.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E., Hartman P. E. Linkage map of Salmonella typhimurium, edition V. Microbiol Rev. 1978 Jun;42(2):471–519. doi: 10.1128/mr.42.2.471-519.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]



