Abstract
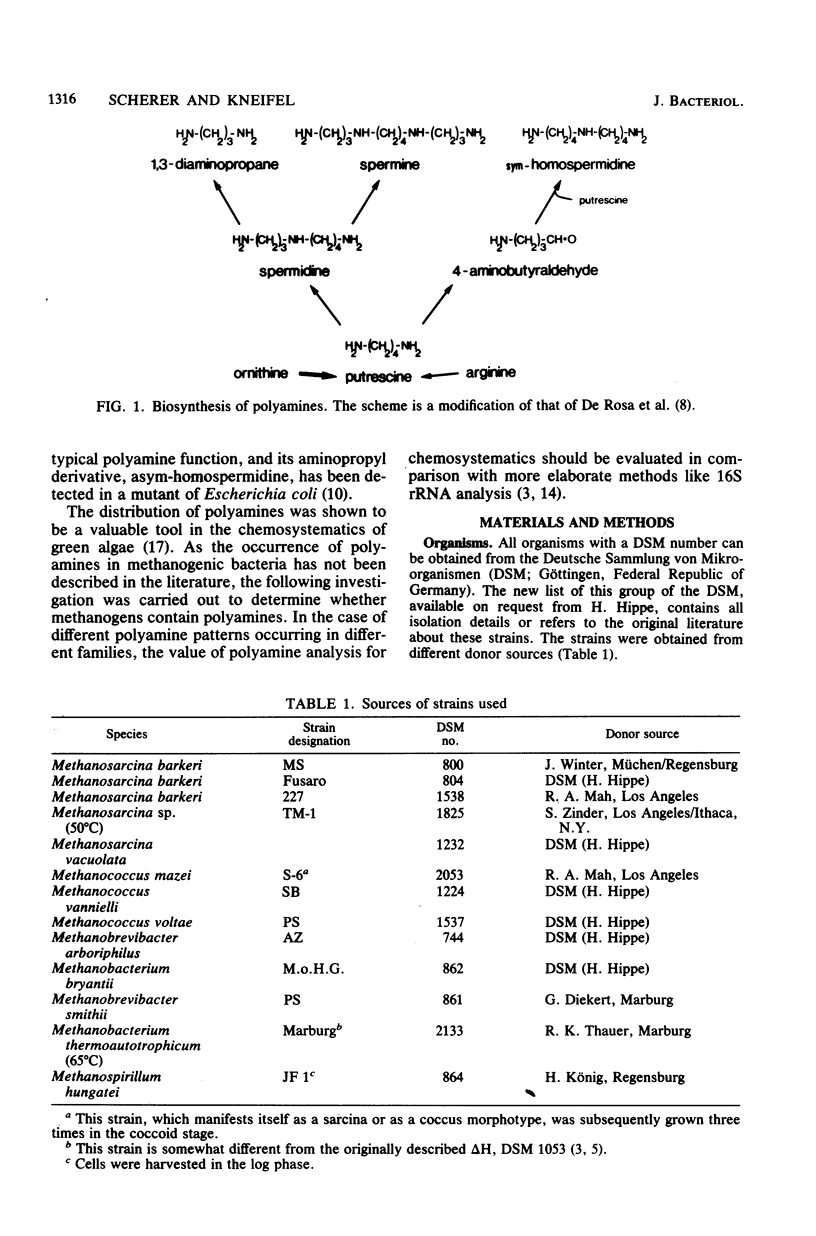
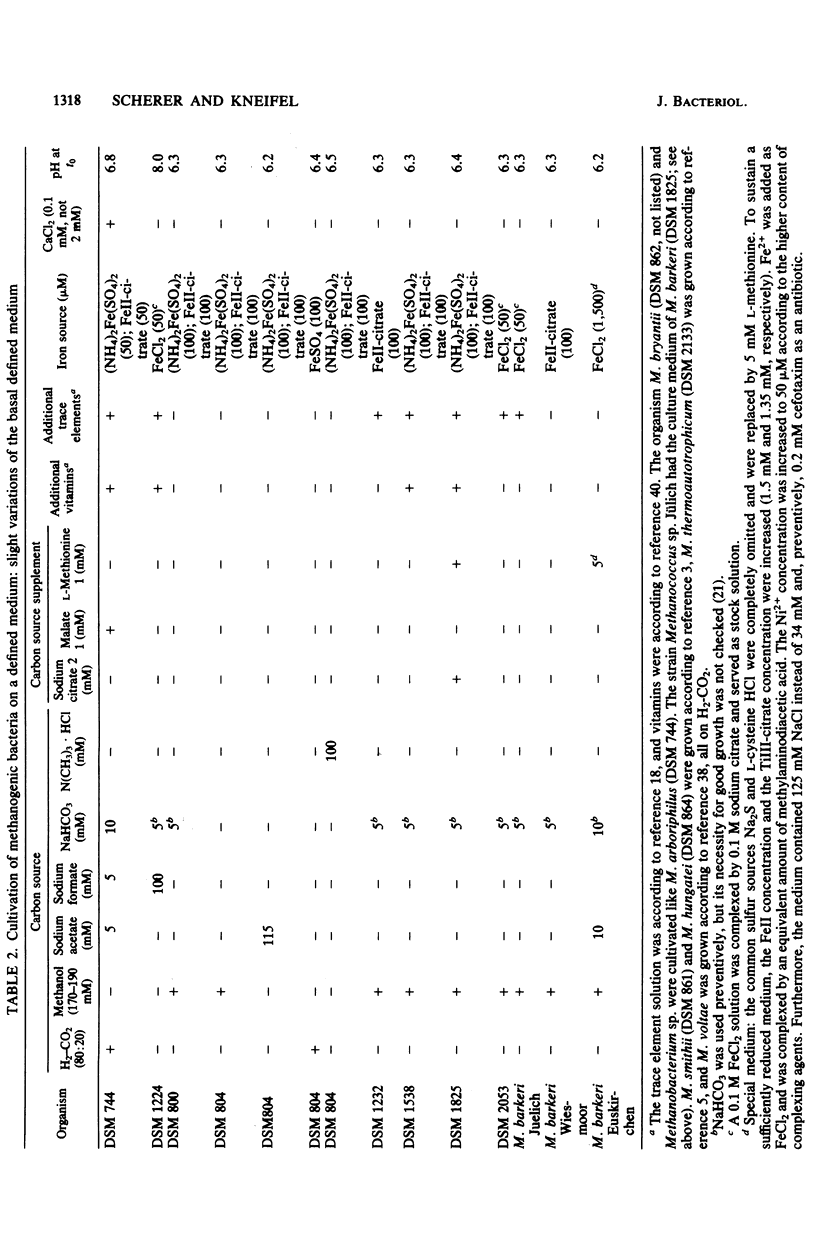
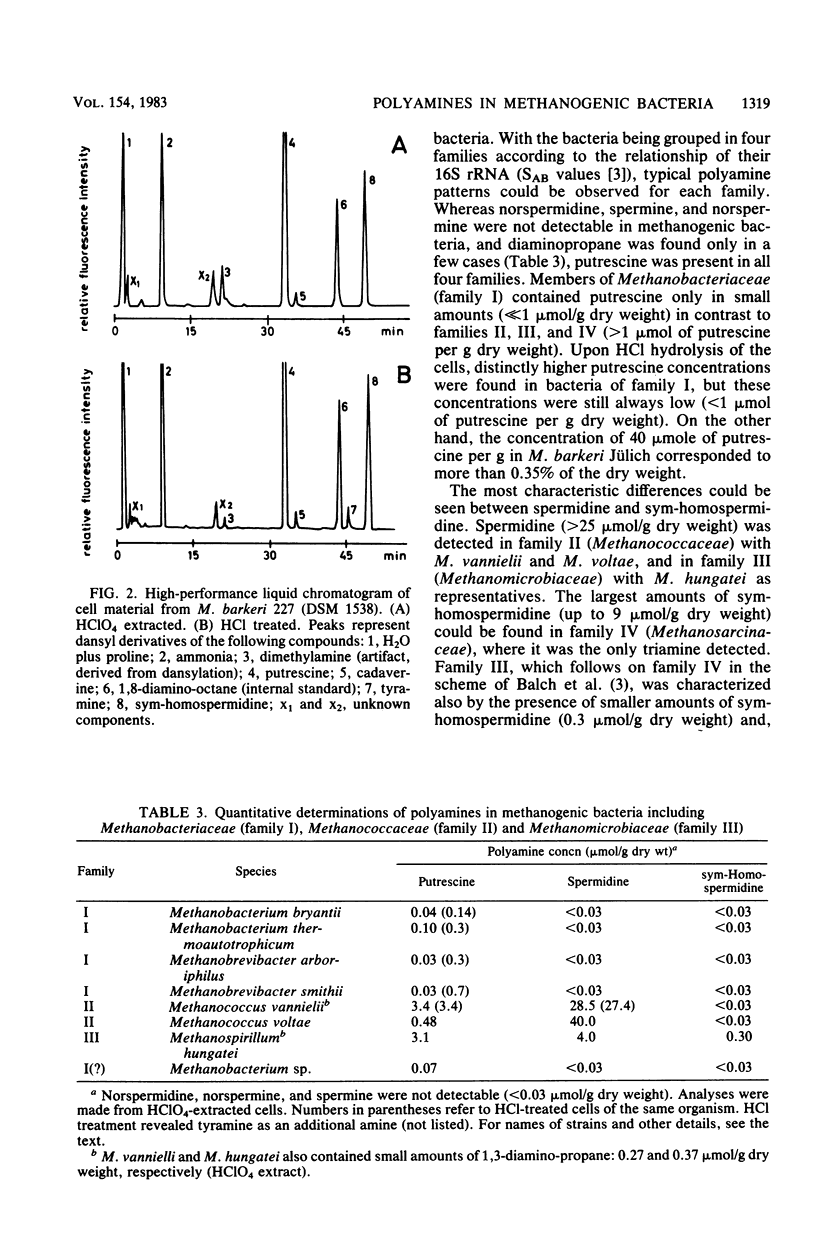
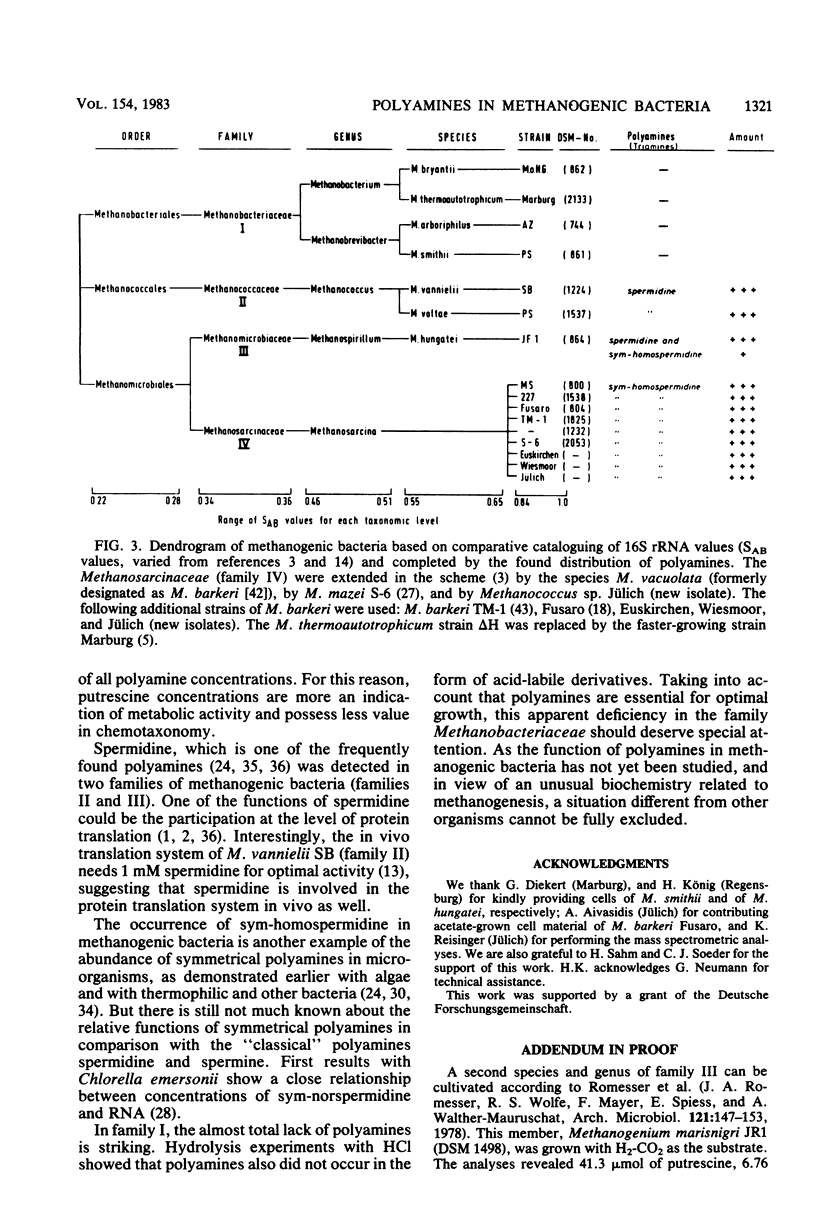
Members of all four families of methanogenic bacteria were analyzed for polyamine concentrations. High-performance liquid chromatography analysis of dansylated cell extracts revealed typical polyamine patterns for each family. Members of Methanobacteriaceae (family I) were characterized by very low polyamine concentrations; members of Methanococcaceae (family II) were characterized by putrescine and high spermidine concentrations; members of Methanomicrobiaceae (family III) were characterized by the presence of putrescine, spermidine, and sym-homospermidine; and members of Methanosarcinaceae (family IV) contained only high concentrations of sym-homospermidine in addition to putrescine. The highest polyamine concentration was found in Methanosarcina barkeri Jülich, with 0.35% putrescine in the dry cell material. The polyamine distribution found coincides with the dendrogram based on comparative cataloguing of 16S rRNA and offers a new, rapid chemotaxonomic method for characterizing methanogenic bacteria. Variation of the growth substrates (H2-CO2, methanol, acetate, and trimethylamine) for M. barkeri resulted in quantitative but not qualitative differences in polyamine composition.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balch W. E., Fox G. E., Magrum L. J., Woese C. R., Wolfe R. S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979 Jun;43(2):260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch W. E., Wolfe R. S. Specificity and biological distribution of coenzyme M (2-mercaptoethanesulfonic acid). J Bacteriol. 1979 Jan;137(1):256–263. doi: 10.1128/jb.137.1.256-263.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway de Macario E., Wolin M. J., Macario A. J. Antibody analysis of relationships among methanogenic bacteria. J Bacteriol. 1982 Jan;149(1):316–319. doi: 10.1128/jb.149.1.316-319.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekert G., Konheiser U., Piechulla K., Thauer R. K. Nickel requirement and factor F430 content of methanogenic bacteria. J Bacteriol. 1981 Nov;148(2):459–464. doi: 10.1128/jb.148.2.459-464.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion A. S., Cohen S. S. Polyamine stimulation of nucleic acid synthesis in an uninfected and phage-infected polyamine auxotroph of Escherichia coli K12 (arginine-agmatine ureohydrolase-putrescine-spermidine-lysine-cadaverine). Proc Natl Acad Sci U S A. 1972 Jan;69(1):213–217. doi: 10.1073/pnas.69.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doddema H. J., Vogels G. D. Improved identification of methanogenic bacteria by fluorescence microscopy. Appl Environ Microbiol. 1978 Nov;36(5):752–754. doi: 10.1128/aem.36.5.752-754.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirich L. D., Vogels G. D., Wolfe R. S. Distribution of coenzyme F420 and properties of its hydrolytic fragments. J Bacteriol. 1979 Oct;140(1):20–27. doi: 10.1128/jb.140.1.20-27.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox G. E., Magrum L. J., Balch W. E., Wolfe R. S., Woese C. R. Classification of methanogenic bacteria by 16S ribosomal RNA characterization. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4537–4541. doi: 10.1073/pnas.74.10.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godsy E. M. Isolation of Methanobacterium bryantii from a Deep Aquifer by Using a Novel Broth-Antibiotic Disk Method. Appl Environ Microbiol. 1980 May;39(5):1074–1075. doi: 10.1128/aem.39.5.1074-1075.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamana K., Matsuzaki S. Widespread occurrence of norspermidine and norspermine in eukaryotic algae. J Biochem. 1982 Apr;91(4):1321–1328. doi: 10.1093/oxfordjournals.jbchem.a133818. [DOI] [PubMed] [Google Scholar]
- Hippe H., Caspari D., Fiebig K., Gottschalk G. Utilization of trimethylamine and other N-methyl compounds for growth and methane formation by Methanosarcina barkeri. Proc Natl Acad Sci U S A. 1979 Jan;76(1):494–498. doi: 10.1073/pnas.76.1.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten T. J., Bongaerts H. C., van der Drift C., Vogels G. D. Acetate, methanol and carbon dioxide as substrates for growth of Methanosarcina barkeri. Antonie Van Leeuwenhoek. 1980;46(6):601–610. doi: 10.1007/BF00394016. [DOI] [PubMed] [Google Scholar]
- Kandler O., König H. Chemical composition of the peptidoglycan-free cell walls of methanogenic bacteria. Arch Microbiol. 1978 Aug 1;118(2):141–152. doi: 10.1007/BF00415722. [DOI] [PubMed] [Google Scholar]
- Makula R. A., Singer M. E. Ether-containing lipids of methanogenic bacteria. Biochem Biophys Res Commun. 1978 May 30;82(2):716–722. doi: 10.1016/0006-291x(78)90933-6. [DOI] [PubMed] [Google Scholar]
- Stevens L., Winther M. D. Spermine, spermidine and putrescine in fungal development. Adv Microb Physiol. 1979;19:63–148. doi: 10.1016/s0065-2911(08)60198-8. [DOI] [PubMed] [Google Scholar]
- Tabor C. W., Tabor H. 1,4-Diaminobutane (putrescine), spermidine, and spermine. Annu Rev Biochem. 1976;45:285–306. doi: 10.1146/annurev.bi.45.070176.001441. [DOI] [PubMed] [Google Scholar]
- WOLIN E. A., WOLIN M. J., WOLFE R. S. FORMATION OF METHANE BY BACTERIAL EXTRACTS. J Biol Chem. 1963 Aug;238:2882–2886. [PubMed] [Google Scholar]
- Whitman W. B., Ankwanda E., Wolfe R. S. Nutrition and carbon metabolism of Methanococcus voltae. J Bacteriol. 1982 Mar;149(3):852–863. doi: 10.1128/jb.149.3.852-863.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehnder A. J., Wuhrmann K. Titanium (III) citrate as a nontoxic oxidation-reduction buffering system for the culture of obligate anaerobes. Science. 1976 Dec 10;194(4270):1165–1166. doi: 10.1126/science.793008. [DOI] [PubMed] [Google Scholar]
- Zinder S. H., Mah R. A. Isolation and Characterization of a Thermophilic Strain of Methanosarcina Unable to Use H(2)-CO(2) for Methanogenesis. Appl Environ Microbiol. 1979 Nov;38(5):996–1008. doi: 10.1128/aem.38.5.996-1008.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]


