Abstract
The current paradigm in designing biomaterials is to optimize material chemical and physical parameters based on correlations between these parameters and downstream biological responses, whether in vitro or in vivo. Extensive developments in molecular design of biomaterials have facilitated identification of several biophysical and biochemical variables (e.g. adhesion peptide density, substrate elastic modulus) as being critical to cell response. However, these empirical observations do not indicate whether different parameters elicit cell responses by modulating redundant variables of the cell-material interface (e.g. number of cell-material bonds, cell-matrix mechanics). Recently, a molecular fluorescence technique, Fluorescence Resonance Energy Transfer (FRET) has been applied to quantitatively analyze parameters of the cell-material interface for both two and three-dimensional adhesion substrates. Tools based on FRET have been utilized to quantify several parameters of the cell-material interface relevant to cell response, including molecular changes in matrix proteins induced by interactions both with surfaces and cells, the number of bonds between integrins and their adhesion ligands, and changes in the crosslink density of hydrogel synthetic extracellular matrix analogs. As such techniques allow both dynamic and 3D analyses they will be useful to quantitatively relate downstream cellular responses (e.g. gene expression) to the composition of this interface. Such understanding will allow bioengineers to fully exploit the potential of biomaterials engineered on the molecular scale, by optimizing material chemical and physical properties to a measurable set of interfacial parameters known to elicit a predictable response in a specific cell population. This will facilitate the rational design of complex, multi-functional biomaterials used as model systems for studying diseases or for clinical applications.
Keywords: Extracellular matrix (ECM), Integrin, Fibronectin (FN), Fluorescence, Green Fluorescent Protein (GFP)
Biomaterials Science: Correlating Material Properties to Biological Responses
Research on cell-material interactions typically focuses either on materials chemistry, fabrication and evaluation or on the biological response to the material in animal models or tissue culture. Physical and chemical materials properties and biological responses can be characterized rigorously using a variety of available methods. In contrast, there are few tools available to directly quantify cell-material interactions at the molecular level (e.g. the number of bonds between cells and adhesion ligands). This is especially true when cells are encapsulated into three-dimensional microenvironments (e.g. hydrogels), which are more physiologically relevant to the biology of most cell types than standard monolayer culture (Cukierman 2001). Thus, the biological response, as measured through changes in adhesion, gene expression or other phenotypic factors, is often indirectly correlated with materials chemistry or physical properties (e.g. elastic modulus) rather than related directly to parameters of the cell-material interface (e.g. bond number). This approach is complicated by the large number of inputs by sources other than the material (e.g. soluble stimuli) that may alter the cell response to material cues (Figure 1). More importantly, it assumes that the cell-material interface is static, despite evidence that cells actively rearrange the structure of the extracellular matrix (ECM) during adhesion (Baneyx 2002, Kong 2005a, Krammer 1999, Discher 2005) and migration (Ohashi 1999, Harris 1981).
Figure 1. Biophysical and Biochemical Variables Affecting Cell Phenotype.
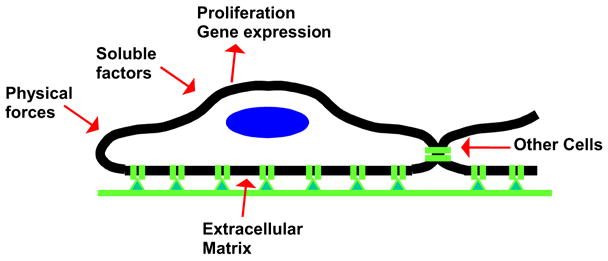
Cell responses (e.g. proliferation, gene expression) are modulated by the interface between cell and ECM, which is affected by the presentation of adhesion ligands and the physical properties of the matrix (e.g. compliance). However, cell responses are also affected by soluble chemical factors (e.g. cytokines, growth factors), physical forces (e.g. cyclic strain) and signals from other cells, which may be mechanical or chemical (e.g. paracrine growth factors) in nature. Signals from one source (e.g. soluble chemicals) influence the manner in which signals from other sources (e.g. ECM) are integrated by cells, further complicating correlations between a single material chemical or physical parameter and complex cellular responses in vitro or in vivo.
Cells interact with adhesion substrates through a variety of mechanisms, including receptor-ligand binding (Leckband 2001). Perhaps the best characterized set of receptors governing receptor mediated cell adhesion to the extracellular matrix (ECM) is the integrin family (Hynes 2002), which interacts specifically with adhesion epitopes including the Arg-Gly-Asp (RGD) sequence found in many natural ECM molecules (e.g. FN, Von Willebrand Factor; Wilson 2005, Pierschbacher 1984, Ruoslahti 1996). It is understood that biophysical and biochemical parameters (e.g. surface charge, density of adhesion epitopes) of materials acting as cell adhesion substrates affect a variety of biological responses, including cell shape, proliferation, migration and gene expression (Folkman 1978, Lee 2001, Lutolf 2005, Cavalcanti 2006, Harbers 2005, Kato 2004, Lee 2004, Maheshwari 2000, Massia 1991, Discher 2005, Engler 2004, Kong 2005a, Kong 2005b, Pelham 1997). However, because of the lack of tools available to characterize the cell-material interface, it is unclear whether the aforementioned chemical variables underlying these behaviors elicit cellular responses through common changes of interfacial parameters.
Current work on biomaterials design typically focuses on modulating a specific material characteristic (e.g. surface charge) and observing a biological parameter of interest (e.g. proliferation), and this approach essentially treats the quantitative aspects of the cell-material interfacial interaction as a black box. Understanding cell response in terms of quantitative parameters of this interface between cell and ECM likely requires measurements of the number of bonds found between receptors and adhesion peptide. It may also require quantitative data on nanoscale rearrangement of the ECM via cellular contractile forces. Given a trend in biomaterials fabrication toward increasing complexity and multi-functionality to exploit various control points of the cell response to biomaterials chemistry (Lutolf 2005), such understanding is likely essential in developing criteria for rational design. Methods for quantifying the cell-material interface are especially important toward the design of three-dimensional microenvironments (e.g. stem cell niches), for which ligand presentation can be controlled but understanding of receptor-ligand interactions can currently only be done in a qualitative fashion. In this article, we first review available approaches for engineering peptide presentation by synthetic and natural ECM and current work to devise probes to characterize the cell-material interface for 2-D systems. We then focus on the use of molecular fluorescence phenomena, including fluorescence resonance energy transfer (FRET), which enable measurement of various cell-ligand interaction parameters in three-dimensional systems.
Characterizing Ligand Presentation by Biomaterials
Tailoring Ligand Presentation on Cell Adhesion Matrices
The simplest way to present adhesion peptides from a biomaterial is to allow natural ECM proteins to adsorb non-specifically to its surface. Cellular response (e.g. focal adhesion assembly, proliferation, differentiation) is modulated both by the concentration of adsorbed protein and the surface chemistry of the substrate (Garcia 1999, Keselowsky 2005, Gallant 2005, Mooney 1995). The effects of surface chemistry on the conformation and adhesiveness of adsorbed ECM proteins have been studied extensively and are reviewed elsewhere (Wilson 2005, Horbett 2004). The thickness and mass of the adsorbed layer can be quantified via ellipsometry, and the chemical composition measured using X-ray Photoelectron Spectroscopy (XPS; Horbett 2004). Presentation of adhesion epitopes by the adsorbed protein can also be estimated using time-of-flight secondary ion mass spectrometry (TOFSIMS; Wang 2004). TOFSIMS can be combined with imaging to quantify the spatial distribution of distinct chemical epitopes on materials that are deliberately engineered to present patterned surface chemistry to vary protein adsorption (Thomas 2002).
Native matrix proteins may also be presented to cells as three-dimensional hydrogels, mimicking many tissues found in vivo. Using ECM derived from fibroblasts, Cukierman et al. showed that the structure of adhesion plaques in adherent cells depended on the dimensionality of these natural matrices: the adhesion plaques formed by cells adherent to cell-derived hydrogel matrices were similar to the plaques found in tissues, whereas presentation of the same chemical epitopes after mechanical flattening to make a 2-D matrix altered the composition and structure of adhesion plaques (Cukierman 2001). Natural ECM are used extensively for clinical applications (e.g. cosmetic surgery) and as substrates for cell adhesion and migration studies. Some of the material properties relevant to cell response, such as fiber alignment, can be quantified in such systems via scanning electron microscopy (SEM; Baker 2006).
There are significant drawbacks to the use of non-specific protein adsorption for studying cell-biomaterial interactions in 2D. First, the initial conformation and density of adsorbed protein is difficult to control. Second, the relative abundance of different proteins in the layer changes with time due to competition amongst proteins for a limited number of surface binding sites (e.g. the Vroman effect; Wilson 2005, Horbett 2004). Finally, cells may alter the protein content of the absorbed layer by mechanically rearranging its contents and synthesizing their own ECM molecules. The initial composition of adsorbed protein layers can be controlled by coating surfaces with purified ECM components such as FN (Ingber 1990, Mooney 1995). The density of the coated protein can be quantified by solvating protein labeled with radioactive isotopes (typically 125I, Ingber 1990), or via immunoblotting (Wilson 2005, Garcia 1999). Exquisite control over surface chemistry independent from substrate mechanics is provided by self assembled monolayers (SAMs) (Prime 1991); the use of these engineered surfaces has suggested that surface charge may modulate adsorbed FN conformation and interaction with integrins (Keselowsky 2005, Gallant 2005). Similar concerns regarding compositional variation in the naturally derived 3D matrices has motivated the development of more well-defined systems to selectively combine defined ECM molecules (e.g. FN and fibrinogen) to make three-dimensional matrices for adhesion and migration studies. (Midwood 2002).
Ligand presentation can be engineered in a more precise fashion using synthetic analogs of the ECM. For example, biomimetic adhesion peptides can be adsorbed to surfaces to facilitate receptor-mediated cell adhesion; studies using this approach have indicated that the RGD peptide alone is insufficient for promoting spreading of some cell types (e.g. hepatocytes) that spread and proliferate when adherent to FN (Hansen 1995). Adhesion peptides may also be grafted to proteins that do not present adhesion epitopes but prevent surface adsorption of ECM molecules (e.g. bovine serum albumin, BSA; Pierschbacher 1984), or by substrates that are chemically modified to prevent protein deposition (e.g. glycophase glass; Massia 1991). Peptides and matrix proteins can also be presented from well defined surfaces formed using SAMs; if one of the components of the SAM is covalently attached to biomimetic peptides, the surface resists both deposition of matrix protein and rearrangement of existing adhesion epitopes by cells, making the correlation between initial surface properties and cell response more direct (Kato 2004, Roberts 1998).
Synthetic Extracellular Matrices based on Polymer Hydrogels
A popular approach in designing materials for medical applications (e.g. drug delivery and tissue regeneration) is to engineer synthetic extracellular matrices (sECM) by combining polymeric materials with biomimetic, integrin-binding peptides. Hydrogels are of particular interest in this approach, as they resist non-specific protein adsorption, allowing integrin-mediated cell adhesion to be specific to the peptides of interest (Lutolf 2005, Lee 2001, Rowley 1999). An additional advantage of using sECM based on hydrogel forming polymers such as poly(acrylamide), poly(ethylene glycol) (PEG) and sodium alginate is that substrate mechanical properties can be modulated independent of adhesion epitope presentation (Augst 2006, Lutolf 2005, Pelham 1997, Kong 2005a, Kong 2005b). The relationships between mechanical properties, including elastic modulus and microstructural parameters such as crosslink density are well understood and can be predicted based on well-established theories (e.g. rubber elasticity) and tested with rheology (Lee 2001, Anseth 1996). Independent of hydrogel mechanical properties, adhesion epitope type and structure can be modulated during peptide synthesis (Harbers 2005, Kato 2004, Massia 1991). Thus, sECM can be used as model systems to link the biophysical and biochemical properties of the ECM to cell responses such as adhesion, migration, shape, proliferation and differentiation
Effects of Matrix Physical and Chemical Properties on Cellular Responses
Studies with synthetic and natural ECM have shown that these cellular responses are influenced by several peptide presentation variables, including the density, type and structure of the ligands, as well as their nanoscale organization (Cavalcanti 2006, Harbers 2005, Kato 2004, Lee 2004, Maheshwari 2000, Massia 1991). The cell response has also been linked to the mechanical properties of the sECM (Discher 2005, Engler 2004, Kong 2005a, Kong 2005b, Pelham 1997) (Figure 2). Microscale parameters of substrate organization (e.g. fiber orientation) also play a role in cell responses including migration (Guido 1993). Finally, soluble byproducts of biomaterial fabrication (e.g. divalent cations used as crosslinkers) may effect cell responses by modulating the activation state of integrins involved in adhesion to RGD (Takagi 2002, Suehiro 2000, Rowley 2002). Nanoscale peptide organization (e.g. spacing between epitopes) can be controlled in a statistically averaged fashion by varying the bulk density of peptide grafted to substrate. Bulk peptide density can be decoupled from nanoscale organization by mixing multivalent polymers modified with multiple peptides with unmodified polymer (Maheshwari 2000, Lee 2004). Recently, nanopatterning methods have been developed that allow more precise control over the nanoscale topography of adhesion epitopes (Cavalcanti 2006, Lee 2002, Cherniavskaya 2005, Gu 2004).
Figure 2. Controlling Peptide Presentation Variables with Materials Chemistry.
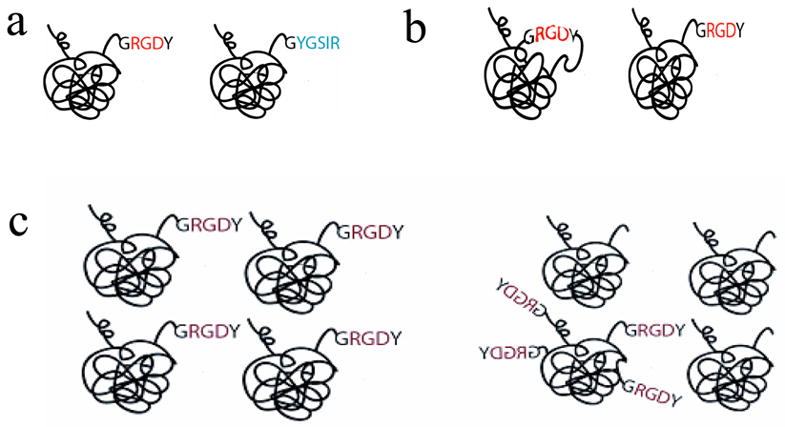
2.a: Adhesion epitope can be varied by changing the amino acid sequence of the biomimetic peptide (e.g. RGD vs. YGSIR). 2.b: Peptide structure can also be varied during synthesis, for example to present the same peptide sequence as either a linear or cyclic peptide. The cyclic peptide more closely resembles the conformation of the RGD sequence found in FN. 2.c: The nanoscale organization of the peptide can be isotropic, as shown on the left, or anisotropic, such that the adhesion epitopes (e.g. RGD) is presented in concentrated clusters rather than evenly distributed on the nanoscale. Anisotropic ligand presentation can accomplished by mixing a multivalent polymer attached to multiple epitopes with unmodified polymer. Presenting peptides in an anisotropic fashion allows one to decouple the nanoscale spacing between individual peptides and their microscale density.
Current Approaches for Characterizing the Cell-Material Interface
Measuring the Strength and Number of Bonds between Adhesion Ligand and Receptors
Variables in peptide presentation by ECM (e.g. peptide structure, nanoscale organization) and substrate mechanical properties may differentially elicit cellular responses through interfacial parameters (e.g. the number and strength of integrin-adhesion ligand bonds). The response may further involve cellular traction forces, generated during rearrangement of ECM ligands, that potentially correspond to intracellular stresses generated through the process of forming and stabilizing focal adhesions (FA; Discher 2005, Kong 2005, Ingber 1990, Ingber 1994, Stupack 2002). One way to indirectly estimate bond number and strength is to measure the relative force required to detach a population of cells from a biomaterial subjected to centrifugation or shear (Harbers 2005, Garcia 1999, Gallant 2005, Maheshwari 2000). An excellent overview on the use of physical techniques to analyze cell adhesion is provided by Curtis and Lackie (Curtis 1991). Recently, the ability of cells to act as crosslinks for peptide-modified hydrogels has been exploited to estimate the number of bonds between RGD and integrin in three-dimensional alginate sECM crosslinked by pre-osteoblasts (Lee 2003). However, in this case, only the subset of polymer chains that bind to multiple cells cause a detectable change in the elastic response of the cell-hydrogel composite.
Estimating either bond strength or number using mechanical approaches is complicated by the complex, coupled relationship between bond number, bond strength and critical detachment force. Theoretical prediction indicates that the kinetics of bond dissociation (koff) are sensitive to forces applied to the bonds (Bell 1978). Furthermore, detachment of adherent cells depends on collective interactions of many ligand-receptor pairs and dynamic changes in these interactions during adhesion and detachment (Ward 1993, Merkel 1999, Dembo 1988). Interactions between pairs of receptors (e.g. integrin clustering) also affect detachment resistance (Gallant 2005). Measurement of adhesion in live cells using mechanical means is further complicated by a strong dependence of integrin-mediated adhesion strength on cell state and integrin activation (both being functions of soluble chemical stimuli as well as the biomaterial substrate; Ingber 1994, Takagi 2002, Suehiro 2000).
In some cases, it is possible to quantify the amount of ligand actually bound to integrin through biochemical means. Biochemical techniques that rely on antibody detection of integrins are especially useful for these measurements due to the availability of antibodies engineered to be specific to conformations of the integrin with bound ligand (Takagi 2002, Paszek 2005). Through the use of crosslinking molecules that link integrin to ECM, and the selective use of detergents that solubilize cellular but not ECM proteins, it is possible to quantify bound integrin by Western blot or ELISA (Garcia 1999, Keselowsky 2005, Gallant 2005). This technique is confined, however, to adhesion substrates in which diffusion of crosslinker into cell-matrix contacts is not limited by pore size or reaction with scaffold, and may not be suitable to analyze cell-ligand interactions in three-dimensional cell culture. This is also a destructive technique that does not allow dynamic measurements. To simplify the measurement of bond strength, a surface force apparatus has been used to measure the detachment force of a highly defined system of receptors and ligands attached to alternate surfaces (Leckband 1995, Leckband 2001). The strength of single receptor-ligand bonds can also be probed in live cells using atomic force microscopy (AFM; Kim 2003, Lehenkari 1999).
Analyzing and Manipulating Cell-Matrix Mechanics
Whereas techniques have been developed to estimate the number of cell-substrate bonds, it has become increasingly apparent that cell-matrix mechanics play an active role in the cell response to materials (Ingber 1994, Chen 1997, Discher 2005). Although it is currently unclear how exactly these mechanical interactions are transduced into chemical signals within cells, several potential mechanisms have been identified. It is also possible that cell-matrix mechanics may influence the stability of cell-material bonds.
In addition to measurements of bond strength, AFM can also be used to probe the mechanics of cellular and ECM deformation, and has been used in conjunction with molecular dynamics simulations to suggest a role for force-induced unfolding of FN in cellular mechanotransduction. (Oberhauser 2002, Krammer 1999, Bao 2003). Other tools can also be used to quantify cell responses to extracellular mechanical forces, both to determine parameters for physical models of cells and to probe effects of cell-matrix mechanics on intracellular signaling (Bao 2003). A subset of these techniques involves external force application to the integrins using beads coated with FN or RGD. Force can be applied to the beads using optical tweezers or magnetic fields (Wang 1993, Jiang 2006). Experiments enabled by these tools have suggested a role for intracellular stresses in eliciting cellular responses (Tamada 2004, Ingber 1994).
Intracellular stresses can also be varied on a grosser scale by applying defined strains to the adhesion substrate (Cunningham 2002, Putnam 1998); they can also be modulated indirectly by varying the projected area over which cell spreading is permitted (Chen 1997, Singhvi 1994, Thomas 2001) or the compliance of the substrate (Perlham 1997, Kong 2005a, Kong 2005b, Brown 2005, Engler 2004). Varying intracellular stresses (and cell traction forces) indirectly with these approaches affects cell migration, viability and gene expression. However, in all these examples it is unclear if bond number is also changing, or is perhaps responsible for some or all of the cellular response.
Because of complications in estimating cell-ligand bond strength and number via mechanical detachment assays, engineering tools have been developed to facilitate non-invasive observation of cell-mediated contractile forces exerted onto synthetic, two-dimensional substrates. These measurements are presumably correlated to both the number of bonds and the mechanics of the cell-matrix interface. The simplest of these tools, traction force microscopy (TFM), involves embedding fluorescent beads into a material such as silicone rubber and then observing displacement of the beads mediated by contractile activity of cells growing on top of the substrate (Munevar 2001). Variants of TFM include microfabricated cantilevers systems, micro-scale posts of adhesion ligands, or elastomers patterned with fluorescent labels (Galbraith 1997, Balaban 2001, Tan 2003).
Characterizing Cell-Material Interactions with Fluorescence
Fluorescence is widely used to study cell signaling and protein-protein interactions. Many intracellular processes relevant to cell-material interactions, including focal adhesion (FA) and cytoskeletal assembly, are studied using fluorescence microscopy (Lichtman 2005). Fluorescence is also useful to quantify adsorption of ECM components to surfaces (Model 1999). Implicit advantages of fluorescence over other methods for quantifying changes in ECM and cellular proteins associated with cell-ECM interactions is the ability of fluorescence microscopy to observe a wide variety of processes, and the simplicity involved in using fluorescence instrumentation. These advantages have made such instruments (e.g. epifluorescence microscopes) nearly ubiquitous in biomaterials and cell biology laboratories, in contrast to more specialized equipment like AFM and surface force apparatus. Fluorescence is an optical phenomenon through which energy imparted onto valence electrons in a molecule by photon excitation decays via emission of lower energy (high wavelength) photons (Lakowicz 2006, Lichtman 2005). Typically, the fluorescent probes used in biological studies are small organic molecules (e.g. fluorescein, rhodamine); however, some proteins exhibit sufficient fluorescence to be used as probes (e.g. green fluorescent protein, GFP) (Giepmans 2006). Recently, the unique quantum properties of semiconductor nanoparticles (e.g. quantum dots) have been applied to develop novel fluorescent probes with high photoluminescence and emission stability (Chan 1998).
In conventional fluorescence microscopy (useful for making observations of 2D systems), objects in the x-y plane are separated with a resolution up to about 200nm, whereas objects in the z-plane are convolved together. In confocal microscopy, each z-plane slice can be resolved and the optical sections reconstructed to form the object using appropriate image processing software, allowing analysis of relevant processes in cells studied in 3D microenvironments (e.g. hydrogels). Because of the high Signal-to-Noise Ratio (SNR) and working range of fluorescence, fluorescence microscopy can be combined with computational image processing techniques, allowing this modality to be used to produce both qualitative and quantitative data on the location, activation kinetics and transport of biomolecules (Miura 2005).
Immunofluorescence
In immunofluorescence (IF), proteins of interest are imaged via fluorescently-tagged antibodies or other molecules with which the protein interacts specifically and with high affinity (e.g. rhodamine labeled phalloidin). Because it is antibody-based, IF facilitates analysis of the conformation and topography of adsorbed proteins and of the bio-availability of antibody-binding epitopes (Wilson 2004, Grinnel 1982). Characterizing adhesion epitope bioavailability in this fashion may be particularly useful in understanding the synergistic effects of non-adhesive proteins such as osteopontin in promoting integrin mediated adhesion to epitopes of FN (Koblinski 2005). IF is used extensively to characterize both the behavior of proteins involved in FA assembly, including integrins, as well as downstream signaling (e.g. changes in gene expression). Like biochemical techniques (e.g. Western Blot, ELISA) involving antibodies, IF can distinguish between integrin activation states or structural changes in other molecules relevant to signaling (e.g. phosphorylation) (Galbraith 2002, Paszek 2005, Takagi 2002).
A drawback to traditional IF is that it provides static snapshots of cells and ECM, which may underestimate the kinetics involving FA formation, integrin clustering and cell-mediated ECM rearrangement – more specifically, IF can capture the evolution of cell behavior (e.g. net displacement in cell migration assays) but is not useful for observing dynamic perturbations that provide useful information regarding cell-ECM interactions. Dynamic analyses of these phenomena are facilitated by non-invasive imaging modalities, including optical coherence tomography and second-harmonic generation imaging (Friedl 2004). In some cases it is possible to perform live IF by introducing proteins exogenously labeled with synthetic fluorophores, although there is the potential that this method of labeling may interfere with adhesion epitope presentation (Keppler 2004). This strategy has been used to analyze recruitment of rhodamine-labeled vincullin into FA by smooth muscle cells as a result of cyclic strain (Cunningham 2002). It is also possible to probe the kinetics of receptor-ligand binding in solution by combining fluorescently labeled ligands with flow assisted cell sorting (FACs; Chigaev 2001, 2003). This strategy has been used to demonstrate the ability of divalent cations (e.g. Ca2+ and Mn2+) to modulate the activation state of α4-integrin in leukocytes (Chigaev 2001), which affects their affinity (e.g. KD) for adhesion peptides. Some fluorescent molecules have been engineered to change their excitation/emission characteristics in response to Ca2+ concentrations; these dyes have been used to study the effects of applied force on the activity of mechanosensitive calcium-ion channels (Munevar 2003).
Genetic Labeling with Green Fluorescent Protein Chimeras
To overcome potential limitations associated with synthetic fluorophores, signaling or target proteins can be genetically labeled with GFP using vectors that introduce DNA expressing the GFP-fusion chimera into cells of interest (Giepmans 2006). GFP-tagging has been used extensively to monitor dynamics of intracellular FA components (Kirchner 2003, Wiseman 2004, Laukaitis 2001) and of ECM components responding to cell adhesion and migration (Ohashi 1999). GFP can also be used as a marker to quantify cells and identify morphological features; this may be particularly useful for studying cell migration in three-dimensional engineered tissues, wherein cells may be difficult to identify using light microscopy (Blum 2004, Lee 2005). GFP-chimeras for a variety of proteins involved in cell-material interaction and FA formation, including α5-integrin, paxillin and vincullin have been developed (Laukaitis 2001, Zamir 1999). Another advantage of genetic labeling is that GFP-tagged proteins can be combined with engineered adhesion matrices to simultaneously observe cell-mediated changes in ECM structure and ECM-mediated changes in the dynamics of FA formation; for example, patterned elastomers can be combined with GFP-vinculin chimeras to correlate cell traction forces with FA localization, size and orientation (Balaban 2001). The same GFP chimera was used to show that FA localization and formation was correlated to mechanical forces applied to integrin-bound beads via optical tweezers (Galbraith 2002). GFP-tagged α5-integrin, paxillin and α-actinin have also been used together with a statistical image processing technique, image correlation spectroscopy (ICS), to analyze intracellular transport of these FA components from the trailing to the leading edge of migrating fibroblasts; this work indicated that α5-integrin and paxillin existed in a complex throughout those cells (Wiseman 2004).
Using FRET as a Molecular Ruler
Although conventional microscopy provides quantitative information regarding both cells and their matrix, the spatial resolution limit of 200nm prohibits protein-protein interaction from being directly observed with conventional microscopy; these nanoscale interactions can be observed through optical phenomena including fluorescence quenching and fluorescence resonance energy transfer (FRET). Because these fluorescence phenomena provide molecular information through relatively common instrumentation, FRET and fluorescence quenching are widely used by cell biologists to study intermolecular interactions (Lichtman 2005, Lakowicz 2006); these phenomena also have the potential to be useful in the analysis of cell-material interfacial parameters. By providing a means to quantify cell-material interactions, FRET and other optical phenomena have the potential to illuminate what has formerly been a black box in biomaterials design.
FRET is a form of non-radiative energy transfer between two fluorescent molecules (Lakowicz 2006). The potential for FRET to occur depends in a non-linear fashion on the distance between these molecules (Box 1) and can be used as a “molecular ruler” to quantify this nanoscale distance (useful for mapping protein structures; Lakowicz 2006, Stryer 1967). FRET is experimentally useful for two reasons: first, the quantum mechanical and optical properties of many synthetic fluorophores and GFP variants (i.e. CFP, YFP) dictate a Förster radius, where the efficiency of energy transfer is 50% efficient (Box 1), in the range of 20–100 Å, the distance over which molecular interactions occur. Second, because the range over which FRET can be observed is very narrow (due to the non-linear dependence of energy transfer efficiency on the 6th power of the distance between molecules involved in the energy transfer), positive observation of FRET can be used to track protein-protein interactions in a semi-quantitative fashion. Unlike IF measurements of co-localization, FRET has sufficient resolution to allow definitive observation of protein interactions, rather than mere proximity. Unlike other optical phenomena like quenching and excimer formation, FRET occurs over a small enough distance that it does not involve the solvent shell around donor and acceptor, and hence is relatively insensitive to many solvent properties as well as sample geometry (Lakowicz 2006). Techniques for quantifying FRET based on analysis of fluorescence spectra or images are discussed in Box 2.
Box 1: Principles of FRET Relevant to Cell-Biomaterials Interactions
In FRET, the energy associated with photon excitation in one molecule (the “donor”) is shared with a nearby molecule (the “acceptor”) through dipole-dipole coupling. The energy shared by dipole coupling is proportional to the cube of the distance, r, between dipoles, and the probability that this energy is transferred from donor to acceptor is proportional to the square of the energy (Förster 1948, 1959). Thus, the rate of FRET (kT) between donor and acceptor is proportional to r6. The prefactor for determining kT(r6) is related to several quantum mechanical and optical properties of the two molecules, including the relative orientation of the dipole moments in each molecule and the refractive index of the medium: (Förster 1948, 1959).
| (Equation 1.1) |
Where κ describes the relative orientation of dipoles, n the refractive index of the medium, Q the quantum yield of the donor in the absence of acceptor, τD the lifetime of the excited state of the donor in absence of the acceptor), and J(λ) describes the degree of spectral overlap between donor emission and acceptor excitation
| (Equation 1.2) |
Where λ is the wavelength (nm), FD(λ) the donor emission as a function of λ, εA(λ) the extinction coefficient of the acceptor, and N Avagadro’s number. The physical parameters of the fluorophores can be represented by the Förster radius R0, defined experimentally as the separation distance at which energy transfer is 50% efficient; this simplifies equation 1.1 (Equation 1.3).
| (Equation 1.3) |
The efficiency of energy transfer is described by Equation 1.4:
| (Equation 1.4) |
Qualitatively, during FRET, one should observe a decrease in emission intensity of the donor and an increase in the intensity of the acceptor (Figure 1.1). Quantitatively, the simplest way to estimate Ψ is by the reduction in the lifetime of the excited state of the donor (D, often measured via reduction in steady-state donor emission intensity) when acceptor is present as compared to donor emission in a control sample that does not contain acceptor (D0) (Equation 1.5).
| (Equation 1.5) |
This calculation assumes that decay of donor emission through means other than FRET is equally probable in the donor control sample and the experimental sample. FRET can also be estimated by comparing the ratio of emission from the acceptor to emission from the donor (Baneyx 2002), or through the increase in acceptor emission in the presence of the donor (Lakowicz 1999/2006). When analyzing FRET data obtained from cells interacting with materials, it is especially important that each experimental condition be matched with a control involving the exact same conditions (except for the presence of acceptors), as variables used to alter the parameters of interest (e.g. crosslink density) may also affect the R0 by changing the refractive index of the medium; further, various drug treatments may affect the solvent environment of the donor and thereby change the lifetime of its excited state. However, if the appropriate control experiments are performed, changes in energy transfer are insensitive to solvent affects (Lakowicz 2006).
Because of the requirement that the spectral overlap between donor and acceptor, J(λ), be significant (Equations 1.1 and 1.2), FRET imaging is made more difficult by significant spectral bleedthrough (excitation of the acceptor by the wavelength of light used to excite donor, and emission of the donor at wavelengths associated with acceptor emission). To quantify the molecular interactions in the face of this complication, one can employ data processing techniques such as linear unmixing (Box 2). Another option is to directly measure the lifetime of the excited state of the donor by fluorescence lifetime imaging (FLIM). Lifetime measurements are independent of both fluorophore concentration and excitation intensity (Periasamy 2001, Lakowicz 2006). Instead, excited state lifetimes are sensitive to fluorescence lifetime, which depends on the solvent environment of the fluorophore and intermolecular interactions (e.g. FRET; Bacskai 2003).
Box 2: Interpretation and Analysis of FRET Experiments
Although all the metrics described in Box 1 reflect what should theoretically be observed when FRET occurs, it is often difficult in practice to isolate the FRET component of acceptor emission. The requirement for spectral overlap between donor and acceptor fluorophores (Equations 1.1 and 1.2) necessitates significant spectral bleedthrough (excitation of the acceptor by the wavelength of light used to excite donor, and emission of the donor at wavelengths associated with acceptor emission). There are two sets of strategies available for separating the components of donor and acceptor emission due to FRET from those occurring due to spectral overlap or quenching. One set of strategies, developed by Gordon et al., involves subtracting out the bleed-through components and estimating Ψ based on the emission of control samples (containing only donors or only acceptors); since it involves an empirical calibration specific to the particular instrument used for data acquisition, the Gordon method has the advantage of being useful in conventional (as well as confocal) fluorescence microscopes (Gordon 1998).
A more sophisticated means of estimating Ψ, which may be more useful in situations where the expected SNR is lower, is spectral unmixing. The emission and excitation spectrum of any sample is assumed to be a function of the spectra of its constituents; under ideal conditions of low concentration, it is typical to assume that the output is a linear combination of each constituent, thus allowing the use of relatively simple algorithms to estimate the relative concentration of each fluorescent constituent in a sample (i.e. linear unmixing, Zimmermann 2005, Neher 2004, Ecker 2004); for FRET measurements, Ψ could be inferred by a relative decrease in relative concentration of donor in the presence of acceptor. Since it is based on the spectral content of fluorophores, linear unmixing requires an instrument capable of isolating spectral signatures from fluorophores (e.g. a spectroscope or a microscope equipped with spectral imaging hardware and software). Whereas the Gordon method discards much of the information on donor and acceptor emission, linear unmixing uses more energy of the signal to calculate Ψ and therefore is more sensitive.
Traditionally, the single distance model (first proposed by Förster, Equation 1.4) is used to estimate the separation between donor and acceptor based on estimated degree of energy transfer. Strictly speaking, this equation is valid only under the following conditions: 1) distance between donor and acceptor is constant for the duration of the measurement; 2) fixed distance between donor and acceptor within all molecules from a measured population; 3) no energy transfer between donors; 4) the number of excited acceptors is much smaller than the number of acceptors in the ground state. When these conditions are met, the net rate of energy transfer is the linear sum of the energy transfer for all donor-acceptor pairs. In practice, it is difficult to satisfy all these criteria, particularly when the desired application involves live-cell imaging. Conditions 1 and 2 are strictly satisfied only in situations where donor and acceptor are attached to specific points of a rigid molecule. Condition 3 is violated by many common fluorophores, including fluorescein and rhodamine (Lakowicz 2003). Since energy transfer between donors, or autoRET, exhibits the same distance dependency as FRET, it may be controlled by optimizing the concentration of donor fluorophore within a particular experimental system (e.g. a lower concentration of donors leads to a diminished probability of autoRET).
Depending on how large the difference between experimental and these ideal conditions, more sophisticated calculations than the single distance model may be appropriate. For example, if FRET occurs between two labels on the same molecule for which the total concentration is relatively high (but not high enough to elicit autoRET), the distance between donor and acceptor in a flexible molecule is assumed to follow a normal distribution. When there is a chance of interaction of multiple donors with multiple acceptors, as is the case for FRET measurements made when both donor and acceptor are embedded into membranes (Lakowicz 2006, Periasamy 2001), it may be necessary to apply some of the more sophisticated analytical and numerical solutions to estimate the degree of energy transfer in two-dimensional systems (Zimet 1995, Fung 1978, Wolber 1979). The Förster radius is typically calculated assuming randomization of the orientation of donor and acceptor dipole moments (κ = 2/3); forces used to create aligned micro-environments (e.g. collagen hydrogels where fibers are poled by magnetic fields; Guido 1993) may exert forces on donor and acceptor fluorophores that yield non-random dipole moment orientations, in addition to exhibiting directionally anisotropic R0 due to changes in refractive index. The Förster radius for fluorophores in an aligned system could be estimated by comparing measurements of energy transfer that were expected to be identical between the aligned gel and a non-aligned gel.
In biomaterials engineering, peptide presentation variables are altered without a direct measurement of the number of bound adhesion ligands; this information would be useful toward understanding how the ECM modulates integrin-ligand interactions. To address this issue, FRET has been used to estimate the number of bonds, Nb between RGD and integrin receptors of pre-osteoblasts encapsulated in three-dimensional alginate sECM (Figure 3) (Kong 2006). These measurements have indicated that the efficiency with which cells bind to adhesion ligand varies not only with regard to ligand density (as one might expect) but also with regard to matrix mechanical properties and cellular contractility, as controlled by drug treatment (Kong 2006, unpublished data). Whereas it is typical to assume that Nb is a simple function of adhesion ligand density, these data suggest that other variables in the cellular microenvironment, including nanoscale peptide presentation and ECM mechanics, modulate availability or binding of ligand to cell receptors.
Figure 3. Fluorescent Resonance Energy Transfer as a probe for cell-material interactions.
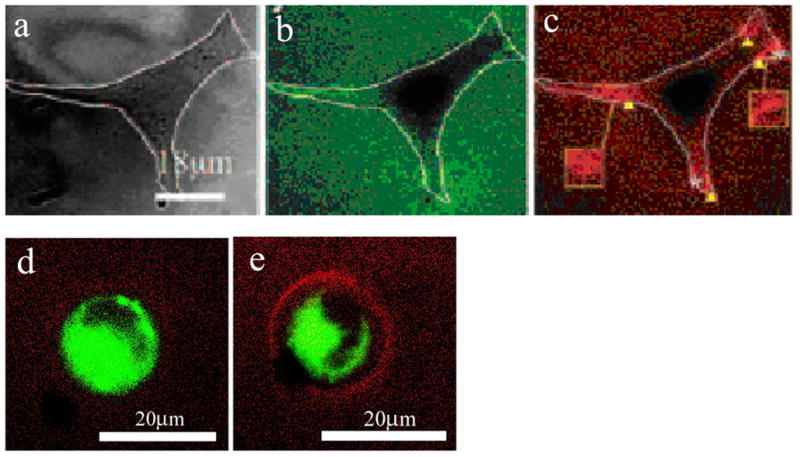
3.a: Mammalian cells adhere to alginate sECM exclusively through biomimetic RGD peptides (DIC image). 3.b–c: RGD peptides are separately coupled to donor (e.g. carboxyfluorescein) or acceptor (e.g. tetramethylrhodamine) succinimidyl esters via a lysine residue on the peptide. The solutions of alginate are mixed at an equal ratio and crosslinked. Cellular clustering of fluorophore-coupled RGD peptides presented by alginate sECM can then be monitored by FRET, through reduction in the emission of the green donor (3.b) and corresponding increase in emission of the acceptor (3.c) given excitation of the donor. The donor emission was estimated by sub-dividing the image into a grid and quantifying the number of pixels (per grid subunit) in the green emission channel exceeding an intensity threshold using ImageJ; the degree of energy transfer was calculated by comparing the estimate of donor emission to the donor emission from a control that lacked tetramethylrhodamine tagged RGD according to Equation 1.5. Regions of cells with the greatest degree of contractility (e.g. edges) were found to have the highest degree of energy transfer. The relative force used to cluster peptides can be estimated from the FRET measurement assuming that the mean initial distance between RGD peptides on separate alginate chains is equal to the radius of gyration of alginate in solution (Kong 2005a). 3.d–e: Integrin-RGD bond formation for cells encapsulated into three-dimensional RGD-modified alginate sECM can be monitored via energy transfer from a membrane-embedded fluorescein-tagged lipid analog (donor) and fluorophore (tetramethylrhodamine) attached to RGD (acceptor). When the acceptor is absent, there is minimal emission of the acceptor (3.d). Energy transfer is also inhibited if integrin receptors are saturated with unlabelled, soluble RGD prior to encapsulation, confirming that energy transfer is specific to RGD-integrin bonds (Kong 2006). In contrast, when the acceptor is present, there is marked reduction in green emission and corresponding increase in red emission at the cell-material interface (3.e) (unpublished data).
Structural biologists have used FRET to map biomacromolecules and determine how the structure of proteins (including FN) is affected by various solvents (Lai 1993). Recently, this approach has been adapted to quantify structural changes of ECM components used as cell adhesion substrates, as semi-quantitative energy transfer measurements can estimate conformational changes in FN upon its interaction with various surfaces (Baneyx 2001, Baugh 2004). This technique can also quantify structural changes in FN stemming from acto-myosin mediated contractility of ECM by adherent cells (Baneyx 2002). These experiments indicated that treatment of cells with Cytochalasin D, an inhibitor of actin filament polymerization, decreased traction-mediated stretching of FN. The nanoscale clustering of RGD by pre-osteoblasts grown on the surface of sECM has also been quantified with FRET. In these studies, the efficiency of energy transfer was used to simultaneously estimate the nanoscale rearrangement of adhesion peptides and the relative force, F, cells exerted to cluster RGD-integrin complexes during FA formation (Figure 3). These force estimates, but not the degree of RGD clustering, were correlated to both cell viability and differentiation (Kong 2005a). Cellular traction forces are correlated to intracellular stresses; these stresses could be quantitatively estimated from F using various models of cellular mechanics (Bao 2003).
Ligand binding is also sensitive to soluble chemical stimuli in the cellular microenvironment, as these signals mediate changes in integrin activation (Takagi 2002). Various cations strongly affect the affinity of α4-integrin for adhesion peptides, as determined using FACs. Recently, this approach was combined with measurements of FRET between fluorescent peptides and cell membrane labeled with a FRET acceptor to estimate integrin activation state in real-time (Chigaev 2003).
FRET is also used to monitor dynamics of protein interactions in live cells via GFP-fusion chimeras with different spectral properties (e.g. YFP or CFP, Giepmans 2006). It has been recently used to monitor tyrosine phosphorylation of FA proteins including focal adhesion kinase (FAK) and paxillin during FA formation (Ballestrom 2006). FRET has also been used as a sensor for mechanotransduction via phosphorylation of Src in endothelial cells subjected to force from optical tweezers (Wang 2005). In this study, a synthetic protein substrate for activated Src containing CFP/YFP domains was developed that underwent homoFRET (FRET within the same molecule) in the native state, but not when the substrate was unfolded following phosphorylation by activated Src. This allowed real-time analysis of a signaling cascade involved in response to the cell-material interaction. FRET is also used extensively to monitor the dynamics and structure of the cell membrane and its protein constituents (Krishnan 2001).
FRET measurements can also be used to measure dissociation of intermolecular complexes, such as those formed by DNA and polycations like poly(ethylene immine) (PEI), which are commonly used as vehicles for non-viral gene delivery (Kong 2005b, Itaka 2002). Although the efficiency of gene transfection can be measured via expression markers (luciferase, GFP), the efficiency of gene expression is affected by factors other than the efficiency with which the vector is taken up by cells; thus, FRET provides direct and distinct measurements of both uptake and complex stability that can be used to calculate the efficiency of gene delivery, irrespective of the capability of the cell to express protein encoded by delivered DNA (Kong 2005b). FRET has also been used to monitor the stability of growth factor in an ex vivo model of arterial tissue to determine whether or not degradation of this molecule is of concern in design of drug eluting stents (Wu 2004). Advancements in biophysical spectroscopy have recently enabled the combination of FRET with fluorescence correlation spectroscopy (FCS), a technique that uses statistical information on particle movements to identify molecular interactions and transport mechanisms (Eggeling 2005, Remaut 2005). Combining FRET with FCS may be advantageous in situations where the concentration of the molecule of interest (e.g. anti-sense DNA) is extremely low (Remaut 2005). Similar approaches to those mentioned here could be used to monitor stability of a propeptide or pro-drug in cells by labeling alternative ends of a single molecule with donor and acceptor; as in the above studies, dissociation of the pro-molecule would decrease the measured energy transfer. In combination with intravital imaging systems, that have recently been used for in vivo FRET and fluorescence liftetime emission (FLIM) imaging (Stockholm 2005, Garcia-Alloza 2004) this approach may be useful for analyzing delivery of complex, macromolecular drugs.
Fluorescence can also be used to track cell migration and cell-mediated degradation of sECM. Dye quenched fluorogenic substrates have been linked to PEG-based hydrogels crosslinked with an MMP-degradable peptide to monitor changes in crosslink density during cell migration (Lee 2005). Monitoring the trajectory of migrating cells in this fashion may provide insight into variables of adhesion ligand presentation and matrix mechanics that affect cellular taxis within three-dimensional matrices; this may be particularly useful if fluorescence dequenching is combined with micropatterning techniques used to create gradient hydrogels (Burdick 2004). Excimer formation, a phenomenon that, like FRET, depends on energy transfer between molecules, has been used to track changes in the proximity of pyrene molecules attached to polymer in hydrogels; these measurements were correlated to the crosslink density of the hydrogel non-invasively. This approach may be useful in evaluating the structure of biomaterials based on self-assembled peptides or polymers and in monitoring cell-mediated hydrogel degradation in situ (Kong 2003, Silva 2004).
In the studies of bond number and adhesion epitope clustering, FRET has proven useful in quantifying cell-material interaction parameters on the scale of single cells and large populations in both 2D and 3D cellular microenvironments. Furthermore, this technique is adaptable to a wide variety of instrumentation, including fluorescence spectroscopes, conventional, total internal reflection (TIRF) and confocal fluorescence microscopes, and FACs. In terms of downstream biological responses such as differentiation or viability, measurements of Nb and F provide direct estimates of the degree of activation of intracellular signaling cascades based respectively on integrin-ligand bond number (Asthagiri 1999, Stupack 2002) and cell-matrix mechanical interactions (Ingber 1994, Discher 2005).
Using Quantitative Measurements of Cell-Material Interactions as Criteria for Biomaterials Engineering
The ultimate goal of directly characterizing receptor-mediated cell-material interactions is to develop criteria for the rational design of biomaterials for tissue engineering and drug delivery on both the micrometer and nanometer scales. Measuring relevant parameters of the cell-material interface (e.g. number cell-sECM bonds, along with cellular rearrangement of the matrix) as functions of biomaterial fabrication parameters (e.g. peptide structure, crosslink density) may lead to a quantitative understanding of how Nb and F, and changes in these quantities with time, may be varied through materials chemistry and physical properties. In turn, optimal cell responses can be determined as mechanistic consequences of parameters such as Nb and F, rather than as empirical correlates to biomaterial fabrication parameters.
Engineering analysis and modeling is becoming increasingly important in understanding complex biological systems and in design of biomaterials for regenerative applications (Lauffenburger 1993). In that vein, several groups have incorporated modeling of peptide presentation by synthetic matrices such as RGD-modified polymer hydrogels to analyze cell adhesion, migration, proliferation and differentiation in terms of the nano-scale distance between adhesion ligands (Comisar 2005, Irvine 2002, Brinkerhoff 2005). Others have used modeling to predict the effects of matrix compliance on cellular organization in the context of tissue formation (Bischofs 2006). In both cases, predictions of bond affinity, structure and intracellular stress sensing are linked to experimentally observed downstream response, rather than initial adhesion. The ability to measure quantities such as Nb and F may lead to unified models of the cell-material interface that incorporate both biochemical (peptide presentation) and biophysical (matrix mechanical properties) information across multiple spatial and temporal scales. Such models would uncover basic knowledge of how cells receive information from their extracellular microenvironment and may facilitate the fabrication of multi-functional biomaterials based on a rational set of design criteria, rather than empirical correlations. In turn, this type of engineering approach may improve the ease of manufacturing and the clinical utility of multi-functional biomaterials for applications in basic science and clinical technology.
Figure 1.1. Indications for and Observations During FRET.
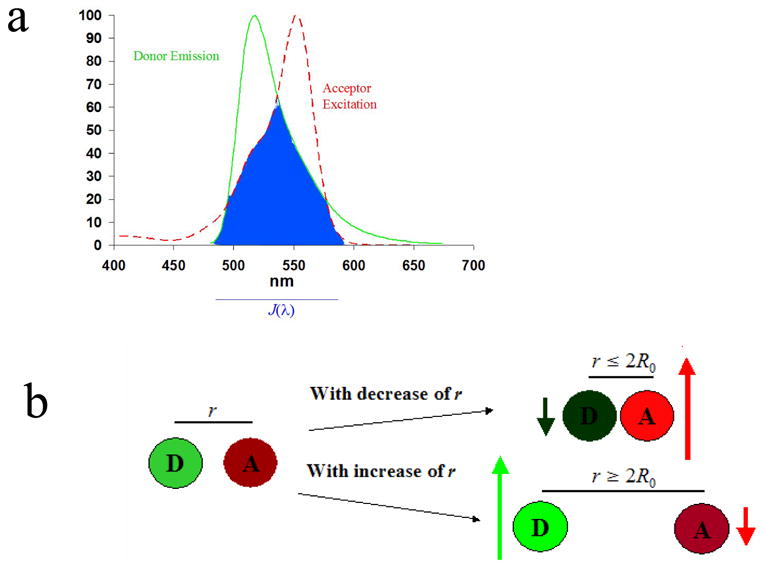
1.1a: Conditions required for FRET: overlap of the emission spectrum of the donor (“D”) and excitation spectrum of the acceptor (“A”) described by overlap interval J(λ). The overlap integral shown is for the fluorescein-rhodamine pair. 1.1b: Qualitative results of FRET are shown: as the distance r separating the donor and acceptor is decreased, the relative emission of the donor decreases while the emission of the acceptor increases.
Acknowledgments
We thank Susan X. Hsiong and Hyun Joon Kong for critical reading of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References List
- 1.Alsberg E, Kong HJ, Hirano Y, Smith MK, Albeiruti A, Mooney DJ. Regulating bone formation via controlled scaffold degradation. J Dent Res. 2003;82(11):903–8. doi: 10.1177/154405910308201111. [DOI] [PubMed] [Google Scholar]
- 2.Anseth KS, Bowman CN, Brannon-Peppas L. Mechanical properties of hydrogels and their experimental determination. Biomaterials. 1996;17(17):1647–57. doi: 10.1016/0142-9612(96)87644-7. [DOI] [PubMed] [Google Scholar]
- 3.Asthagiri AR, Nelson CM, Horwitz AF, Lauffenburger DA. Quantitative relationship among integrin-binding, adhesion and signaling via focal adhesion kinase and extracellular signal-related kinase 2. J Biol Chem. 274(38):27119–27. doi: 10.1074/jbc.274.38.27119. [DOI] [PubMed] [Google Scholar]
- 4.Augst AD, Kong HJ, Mooney DJ. Alginate Hydrogels as Biomaterials. Macromol Biosci. 2006;6(8):623–33. doi: 10.1002/mabi.200600069. [DOI] [PubMed] [Google Scholar]
- 5.Bacskai BJ, Skoch J, Hickey GA, Allen R, Hyman BT. Fluorescence resonance energy transfer determinations using multiphoton fluorescence lifetime imaging microscopy to characterize amyloid-beta plaques. J Biomed Opt. 2003;8(3):368–75. doi: 10.1117/1.1584442. [DOI] [PubMed] [Google Scholar]
- 6.Baker SC, Atkin N, Gunning PA, Granville N, Wilson K, Wilson D, Southgate J. Characterization of electrospun polystyrene scaffolds for three-dimensional in vitro biological studies. Biomaterials. 2006;27(16):3136–46. doi: 10.1016/j.biomaterials.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 7.Balaban NQ, Schwarz US, Riveline D, Goichberg P, Tzur G, Sabanay I, Mahalu D, Safran S, Bershadsky B, Addadi L, Geiger B. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3(5):466–72. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 8.Ballestrem C, Erez N, Kirchner J, Kam Z, Bershadsky A, Geiger B. Molecular mapping of tyrosine-phosphorylated proteins in focal adhesions using fluorescence resonance energy transfer. J Cell Sci. 2006;119:866–75. doi: 10.1242/jcs.02794. [DOI] [PubMed] [Google Scholar]
- 9.Banexy G, Baugh L, Vogel V. Coexisting conformations of FN in cell culture imaged using fluorescence resonance energy transfer. Proc Natl Acad Sci USA. 2001;98(25):14464–8. doi: 10.1073/pnas.251422998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baneyx G, Baugh L, Vogel V. FN extension and unfolding within cell matrix fibrils controlled by cytoskeletal tension. Proc Natl Acad Sci USA. 2002;99(8):5139–43. doi: 10.1073/pnas.072650799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bao G, Suresh S. Cell and molecular mechanics of biological materials. Nat Mater. 2003;2(11):715–25. doi: 10.1038/nmat1001. [DOI] [PubMed] [Google Scholar]
- 12.Baugh L, Vogel V. Structural changes of FN adsorbed to model surfaces probed by fluorescence resonance energy transfer. J Biomed Mater Res A. 2004;69(3):525–34. doi: 10.1002/jbm.a.30026. [DOI] [PubMed] [Google Scholar]
- 13.Bell GI. Models for Specific Adhesion of Cells to Cells. Science. 1978;200(4342):618–27. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 14.Bischofs IB, Schwarz US. Collective effects in cellular structure formation mediated by compliant environments: a Monte Carlo study. Act Biomater. 2006;2(3):253–65. doi: 10.1016/j.actbio.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Blum JS, Temenoff JS, Park H, Jansen JA, Mikos AG, Barry MA. Development and characterization of enhanced green fluorescent protein and luciferase expressing cell line for non-destructive evaluation of tissue engineering constructs. Biomaterials. 2004;25:5809–19. doi: 10.1016/j.biomaterials.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 16.Brinkerhoff CJ, Lindermann JJ. Integrin dimerization and ligand organization: key components in integrin clustering for cell adhesion. Tissue Eng. 2005;11(5–6):865–76. doi: 10.1089/ten.2005.11.865. [DOI] [PubMed] [Google Scholar]
- 17.Brown XQ, Ookawa K, Wong JY. Evaluation of polydimethylsiloxane scaffolds with physiologically-relevant elastic moduli: interplay of substrate mechanics and surface chemistry effects on vascular smooth muscle cell response. Biomaterials. 2005;26:3123. doi: 10.1016/j.biomaterials.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Burdick JA, Khademhosseini A, Langer R. Fabrication of gradient hydrogels using a microfluidics/photopolymerization process. Langmuir. 2004;20(13):5153–6. doi: 10.1021/la049298n. [DOI] [PubMed] [Google Scholar]
- 19.Cavalcanti-Adam EA, Micoulet A, Blümmel J, Auernheimer J, Kssler H, Spatz JP. Lateral spacing of integrin ligands influences cell spreading and focal adhesion assembly. Eur J Cell Biol. 2006;85(3–4):219–24. doi: 10.1016/j.ejcb.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 20.Carvalho RS, Kostenuik PJ, Salih E, Bumann A, Gerstenfeld LC. Selective adhesion of osteoblastic cells to different integrin ligands induces osteopontin gene expression. Matrix Biol. 2003;22:241–9. doi: 10.1016/s0945-053x(03)00038-6. [DOI] [PubMed] [Google Scholar]
- 21.Chan WCW, Nie S. Quantum Dot Bioconjugates for Ultrasensitivie Nonisotopic Detection. Science. 1998;281(5385):2016–18. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 22.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric Control of Cell Life and Death. Science. 1997;276:1425–8. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 23.Cherniavskaya O, Chen CJ, Heller E, Sun E, Provezano J, Kam L, Hone J, Sheetz MP, Wind SJ. Fabrication and surface chemistry of nanoscale bioarrays deisgned for the study of cytoskeletal protein binding interactions and their effect on cell motility. J Vac Sci Technol B. 2005;23(6):2972–8. [Google Scholar]
- 24.Chigaev A, Blenc AM, Braaten JV, Kumaraswamy N, Kepley CL, Andrews RP, Oliver JM, Edwards BS, Prossnitz ER, Larson RS, Sklar LA. Real Time Analysis of the Affinity Regulation of α4-Integrin. J Biol Chem. 2001;276(52):48670–8. doi: 10.1074/jbc.M103194200. [DOI] [PubMed] [Google Scholar]
- 25.Chigaev A, Buranda T, Dwyer DC, Prossnitz ER, Sklar LA. FRET Detection of Cellular α4-Integrin Conformational Activation. Biophys J. 2003;85:3951–62. doi: 10.1016/S0006-3495(03)74809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Comisar WA, Hsiong SX, Kong HJ, Mooney DJ, Linderman JJ. Multi-scale modeling to predict ligand presentation with RGD nanopatterned hydrogels. Biomaterials. 2006;27(10):2322–9. doi: 10.1016/j.biomaterials.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 27.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking Cell-Matrix Adhesion to the Third Dimension. Science. 2001;294:1708–12. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 28.Cunningham JJ, Linderman JJ, Mooney DJ. Externally Applied Cyclic Strain Regulates Localization of Focal Contact Components in Cultured Smooth Muscle Cells. Ann Biomed Eng. 2002;30(7):927–35. doi: 10.1114/1.1500408. [DOI] [PubMed] [Google Scholar]
- 29.Curtis ASG, Lackie JM, editors. Measuring Cell Adhesion. West Sussex, England: John Wiley & Sons Ltd; 1991. [Google Scholar]
- 30.Dembo M, Torney DC, Saxman K, Hammer D. The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proc R Soc Lond B Biol Sci. 1988;234(1274):55–83. doi: 10.1098/rspb.1988.0038. [DOI] [PubMed] [Google Scholar]
- 31.Discher DE, Janmey P, Wang YL. Tissue Cells Feel and Respond to the Stiffness of Their Substrate. Science. 2005;310:1139–43. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 32.Ecker RC, Martin R, Steiner GE, Schmid JA. Application of Spectral Imaging Microscopy in Cytomics and Fluorescence Resonance Energy Transfer (FRET) Analysis. Cytometry A. 2004;59(2):172–81. doi: 10.1002/cyto.a.20053. [DOI] [PubMed] [Google Scholar]
- 33.Eggeling C, Kask P, Winkler D, Jäger S. Rapid Analysis of Förster Resonance Energy Transfer by Two-Color Global Fluorescence Correlation Spectroscopy: Trypsin Proteinase Reaction. Biophys J. 2005;89:605–18. doi: 10.1529/biophysj.104.052753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Engler AJ, Griffin MA, Sen S, Bonnemann CG, Sweeny HL, Discher DE. Myotubes differentiate optimally on substrates with tissue-like stifFNess: pathological implications for soft or stiff microenvironments. J Cell Biol. 2004;166(6):877–87. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Folkman J, Moscona A. Role of Cell-Shape in Growth-Control. Nature. 1978;273(5661):345–9. doi: 10.1038/273345a0. [DOI] [PubMed] [Google Scholar]
- 36.Förster Th. Intermolecular energy migration and fluorescence. Annalen Der Physik. 1948;2:55–75. in German. [Google Scholar]
- 37.Förster Th. 10th Spiers memorial lecture – transfer mechanisms of electronic excitation. Discuss Faraday Soc. 1959;27:7–17. [Google Scholar]
- 38.Friedl P. Dynamic imaging of cellular interactions with extracellular matrix. Histochem Cell Biol. 2004;122:183–90. doi: 10.1007/s00418-004-0682-0. [DOI] [PubMed] [Google Scholar]
- 39.Fung BKK, Stryer L. Surface Density Determination in Membranes by Fluorescence Energy Transfer. Biochemistry. 1978;17(24):5241–8. doi: 10.1021/bi00617a025. [DOI] [PubMed] [Google Scholar]
- 40.Galbraith CG, Sheetz MP. A micromachined device provides a new bend on fibroblast traction forces. Proc Natl Acad Sci USA. 1997;94(17):9114–8. doi: 10.1073/pnas.94.17.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia AJ, Vega MD, Boettiger D. Modulation of Cell Proliferation and Differentiation through Substrate-dependent Changes in FN Conformation. Mol Biol Cell. 1999;10:785–98. doi: 10.1091/mbc.10.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia-Alloza M, Bacskai BJ. Techniques for brain imaging in vivo. Neuromolecular Med. 2004;6(1):65–78. doi: 10.1385/NMM:6:1:065. [DOI] [PubMed] [Google Scholar]
- 43.Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. The Fluorescent Toolbox for Assessing Protein Location and Function. Science. 2006;312:217–24. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 44.Gordon GW, Berry G, Liang XH, Levine B, Herman B. Quantitative fluorescence resonance energy transfer measurements using fluorescent microscopy. Biophys J. 1998;74(5):2702–13. doi: 10.1016/S0006-3495(98)77976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grinnell F, Feld MK. Fibronectin Adsorption on Hydrophillic and Hydrophobic Surfaces Detected by Antibody Binding and Analyzed during Cell Adhesion in Serum-containing Medium. J Biol Chem. 1982;257(9):4888–93. [PubMed] [Google Scholar]
- 46.Gu J, Yam CM, Cai C. Nanometric Protein Arrays on Protein-Resistant Monolayers on Silicon Surfaces. J Am Chem Soc. 2004;126(26):8098–9. doi: 10.1021/ja048405x. [DOI] [PubMed] [Google Scholar]
- 47.Guido S, Tranquillo RT. A methodology for the systematic and quantitative study of cell contact guidance in oriented collagen gels. Correlation of fibroblasts orientation and gel birefringence. J Cell Sci. 1993;105:317–31. doi: 10.1242/jcs.105.2.317. [DOI] [PubMed] [Google Scholar]
- 48.Hansen LK, Mooney DJ, Vacanti JP, Ingber DE. Integrin Binding and Cell Spreading on Extracellular Matrix Act at Different Points in the Cell Cycle to Promote Hepatocyte Growth. Mol Biol Cell. 1994;5:967–75. doi: 10.1091/mbc.5.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harbers GM, Healy KE. The effect of ligand type and density on osteoblast adhesion, proliferation, and matrix mineralization. J Biomed Mater Res A. 2005;75(4):855–69. doi: 10.1002/jbm.a.30482. [DOI] [PubMed] [Google Scholar]
- 50.Harris AK, Stopak D, Wild P. Fibrblast traction as a mechanism for collagen morphogenesis. Nature. 1981;290(5803):249–51. doi: 10.1038/290249a0. [DOI] [PubMed] [Google Scholar]
- 51.Horbett TA. Proteins: Structure, Properties, and Adsorption to Surfaces. Chapter 3.2. In: Ratner BJ, Hoffman AS, Schoen FJ, Lemons JE, editors. Biomaterials Science: An Introduction to Materials in Medicine. 2. New York: Academic Press; 2004. [Google Scholar]
- 52.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110(6):673–87. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 53.Ingber DE. Fibronectin controls capillary endothelial cell growth by modulating cell shape. Proc Natl Acad Sci USA. 1990;87:3579–83. doi: 10.1073/pnas.87.9.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ingber DE, Dike L, Hansen L, Liley H, Maniotis A, McNamee H, Mooney D, Plopper G, Sims J, Wang N. Cellular tensegrity: exploring how mechanical changes in the cytoskeleton regulate cell growth, migration, and tissue pattern during morphogenesis. Int Rev Cytol. 150:173–224. doi: 10.1016/s0074-7696(08)61542-9. [DOI] [PubMed] [Google Scholar]
- 55.Irvine DJ, Hue KA, Mayes AM, Griffith LG. Simulations of cell-surface integrin binding to nanoscale-clustered adhesion ligands. Biophys J. 2002;82:120–32. doi: 10.1016/S0006-3495(02)75379-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Itaka K, Harada A, Nakamura K, Kawaguchi H, Katakoa K. Evaluation by fluorescence resonance energy transfer of the stability of nonviral gene delivery vectors under physiological conditions. Biomacromol. 2002;3:841–5. doi: 10.1021/bm025527d. [DOI] [PubMed] [Google Scholar]
- 57.Jiang G, Huang AH, Cai Y, Tanase M, Sheetz MP. Rigidity sensing at the leading edge through αVβ3 integrins and RPTPα. Biophys J. 2006;90(5):1804–9. doi: 10.1529/biophysj.105.072462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson AE. Fluorescence Approaches for Determining Protein Conformations, Interactions and Mechanisms at Membranes. Traffic. 2005;6:1078–92. doi: 10.1111/j.1600-0854.2005.00340.x. [DOI] [PubMed] [Google Scholar]
- 59.Kato M, Mrksich M. Using model substrates to study the dependence of focal adhesion formation on the affinity of integrin-ligand complexes. Biochemistry. 2004;43(10):2699–707. doi: 10.1021/bi0352670. [DOI] [PubMed] [Google Scholar]
- 60.Keppler A, Pick H, Arrivoli C, Vogel H, Johnsson K. Labeling of fusion proteins with synthetic fluorophores in live cells. Proc Natl Acad Sci USA. 2004;101(27):9955–9. doi: 10.1073/pnas.0401923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keselowsky BG, Collard DM, Garcia AJ. Integrin binding specificity regulates biomaterial surface chemistry effects on differentiation. Proc Natl Acad Sci USA. 2005;102(17):5953–7. doi: 10.1073/pnas.0407356102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim H, Arakawa H, Osada T, Ikai A. Quantification of cell adhesion force with AFM: distribution of vitronectin receptors on a living MC3T3-E1 cell. Ultramicroscopy. 2003;97(1–4):359–63. doi: 10.1016/S0304-3991(03)00061-5. [DOI] [PubMed] [Google Scholar]
- 63.Kirchner J, Kam Z, Tzur G, Bershadsky AD, Geiger B. Live-cell monitoring of tyrosine phosphorylation in focal adhesions following microtubule disruption. J Cell Sci. 2002;116:975–86. doi: 10.1242/jcs.00284. [DOI] [PubMed] [Google Scholar]
- 64.Koblinski JE, Wu M, Demeler B, Jacob K, Kleinman HK. Matrix cell adhesion activated by non-adhesion proteins. J Cell Sci. 2005;118:2965–74. doi: 10.1242/jcs.02411. [DOI] [PubMed] [Google Scholar]
- 65.Kong HJ, Lee KY, Mooney DJ. Nondestructively probing the cross-linking density of polymeric hydrogels. Macromolecules. 2003;36(20):7887–90. [Google Scholar]
- 66.Kong HJ, Polte TR, Alsberg E, Mooney DJ. FRET measurements of cell-traction forces and nano-scale clustering of adhesion ligands varied by substrate stiffness. Proc Natl Acad Sci USA. 2005;102(12):4300–5. doi: 10.1073/pnas.0405873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kong HJ, Liu J, Riddle K, Matsumoto T, Leach K, Mooney DJ. Non-viral gene delivery regulated by stiffness of cell adhesion substrates. Nat Mater. 2005;4:460–4. doi: 10.1038/nmat1392. [DOI] [PubMed] [Google Scholar]
- 68.Kong HJ, Boontheekul T, Mooney DJ. Quantifying the relation between ligand-receptor bond formation and cell phenotype. Proc Natl Acad Sci USA. 2006;103(49):18534–9. doi: 10.1073/pnas.0605960103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kramer A, Lu h, Isralewitz B, Schulten K, Vogel V. Forced unfolding of the FN type III module reveals a tensile molecular recognition switch. Proc Natl Acad Sci USA. 1999;96:1351–6. doi: 10.1073/pnas.96.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krishnan RV, Varma R, Mayor S. Fluorescence Methods to Probe Nanometer-Scale Organization of Molecules in Living Cell Membranes. J Fluoresc. 2001;11(3):211–26. [Google Scholar]
- 71.Lai CS, Wolff CA, Novello D, Griffone L, Cuniberti C, Molina F, Rocco M. Solution Structure of Human Plasma FN Under Different Solvent Conditions. J Mol Biol. 1993;230:625–40. doi: 10.1006/jmbi.1993.1174. [DOI] [PubMed] [Google Scholar]
- 72.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3. New York: Springer; 2006. [Google Scholar]
- 73.Lauffenburger D, Linderman J. Receptors: models for binding, trafficking and signaling. New York: Oxford University Press; 1993. [Google Scholar]
- 74.Leckband D, Israelachvili J. Intermolecular forces in biology. Quart Rev Biophys. 2001;34(2):105–267. doi: 10.1017/s0033583501003687. [DOI] [PubMed] [Google Scholar]
- 75.Leckband D. The surface force apparatus – a tool for probing molecular protein interactions. Nature. 1995;376(6541):617–8. doi: 10.1038/376617a0. [DOI] [PubMed] [Google Scholar]
- 76.Lee KB, Park SJ, Mirkin CA, Smith JC, Mrksich M. Protein nanoarrays generated by dip-pen nanolithography. Science. 2002;295:1702–5. doi: 10.1126/science.1067172. [DOI] [PubMed] [Google Scholar]
- 77.Lee KY, Mooney DJ. Hydrogels for Tissue Engineering. Chem Rev. 2001;101(7):1869–79. doi: 10.1021/cr000108x. [DOI] [PubMed] [Google Scholar]
- 78.Lee KY, Kong HJ, Larson RG, Mooney DJ. Hydrogel Formation via Cell Crosslinking. Adv Mater. 2003;15(21):1828–32. [Google Scholar]
- 79.Lee KY, Alsberg E, Hsiong S, Comisar W, Linderman J, Ziff R, Mooney DJ. Nanoscale Adhesion Ligand Organization Regulates Osteoblast Proliferation and Differentiation. Nanoletters. 2004;4(8):1501–6. doi: 10.1021/nl0493592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee SH, Miller JS, Moon JJ, West JL. Proteolytically Degradable Hydrogels with a Fluorgenic Substrate for Studies of Cellular Proteolytic Activity and Migration. Biotechnolo Prog. 2005;21(6):1736–41. doi: 10.1021/bp0502429. [DOI] [PubMed] [Google Scholar]
- 81.Lehenkari PP, Horton MA. Single integrin molecule adhesion forces in intact cells measured by atomic force microscopy. Biochem Biophys Res Common. 1999;259(3):645–50. doi: 10.1006/bbrc.1999.0827. [DOI] [PubMed] [Google Scholar]
- 82.Lichtman JW, Conchello JA. Fluorescence microscopy. Nature Methods. 2005;12(12):910–920. doi: 10.1038/nmeth817. [DOI] [PubMed] [Google Scholar]
- 83.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol. 2005;23(1):47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 84.Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB, Hubbell JA. Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: engineering cell-invasion characteristics. Proc Natl Acad Sci USA. 2003;100(9):5413–8. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maheshwari G, Brown G, Lauffenburger DA, Wells A, Griffith LG. Cell adhesion and motility depend on nanoscale RGD clustering. J Cell Sci. 2000;113:1677–86. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 86.Massia SP, Hubbell JA. An RGD spacing of 440 nm is sufficient for integrin alpha V beta 3-mediated fibroblast spreading and 140 nm for focal contact and stress fiber formation. J Cell Biol. 1991;114(5):1089–100. doi: 10.1083/jcb.114.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Merkel R, Nassoy P, Leung A, Ritchie K, Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397:50–3. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- 88.Midwood KS, Wierzbicka-Patynowski I, Schwarzbauer JE. Chapter 6 in Methods in Cell Biology. Vol. 69. Elsevier Science Academic Press; San Diego, CA USA: 2002. Preparation and Analysis of Synthetic Multicomponent Extracellular Matrix. [DOI] [PubMed] [Google Scholar]
- 89.Miura K. Tracking Movement in Cell Biology. Adv Biochem Engin/Biotechnol. 2005;95:267–95. doi: 10.1007/b102218. [DOI] [PubMed] [Google Scholar]
- 90.Model MA, Healy KE. Quantification of the surface density of a fluorescent label with the optical microscope. J Biomed Mater Res. 1999;50(1):90–6. doi: 10.1002/(sici)1097-4636(200004)50:1<90::aid-jbm13>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 91.Munevar S, Wang Y, Dembo M. Traction force microscopy of migrating normal and H-ras transformed 3T3 fibroblasts. Biophys J. 2001;80(4):1744–57. doi: 10.1016/s0006-3495(01)76145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Munevar S, Wang YL, Dembo M. Regulation of mechanical interactions between fibroblasts and the substratum by stretch-activated Ca2+ entry. J Cell Sci. 2003;117:85–92. doi: 10.1242/jcs.00795. [DOI] [PubMed] [Google Scholar]
- 93.Neher RA, Neher E. Applying spectral fingerprinting to the analysis of FRET images. Microsc Res Tech. 2004;64(2):185–95. doi: 10.1002/jemt.20078. [DOI] [PubMed] [Google Scholar]
- 94.Oberhauser AF, Badilla-Fernandez C, Carrion-Vazquez M, Fernandez JM. The Mechanical Hierarchies of FN Observed with Single-molecule AFM. J Mol Biol. 2002;319:433–47. doi: 10.1016/S0022-2836(02)00306-6. [DOI] [PubMed] [Google Scholar]
- 95.Ohashi T, Kiehart DP, Erickson HP. Dynamics and elasticity of the FN matrix in living cell culture visualized by FN-green fluorescent protein. Proc Natl Acad Sci USA. 1999;96:2153–8. doi: 10.1073/pnas.96.5.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malingnant phenotype. Cancer Cell. 2005;8:241–54. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 97.Pelham RJ, Wang YL. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA. 1997;94:13661–5. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pierschbacher MD, Ruoslahti E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature. 1984;309(5963):30–3. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 99.Prime KL, Whitesides GM. Self-Assembled Organic Monolayers – Model Systems for Studying Adsorption of Proteins at Surfaces. Science. 1991;252(5009):1164–7. doi: 10.1126/science.252.5009.1164. [DOI] [PubMed] [Google Scholar]
- 100.Putnam AJ, Cunningham JJ, Dennis RG, Linderman JJ, Mooney DJ. Microtubule assembly is regulated by externally applied strain in cultured smooth muscle cells. J Cell Sci. 1998;111:3379–87. doi: 10.1242/jcs.111.22.3379. [DOI] [PubMed] [Google Scholar]
- 101.Remaut K, Lucas B, Braeckmans K, Sanders NN, De Smedt SC, Demeester J. FRET-FCS as a tool to evaluate the stability of oligonucelotide drugs after intracellular delivery. J Control Release. 2005;103(1):259–71. doi: 10.1016/j.jconrel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 102.Roberts C, Chen CS, Mrksich M, Martichonok V, Ingber DE, Whitesides GM. Using Mixed Self-Assembled Monolayers Presenting RGD and (EG)3OH Groups To Characterize Long-Term Attachment of Bovine Capillary Endothelial Cells to Surfaces. J Am Chem Soc. 1998;120:6548–55. [Google Scholar]
- 103.Rowley JA, Madlambayan G, Mooney DJ. Alginate hydrogels as synthetic extracellular matrix materials. Biomaterls. 1999;20(1):45–53. doi: 10.1016/s0142-9612(98)00107-0. [DOI] [PubMed] [Google Scholar]
- 104.Rowley JA, Sun Z, Goldman D, Mooney DJ. Biomaterials to Spatially Regulate Cell Fate. Adv Mater. 2002;14(12):886–9. [Google Scholar]
- 105.Ruoslahti E. RGD and Other Recognition Sequences For Integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 106.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Selective differentiation of neural progenitor cells by high-epitope density nanofibers. Science. 2004;303(5662):1352–5. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 107.Singhvi R, Kumar A, Lopez GP, Stephanopoulos GN, Wang DI, Whitesides GM, Ingber DE. Engineering cell shape and function. Science. 1994;264(5159):696–8. doi: 10.1126/science.8171320. [DOI] [PubMed] [Google Scholar]
- 108.Stockholm D, Bartoli M, Sillon G, Bourg N, Davoust J, Richard I. Imaging Calpain Protease Activity by Multiphoton FRET in Living Mice. J Mol Biol. 2005;346:215–22. doi: 10.1016/j.jmb.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 109.Stryer L, Haugland RP. Energy Transfer – A Spectroscopic Ruler. Proc Natl Acad Sci USA. 1967;58(2):719–26. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stupack DG, Cheresh DA. Get a ligand, get a life: integrins, signaling and cell survival. J Cell Sci. 2002;115:3729–38. doi: 10.1242/jcs.00071. [DOI] [PubMed] [Google Scholar]
- 111.Takagi J, Springer TA. Integrin activation and structural rearrangement. Immunol Rev. 2002;186:141–63. doi: 10.1034/j.1600-065x.2002.18613.x. [DOI] [PubMed] [Google Scholar]
- 112.Tamada M, Sheetz MP, Sawada Y. Activation of a signaling cascade by cytoskeleton stretch. Dev Cell. 2004;7(5):709–18. doi: 10.1016/j.devcel.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 113.Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc Natl Acad Sci USA. 2003;100(4):1484–9. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Thomas CH, Collier JH, Sfeir CS, Healy KE. Engineering gene expression and protein synthesis by modulation of nuclear shape. PNAS. 2002;99:1972–7. doi: 10.1073/pnas.032668799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang H, Castner DG, Ratner BD, Jiang S. Probing the orientation of surface-immobilized immunoglobulin G by time-of-flight secondary ion mass spectrometry. Langmuier. 2004;20(5):1877–87. doi: 10.1021/la035376f. [DOI] [PubMed] [Google Scholar]
- 116.Wang N, Butler JP, Ingber DE. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 1993;260(5111):1124–7. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- 117.Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, Tsien RY, Chien S. Visualizing the mechanical activation of Src. Nature. 2005;434:1040–5. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- 118.Ward MD, Hammer DA. A theoretical analysis for the effect of focal contact formation on cell-substrate attachment strength. Biophys J. 1993;64:936–59. doi: 10.1016/S0006-3495(93)81456-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wilson CJ, Clegg RE, Leavesley DL, Pearcy MJ. Mediation of biomaterial-cell interactions by adsorbed proteins: a review. Tissue Eng. 2005;11(1–2):1–18. doi: 10.1089/ten.2005.11.1. [DOI] [PubMed] [Google Scholar]
- 120.Wiseman PW, Brown CM, Webb DJ, Hebert B, Johnson NL, Squier JA, Ellisman MH, Horwitz AF. Spatial mapping of integrin interactions and dynamics during cell migration by image correlation microscopy. J Cell Sci. 2004;117(23):5521–34. doi: 10.1242/jcs.01416. [DOI] [PubMed] [Google Scholar]
- 121.Wolber PK, Hudson BS. An Analytic Solution to the Förster Energy Transfer Problem in Two Dimensions. Biophys J. 1979;28:197–210. doi: 10.1016/S0006-3495(79)85171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu D, Edelman ER. Resonance Energy Transfer for Assessing the Molecular Integrity of Proteins for Local Delivery. Biotechnol Bioeng. 2004;85(4):406–12. doi: 10.1002/bit.10902. [DOI] [PubMed] [Google Scholar]
- 123.Zamir E, Katz BZ, Aota S, Yamada KM, Geiger B, Kam Z. Molecular diversity of cell-matrix adhesions. J Cell Sci. 1999;112:1655–69. doi: 10.1242/jcs.112.11.1655. [DOI] [PubMed] [Google Scholar]
- 124.Zimet DB, Thevenin BJM, Verkman AS, Shohet SB, Abney JR. Caclulation of Resonance Energy Transfer in Crowded Biological Membranes. Biophys J. 1995;68:1592–603. doi: 10.1016/S0006-3495(95)80332-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zimmermann T. Spectral Imaging and Linear Unmixing in Light Microscopy. Adv Biochem Engin/Biotechnol. 2005;95:245–65. doi: 10.1007/b102216. [DOI] [PubMed] [Google Scholar]


