Abstract
Rationale: Plasminogen activator inhibitor (PAI)-1 inhibits urokinase and tissue plasminogen activator, required for host response to infection. Whether variation within the PAI-1 gene is associated with increased susceptibility to infection is unknown.
Objectives: To ascertain the role of the 4G/5G polymorphism and other genetic variants within the PAI-1 gene. We hypothesized that variants associated with increased PAI-1 expression would be associated with an increased occurrence of community-acquired pneumonia (CAP).
Methods: Longitudinal analysis (>12 yr) of the Health, Aging, and Body Composition cohort, aged 65–74 years at start of analysis.
Measurements and Main Results: We genotyped the 4G/5G PAI-1 polymorphism and six additional single nucleotide polymorphisms. Of the 3,075 subjects, 272 (8.8%) had at least one hospitalization for CAP. Among whites, variants at the PAI4G,5G, PAI2846, and PAI7343 sites had higher risk of CAP (P = 0.018, 0.021, and 0.021, respectively). At these sites, variants associated with higher PAI-1 expression were associated with increased CAP susceptibility. Compared with the 5G/5G genotypes at PAI4G,5G site, the 4G/4G and 4G/5G genotypes were associated with a 1.98-fold increased risk of CAP (95% confidence interval, 1.2–3.2; P = 0.006). In whole blood stimulation assay, subjects with a 4G allele had 3.3- and 1.9-fold increased PAI-1 expression (P = 0.043 and 0.034, respectively). In haplotype analysis, the 4G/G/C/A haplotype at the PAI4G,5G, PAI2846, PAI4588, and PAI7343 single nucleotide polymorphisms was associated with higher CAP susceptibility, whereas the 5G/G/C/A haplotype was associated with lower CAP susceptibility. No associations were seen among blacks.
Conclusions: Genotypes associated with increased expression of PAI-1 were associated with increased susceptibility to CAP in elderly whites.
Keywords: pneumonia, inflammatory markers, PAI-1, gene, haplotype
AT A GLANCE COMMENTARY
Scientific Knowledge on the Subject
Few studies have examined whether genetic variants influence susceptibility to community-acquired pneumonia.
What This Study Adds to the Field
Genotypes associated with increased expression of PAI-1 are associated with increased susceptibility to community-acquired pneumonia in elderly whites.
Increased activity of proteins involved in the production of the fibrinolytic protein plasmin is part of the innate immune response to infection (1, 2). Urokinase-like plasminogen activator (uPA) and its receptor (uPAR) promote cell migration and neutrophil activity through plasmin-dependent and plasmin-independent mechanisms (3). The inhibitor of uPA, plasminogen activator inhibitor (PAI)-1, is also activated during infection. Increased concentrations in the systemic circulation and the alveolar compartment have been demonstrated in pneumonia, severe sepsis, and acute lung injury (2, 4–6). PAI-1 interacts with inflammatory cytokines to maintain homeostasis (7). Although localized PAI-1 expression may contain infection, systemic overexpression may impair host response to infection. This may be particularly important in the elderly, who have increased circulating PAI-1 concentrations and increased PAI-1 response to lipopolysaccharide (8–10).
A functional insertion/deletion polymorphism, termed 4G/5G, is found 675 base pairs upstream of the transcriptional initiation site (PAI4G,5G) in the PAI-1 gene (7q21.3-q22) (11, 12). Although both alleles bind a transcriptional activator, the 5G allele reduces transcription by binding a repressor protein, and is thereby associated with lower circulating PAI-1 concentrations (13, 14). The 4G/4G polymorphism is more common in patients with nonspecific interstitial pneumonia (15). Whether it plays a role in infection, particularly community-acquired pneumonia (CAP), is not known. Sequencing data from SeattleSNPs (NHLBI Program for Genomic Applications, http://pga.gs.washington.edu) reveals that several other common single nucleotide polymorphisms (SNPs) and haplotypes of the PAI-1 gene have been identified. We hypothesized that genetic variation within the PAI-1 gene associated with increased circulating PAI-1 concentrations would be associated with increased susceptibility to CAP in the elderly.
METHODS
See the online supplement for more details on methods used.
Design and Population
We used data from the Health ABC cohort, a study designed to examine effects of body composition on morbidity and mortality. Subjects were recruited in Memphis, Tennessee, and Pittsburgh, Pennsylvania. To be enrolled, subjects had to be elderly (aged 70–79 yr) and in good health, defined as an ability to walk 0.25 mile, climb 10 steps, and perform activities of daily living without difficulty. Data regarding hospitalization are available for 5 years before enrollment through the Health Care Financing Administration (HCFA) and subjects had up to 7 years of follow-up after enrollment in the study. All participants gave informed consent and the institutional review boards at the Universities of Pittsburgh and Tennessee approved the study.
Outcome Measure
The primary outcome was CAP requiring hospitalization. Ascertainment of CAP was over two periods. During enrollment in Health ABC, hospitalization was ascertained based on participant self-report and active surveillance by study personnel. Criteria for hospitalization for CAP were established prospectively and an adjudicator at each site adjudicated the outcome during this period. We used a combination of discharge summary, International Classification of Diseases, Ninth Revision (ICD-9) diagnoses, admission history and physical examination, and radiology reports to ascertain CAP. For the 5 years before enrollment in Health ABC, ICD-9 discharge diagnoses alone were used to ascertain CAP.
PAI-1 Gene
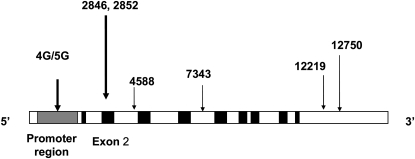
The PAI-1 gene has been completely sequenced at SeattleSNPs and approximately 100 polymorphisms were identified in 2002. We genotyped seven polymorphisms from different haplotype bins (Figure 1 and Table E1A of the online supplement). We first chose the 4G/5G polymorphism because it is functional (13). Nonsynonymous SNPs are more likely to be functional. Five nonsynonymous SNPs were described in the PAI-1 gene. Of these, we chose two SNPs at PAI2846 G/A (Ala14Thr) and PAI2852 G/A (Val17Ile) because the minor allele frequencies were greater than 10%. We then genotyped four synonymous SNPs, two polymorphisms at PAI4588 (C/T) and PAI7343 (A/G) in introns and two polymorphisms at PAI12219 (T/C) and PAI12750 (G/A) in 3′ untranslated regions, to identify common haplotypes.
Figure 1.
Single nucleotide polymorphisms within the plasminogen activator inhibitor (PAI)-1 gene (7q21.3-q22). Gray area denotes promoter region and black areas denote exons. The 12219 and 12750 polymorphisms are in the 3′ untranslated region.
European Ancestry Markers
Blacks were genotyped for 37 ancestry-informative genetic markers, as described elsewhere (16). Using these genotype data, we estimated the proportion of European ancestry for each self-reported black participant (see the online supplement).
Inflammatory Markers
Blood samples were collected for PAI-1, tumor necrosis factor (TNF), and IL-6 protein measurement by venipuncture after an overnight fast, at a mean time of 9:25 a.m., in the absence of infection during enrollment. Cytokines were measured using ELISA (see the online supplement). Cytokine data were missing in fewer than 6% of the cohort.
Whole Blood Ex Vivo Stimulation with Lipopolysaccharide and Peptidoglycan
To evaluate functional significance of PAI-1 polymorphisms associated with higher risk of CAP in the Health ABC cohort, we compared PAI-1, TNF, IL-6, and IL-10 concentrations after ex vivo whole blood stimulation (24 h incubation) using lipopolysaccharide (LPS) from Salmonella minnesota (500 ng/ml) and peptidoglycan (PGN) from Staphylococcus aureus (100 μg/ml) in 23 healthy volunteers from Pittsburgh, Pennsylvania (see online supplement).
Statistical Analyses
We used SAS Genetics (SAS Institute, Cary, NC) to estimate Hardy-Weinberg equilibrium and linkage disequilibrium, to examine association between genotypes and phenotypes, and to construct haplotypes. Associations between genotypes and phenotypes were adjusted to control for false-discovery rate, as described by Hochberg and Benjamini (17).
Nine haplotypes (frequency > 1%) were identified in whites and blacks. The measures of linkage disequilibrium between the SNPs were high in the Health ABC cohort despite choosing these SNPs from different haplotype bins. Therefore, we identified the minimum number of SNPs tagging common haplotypes in whites and blacks separately using BEST software (http://genomethods.org/best/about.htm) (18). SNPs used to tag these haplotypes were also associated with CAP susceptibility and circulating PAI-1 concentrations in univariate analysis. We then used haplotype marker–trait association to ascertain association between haplotypes and CAP susceptibility using exact tests (19).
We used logistic regression analysis to estimate the odds ratio (OR) for genotypes and haplotypes associated with CAP susceptibility. Covariates for this model were chosen based on previously reported risk factors for CAP (20–24), and included age, sex, site, circulating TNF and IL-6 concentrations, history of congestive heart failure, coronary heart disease, smoking, diabetes, FEV1, and serum creatinine. Haplotypes cannot be estimated unequivocally for heterozygotes. Therefore, for the logistic regression model incorporating haplotypes, we first estimated the probability of having each of these haplotypes using haplotype trend analysis (25). These probability scores were then included in the regression model.
The study had adequate power (β > 0.8) to ascertain codominant effects for rare allele frequencies greater than 0.1 and ORs of 2 or greater in blacks and whites separately (26).
RESULTS
Participant Characteristics
Table 1 shows characteristics of the 3,075 participants in Health ABC at enrollment (1997–1998). The mean age of the cohort was 73.6 years. Race was based on self-report and 41.7% of the cohort was black. Although participants were well functioning, comorbid conditions, such as diabetes (15.3%), coronary heart disease (20.8%), and reduced lung function (29.3%), were common.
TABLE 1.
CHARACTERISTICS OF COHORT
| Variable | All Participants (n = 3,075) | Hospitalized for CAP (n = 272) | Never Hospitalized for CAP (n = 2,803) | P Value |
|---|---|---|---|---|
| Demographic characteristics | ||||
| Age, mean ± SD | 73.6 ± 2.9 | 73.9 ± 2.9 | 73.6 ± 2.9 | 0.1 |
| Sex, males, % (n) | 48.5 (1,491) | 53.7 (146) | 48 (1,345) | 0.07 |
| Race, blacks, % (n) | 41.7 (1,281) | 40.4 (110) | 41.8 (1,171) | 0.7 |
| Site, Memphis, % (n) | 50.3 (1,547) | 47.4 (129) | 50.6 (1,418) | 0.3 |
| Comorbid conditions | ||||
| Congestive heart failure, % (n) | 3.1 (95) | 7.0 (19) | 2.7 (76) | <0.0001 |
| Coronary heart disease, % (n) | 20.8 (625) | 30.4 (81) | 19.9 (554) | <0.0001 |
| Diabetes, % (n) | 15.3 (470) | 21.5 (58) | 14.7 (412) | 0.003 |
| Serum creatinine, mg/dl, mean ± SD | 1.1 ± 0.4 | 1.1 ± 0.6 | 1.05 ± 0.4 | 0.003 |
| Predicted FEV1, % (n) | <0.0001 | |||
| FEV1 > 80% | 70.7 (2,175) | 52.2 (142) | 72.5 (2,033) | |
| 80% > FEV1 > 50% | 19.2 (591) | 25 (68) | 14 (523) | |
| FEV1 < 50% | 10.1 (309) | 22.8 (62) | 4.6 (247) | |
| Smoking, % (n) | <0.0001 | |||
| Current | 10.4 (318) | 16.9 (46) | 9.7 (272) | |
| Past | 45.7 (1,404) | 48.9 (133) | 45.4 (1,271) | |
| Pack-years of smoking, mean ± SD | 19.1 ± 28 | 27.4 ± 34 | 18.3 ± 28 | <0.0001 |
| Inflammatory markers, pg/ml, median (IQR) | ||||
| TNF | 3.2 (2.4–4.1) | 3.6 (0.9–16.8) | 3.1 (0.6–29.6) | <0.0001 |
| IL-6 | 1.8 (1.3–2.8) | 2.3 (0.3–13.2) | 1.8 (0.2–16) | <0.0001 |
| PAI-1 | 21 (13–36) | 24 (14–43) | 21 (12–36) | 0.02 |
Definition of abbreviations: CAP = community-acquired pneumonia; IQR = interquartile range; PAI-1 = plasminogen activator inhibitor-1; TNF = tumor necrosis factor.
Fewer than 5% of data are missing for some of the covariates.
Three hundred and four cases of CAP requiring hospitalization occurred over the 12-year ascertainment period from 1992–1993 to 2005. Of these, 106 cases occurred in 106 subjects during the 5 years before enrollment in Health ABC, and 237 cases occurred in 198 subjects after enrollment. Thus, 272 subjects (8.8%) were hospitalized at least once for CAP during the 12-year ascertainment period and 29 participants had more than one episode of CAP hospitalization. The rate of CAP after enrollment in Health ABC was 10.9 cases per 1,000 person-years. Table 1 compares clinical characteristics and inflammatory marker concentrations for participants with and without at least one hospitalization for CAP. Participants with CAP were more likely to have comorbid conditions, such as congestive heart failure, coronary artery disease, and diabetes, elevated serum creatinine concentrations, and reduced lung function; in addition, they were more likely to be or to have been smokers, and to have higher circulating concentrations of TNF and IL-6. We measured PAI-1 concentrations in 3,010 (98%) participants at enrollment, when subjects reported good health and absence of infection. The median PAI-1 concentration was 21 ng/ml. Circulating PAI-1 concentrations were higher among subjects hospitalized for CAP compared with those never hospitalized for CAP (24 vs. 21 ng/ml, respectively; P = 0.02).
Of the 272 participants with hospitalization for CAP, 162 (60%) were white and 110 (40%) were black. Table 2 compares characteristics of participants with and without an episode of CAP stratified by self-reported race. In this stratified analysis, the incidence of CAP was again higher in participants with comorbid conditions and a history of smoking, but results did not reach statistical significance with some of these comorbid conditions. PAI-1 concentrations were higher among whites hospitalized with CAP (25 vs. 21 ng/ml for hospitalized and nonhospitalized subjects, respectively; P = 0.005), but differences were smaller and not statistically significant among blacks who did and did not require hospitalization for CAP (22 vs. 21 ng/ml, respectively; P = 0.7).
TABLE 2.
CHARACTERISTICS OF COHORT STRATIFIED BY SELF-REPORTED RACE
| Whites (n = 1,794)
|
Blacks (n = 1,281)
|
|||
|---|---|---|---|---|
| Variable | Hospitalized for CAP (n = 162) | Never Hospitalized for CAP (n = 1,632) | Hospitalized for CAP (n = 110) | Never Hospitalized for CAP (n = 1,171) |
| Demographic characteristics | ||||
| Age, yr, mean ± SD | 74 ± 2.8 | 73.8 ± 2.9 | 73.8 ± 3 | 73.4 ± 2.9 |
| Sex, males, % (n) | 57.4 (93) | 51.8 (846) | 48.2 (53) | 42.6 (499) |
| Site, Memphis, % (n) | 51.9 (84) | 52.1 (850) | 40.9 (45) | 48.5 (568) |
| Comorbid conditions | ||||
| Congestive heart failure, % (n) | 6.2 (10) | 2.5 (40) | 8.3 (9) | 3.1 (36) |
| Coronary heart disease, % (n) | 24.9 (40) | 20.1 (324) | 39.1 (41) | 19.5 (230) |
| Diabetes, % (n) | 15.5 (25) | 10.4 (170) | 30.3 (33) | 20.7 (242) |
| Serum creatinine, mg/dl, mean ± SD | 1.06 ± 0.6 | 1.01 ± 0.4 | 1.24 ± 0.6 | 1.1 ± 0.4 |
| Predicted FEV1, % (n) | ||||
| FEV1 > 80% | 52.5 (85) | 72.1 (1,176) | 51.8 (57) | 73.2 (857) |
| 80% > FEV1 > 50% | 28.4 (46) | 19.5 (318) | 20 (22) | 17.6 (205) |
| FEV1 < 50% | 19.1 (31) | 8.5 (138) | 28.2 (31) | 9.3 (109) |
| Smoking, % (n) | ||||
| Current | 9.9 (16) | 5.8 (95) | 27.3 (30) | 15.2 (177) |
| Past | 53.7 (87) | 50.3 (819) | 41.8 (46) | 38.7 (452) |
| Pack-years of smoking, mean ± SD | 29.7 ± 35 | 19.9 ± 28.3 | 23.8 ± 30.3 | 15.9 ± 24.8 |
| Inflammatory markers, median (IQR) | ||||
| TNF, pg/ml | 3.7 (2.8–4.8) | 3.3 (2.6–4.2) | 3.3 (2.4–4.4) | 3 (2.3–3.8) |
| IL-6, pg/ml | 2.2 (1.4–3.5) | 1.7 (1.2–2.5) | 2.4 (1.6–3.6) | 2 (1.3–3) |
| PAI-1, ng/ml | 25 (14–53) | 21 (13–36) | 22 (13–36) | 21 (12–36) |
For definition of abbreviations, see Table 1.
Fewer than 5% of data are missing for some of the covariates.
PAI-1 SNPs and Haplotypes
All SNPs were in Hardy-Weinberg equilibrium, except the PAI2852 in whites and PAI4588 and PAI12219 in blacks (Table 3). Compared with whites, blacks are genetically more heterogeneous. Therefore, we analyzed markers for European ancestry among 1,278 (97%) blacks. Because racial admixture could account for Hardy-Weinberg disequilibrium, we estimated Hardy-Weinberg equilibrium among blacks after stratifying for European ancestry markers. All SNPs were in Hardy-Weinberg equilibrium among blacks with less than 10% of markers of European ancestry. Hardy-Weinberg disequilibrium was present for the PAI12219 SNP and those with greater than 25% of markers of European ancestry and for both PAI4588 and PAI12219 SNPs and those with greater than 50% of markers of European ancestry.
TABLE 3.
TEST FOR HARDY-WEINBERG EQUILIBRIUM (EXACT P VALUES) FOR PAI-1 GENOTYPES STRATIFIED BY SELF-REPORTED RACE AND BY PERCENTAGE OF EUROPEAN ANCESTRY FOR BLACKS
| Blacks
|
||||||
|---|---|---|---|---|---|---|
| Polymorphism | Whites | All | <10% (n = 328) | <25% (n = 545) | <50% (n = 332) | <75% (n = 73) |
| PAI4G,5G (rs1799889) | 0.37 | 0.25 | 0.63 | 0.46 | 0.37 | 0.25 |
| PAI2846 (rs6092) | 0.73 | 0.44 | 0.99 | 0.16 | 0.35 | 0.44 |
| PAI2852 (rs6090) | 0.049 | 0.80 | 0.6 | 0.99 | 0.8 | 0.8 |
| PAI4588 (rs2227657) | 0.37 | 0.02 | 0.12 | 0.1 | 0.02 | 0.02 |
| PAI7343 (rs2227674) | 0.39 | 0.056 | 0.67 | 0.72 | 0.1 | 0.06 |
| PAI12219 (rs11178) | 0.70 | 0.04 | 0.64 | 0.04 | 0.04 | 0.03 |
| PAI12750 (rs1050813) | 0.29 | 0.29 | 0.99 | 0.99 | 0.99 | 0.51 |
Measures of linkage disequilibrium between the PAI-1 SNPs are shown in Table E2A. We constructed nine haplotypes (frequency > 1%) for the entire cohort, and the frequency of these haplotypes varied between blacks and whites (Table 4). Although haplotypes A, D, and E accounted for 69.5% of the haplotypes among whites, haplotypes A, B, and C accounted for 76.8% of haplotypes among blacks. Haplotype B was present among 34.1% of black participants, but only 2.2% of whites. In contrast, haplotypes D and E were present in 16.3 and 10.7% of white participants but in only 4.1 and 1.9% of blacks, respectively.
TABLE 4.
FREQUENCY OF HAPLOTYPES BASED ON SEVEN SINGLE NUCLEOTIDE POLYMORPHISMS*
| Haplotype | PAI4G,5G | PAI2846 | PAI2852 | PAI4588 | PAI7343 | PAI12219 | PAI12750 | Frequency (%) |
|---|---|---|---|---|---|---|---|---|
| Whites | ||||||||
| A | 4G | G | G | C | A | C | G | 42.5 |
| F | 4G | G | G | T | A | T | G | 6.7 |
| J | 4G | G | G | C | A | C | A | 3.9 |
| D | 5G | G | G | C | A | T | A | 16.3 |
| E | 5G | A | G | C | G | T | G | 10.7 |
| C | 5G | G | G | C | G | T | G | 9.7 |
| G | 5G | G | G | T | A | T | G | 4.9 |
| B | 5G | G | G | C | A | C | G | 2.2 |
| H | 5G | G | A | T | A | T | G | 1.2 |
| Blacks | ||||||||
| A | 4G | G | G | C | A | C | G | 21.1 |
| I | 4G | G | G | C | G | T | G | 3.4 |
| F | 4G | G | G | T | A | T | G | 2.6 |
| B | 5G | G | G | C | A | C | G | 34.1 |
| C | 5G | G | G | C | G | T | G | 21.6 |
| H | 5G | G | A | T | A | T | G | 5.7 |
| G | 5G | G | G | T | A | T | G | 4.3 |
| D | 5G | G | G | C | A | T | A | 4.1 |
| E | 5G | A | G | C | G | T | G | 1.9 |
Includes haplotypes with frequency greater than 1% in the population. Frequency of I haplotype in whites and J haplotype in blacks is less than 1%.
PAI-1 SNPs and CAP Susceptibility
Among white participants, variants at the PAI4G,5G, PAI2846, and PAI7343 sites had higher risk of CAP (P = 0.018, 0.021, and 0.021, respectively; Table 5). These associations remained significant after adjusting for false discovery. A codominant effect was seen for the 4G allele at the PAI4G,5G site (frequency of CAP = 9.9% vs. 5.3% for the combined 4G/4G and 4G/5G genotypes compared with 5G/5G genotypes, P = 0.005), and those with at least one 4G allele had a 1.98-fold odds of CAP hospitalization (95% confidence interval [CI], 1.2–3.2; P = 0.006). The odds of CAP for white participants with the 4G/4G and 4G/5G genotypes remained unchanged (OR, 2.1; 95% CI, 1.3–3.6) in the multivariable analysis. Similarly, a codominant effect was seen for the AG and GG genotypes at the PAI7343 site (frequency of CAP = 6.5% vs. 10.5%, for the AG and GG genotypes compared with AA genotypes, P = 0.006). Subjects with at least one G allele at the PAI7343 site had a lower risk of CAP in univariate and multivariable analyses (OR, 0.6; 95% CI, 0.4–0.9; P = 0.006; and OR, 0.5; 95% CI, 0.4–0.8; P = 0.001; in univariate and multivariable analyses). A log-additive effect was seen for the genotypes at the PAI2846 site (Table 5) with the highest frequency of CAP for subjects with the GG genotype (9.9%), an intermediate frequency for those with the GA genotype (5.6%), and the lowest frequency for the AA genotype (0%) (P = 0.01), but the overall frequency of subjects with the AA genotype was very low (1.4%).
TABLE 5.
ASSOCIATION BETWEEN GENOTYPE AND PHENOTYPE (PAI-1 AND COMMUNITY-ACQUIRED PNEUMONIA)
| Whites (n = 1,794)
|
Blacks (n = 1281)
|
|||||
|---|---|---|---|---|---|---|
| SNP | Genotype Frequency % (n) | PAI-1 Concentrations* | Frequency of CAP % (n) | Genotype Frequency % (n) | PAI-1 Concentrations* | Frequency of CAP % (n) |
| PAI4G,5G | ||||||
| 5G/5G | 21.8 (380) | 20 (11–31) | 5.3 (20) | 8.5 (104) | 20 (11–34) | 8.5 (9) |
| 5G/4G | 48.6 (846) | 22 (13–37) | 10.1 (85) | 39 (477) | 21 (13–37) | 8.8 (42) |
| 4G/4G | 29.6 (515) | 23 (13–43) | 9.7 (50) | 52.5 (642) | 23 (13–42) | 8.6 (55) |
| P value† | — | 0.0001 | 0.018 (0.049) | — | 0.0001 | 0.99 (0.99) |
| PAI2846 | ||||||
| A/A | 1.4 (25) | 22.5 (13–39) | 0 (0) | 0.1 (1) | 11 | 0 (0) |
| A/G | 20.4 (355) | 20 (12-32.5) | 5.6 (20) | 4.1 (51) | 24 (15–39) | 15.7 (8) |
| G/G | 78.2 (1,359) | 30 (14–44) | 9.9 (135) | 95.8 (1,179) | 21 (12–36) | 8.3 (98) |
| P value† | — | 0.03 | 0.012 (0.049) | — | 0.4 | 0.176 (0.617) |
| PAI2852 | ||||||
| A/A | 0.1 (2) | 15 | 50.0 (1) | 0.4 (5) | 15 (7–39) | 20 (1) |
| A/G | 2.7 (47) | 20 (13–30) | 8.5 (4) | 11.4 (140) | 19 (11–32) | 9.3 (13) |
| G/G | 97.2 (1,692) | 22 (13–38) | 8.8 (149) | 88.2 (1,084) | 21 (13–36) | 8.4 (91) |
| P value† | — | 0.40 | 0.122 (0.171) | — | 0.20 | 0.616 (0.99) |
| PAI4588 | ||||||
| C/C | 76 (1,224) | 21 (13–37) | 8.5 (104) | 76.8 (883) | 21 (12–36) | 9.3 (82) |
| C/T | 22 (353) | 22 (12–39) | 9.1 (32) | 20.8 (239) | 22 (12–36) | 8.4 (20) |
| T/T | 2 (31) | 18 (10–25) | 6.5 (2) | 2.4 (28) | 19 (12.5–35.5) | 7.1 (2) |
| P value† | — | 0.10 | 0.863 (0.863) | — | 0.80 | 0.853 (0.99) |
| PAI7343 | ||||||
| A/A | 62.4 (1,069) | 22 (13–41) | 10.5 (112) | 52.2 (629) | 20 (12–36) | 9.1 (57) |
| A/G | 32.8 (561) | 20 (12–32) | 6.4 (36) | 41.6 (501) | 21.5 (12.5–34.5) | 8 (40) |
| G/G | 4.8 (83) | 23 (14–35) | 7.2 (6) | 6.2 (74) | 22 (13–42.5) | 10.8 (8) |
| P value† | — | 0.03 | 0.021 (0.049) | — | 0.70 | 0.657 (0.99) |
| PAI12219 | ||||||
| C/C | 19.8 (339) | 22 (13–43) | 8.3 (28) | 24.4 (293) | 21 (12–36) | 9.6 (28) |
| C/T | 8.9 (835) | 23 (13–38) | 10.2 (85) | 46.9 (562) | 21 (12–35) | 8 (45) |
| T/T | 31.3 (535) | 20 (11–13) | 6.9 (37) | 28.7 (344) | 21 (13–37) | 9 (31) |
| P value† | — | 0.004 | 0.106 (0.171) | — | 0.78 | 0.721 (0.990) |
| PAI12750 | ||||||
| A/A | 4.5 (75) | 14.5 (9–26) | 10.7 (8) | 0.3 | 56 (17.5–111.5) | 50 (2) |
| A/G | 31.4 (521) | 23 (14–39) | 9.4 (49) | 8.4 | 27 (16–47) | 6.1 (6) |
| G/G | 64.1 (1,062) | 21 (13–37) | 8.6 (91) | 91.3 | 21 (12–35) | 9.5 (97) |
| P value† | — | 0.0004 | 0.743 (0.863) | — | 0.003 | 0.009 (0.066) |
Definition of abbreviations: CAP = community-acquired pneumonia; PAI-1 = plasminogen activator inhibitor-1; SNP = single nucleotide polymorphism.
Genotype data were missing for some patients.
Median PAI-1 concentrations (with interquartile range) in ng/ml.
P values adjusted for false-discovery rate are presented in parenthesis.
In contrast to white participants, only genotypes at the PAI12750 site were associated with CAP susceptibility. Subjects with the AA genotype were associated with the highest frequency of CAP (50%) compared with AG and GG genotype (6.1 and 9.5%, P = 0.01), but only 0.3% of black participants had the AA genotype.
PAI-1 SNPs and Circulating PAI-1 Concentrations
In general, genotypes associated with higher risk of CAP among whites had higher circulating PAI-1 concentrations (Table 5). For example, those with the 4G/4G and 4G/5G genotypes had higher PAI-1 concentrations compared with the 5G/5G genotypes (23 vs. 20 ng/ml, P < 0.0001). Similarly, subjects with the AA genotypes at the PAI7343 site had higher circulating concentrations compared with the AG and GG genotypes (22 vs. 20 ng/ml, P = 0.02).
Population Stratification
We stratified our analyses by site to assess for population stratification (Table 6). Again, genotypes associated with higher risk of CAP at Memphis and Pittsburgh sites were associated with higher circulating PAI-1 concentrations. Although the direction of association was similar at both sites, the magnitude varied. For instance, genotypes associated with higher circulating PAI-1 concentrations at the PAI4G,5G and PAI12219 sites were associated with higher risk of CAP at both sites, but the associations were statistically significant only at the Memphis site. Similarly, genotypes associated with higher circulating concentrations at the PAI2846 and PAI7343 sites were associated with higher risk of CAP at both sites, but the associations were statistically significant at the Pittsburgh site only.
TABLE 6.
ASSOCIATION BETWEEN GENOTYPE AND PHENOTYPE (PAI-1 AND COMMUNITY-ACQUIRED PNEUMONIA) IN WHITES STRATIFIED BY SITE
| Memphis (n = 935)
|
Pittsburgh (n = 859)
|
||||||
|---|---|---|---|---|---|---|---|
| Locus | Genotypes | Risk (%) for CAP | P Value | PAI-1 Concentration (ng/ml) | Risk (%) for CAP | P Value | PAI-1 Concentration (ng/ml) |
| 4G,5G | 4G,4G 4G,5G | 10.5 | 0.0006 | 19 | 9.2* | 0.6 | 26 |
| 5G,5G | 2.6 | 16 | 7.9 | 23 | |||
| 2846 | AA AG | 5.9 | 0.11 | 17 | 4.4 | 0.015 | 23 |
| GG | 9.6 | 19 | 10.3 | 26 | |||
| 2852 | AA AG | 0 | 0.18 | 13.5 | 16.1 | 0.17 | 22 |
| GG | 8.8 | 19 | 8.8 | 25 | |||
| 4588 | TT CT | 8.8 | 0.72 | 16 | 9.9 | 0.48 | 26 |
| CC | 8 | 19 | 8.2 | 25 | |||
| 7343 | GG AG | 6.6 | 0.09 | 17 | 6.4 | 0.026 | 23 |
| AA | 10 | 19 | 11 | 26 | |||
| 12219 | CC CT | 10.3 | 0.004 | 20 | 9* | 0.76 | 26 |
| TT | 4.6 | 17 | 9.6 | 23 | |||
| 12750 | AA AG | 8.8 | 0.93 | 20.5 | 10.4 | 0.38 | 25 |
| GG | 8.6 | 17 | 8.5 | 25 | |||
Definition of abbreviations: CAP = community-acquired pneumonia; PAI-1 = plasminogen activator inhibitor-1.
Interaction P < 0.05 between genotype and site for CAP susceptibility.
PAI-1 SNPs and Cytokine Expression in Ex Vivo Whole Blood Stimulation
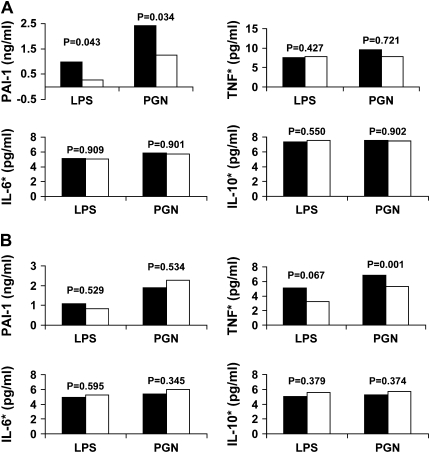
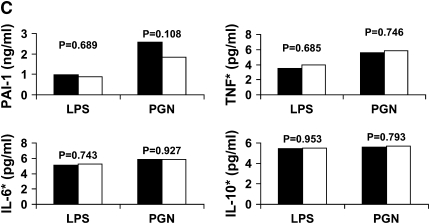
We assessed the functional significance of PAI4G,5G, PAI2846, and PAI7343 genotypes using ex vivo whole blood stimulation with LPS and PGN in 23 healthy white volunteers because these genotypes were associated with CAP susceptibility among white participants (Figure 2). Only the PAI4G,5G polymorphism was associated with increased PAI-1 expression. Stimulation of the whole blood with LPS and PGN resulted in a 3.3- and 1.9-fold higher PAI-1 expression when 4G/4G and 4G/5G genotypes were compared with 4G/4G genotypes at the PAI 4G,5G site (1 vs. 0.3 ng/ml, P = 0.043, and 2.4 vs. 1.3 ng/ml, P = 0.034, for LPS and PGN, respectively) (Figure 2A). However, no differences were seen in TNF, IL-6, and IL-10 expression for the PAI4G,5G genotypes. Stimulation with PGN resulted in higher TNF expression for the AA and AG genotypes when compared with the GG genotypes at PAI2846 site (P = 0.001), but no differences were seen in TNF expression after LPS stimulation and in PAI-1, IL-6, and IL-10 expression after both LPS and PGN stimulation (Figure 2B). No associations were seen between PAI7343 genotypes and either cytokine expression after LPS and PGN stimulation (Figure 2C).
Figure 2.
Changes in plasminogen activator inhibitor (PAI)-1, tumor necrosis factor (TNF), IL-6, and IL-10 concentrations after ex vivo whole blood stimulation with lipopolysaccharide (LPS) and peptidoglycan (PGN) for PAI4G,5G (A), PAI2846 (B), and PAI7343 (C) genotypes. Values for TNF, IL-6, and IL-10 are log converted. PAI4G,5G and PAI7343 genotype data were missing for two subjects and one subject. *Log-converted cytokine concentrations. (A) Solid bars, 4G/4G, 4G/5G (n = 16); open bars, 5G/5G (n = 5). (B) Solid bars, AG, AA (n = 4); open bars, GG (n = 19). (C) Solid bars, AA (n = 14); open bars AG or GG (n = 8).
PAI-1 Haplotypes and Phenotypes
Although we genotyped all seven SNPs in all subjects, the PAI4G,5G, PAI2846, PAI4588, and PAI 7343 SNPs could tag the five common haplotypes (A, C, D, E, F) in whites. Of the 53% of white participants with a 4G allele, 46% had the 4G/G/C/A haplotype, which was associated with higher odds of CAP (P = 0.004) (Table 7). Of the remaining 47% of white participants with the 5G allele, only the 5G/A/C/G haplotype was associated with a lower frequency of CAP and this haplotype was only observed among 10.7% of white participants. Higher probability of the 5G/A/C/G haplotype was associated with lower risk of CAP (P = 0.004). However, higher probability of the 4G/G/C/A haplotype was not associated with higher risk of CAP (P = 0.06). A combination of three SNPs could tag the four common haplotypes (A, B, C, H) with a frequency greater than 5% in blacks (PAI4G,5G/PAI2846/PAI12750) (Table 7), but neither haplotype was associated with CAP susceptibility (P = 0.33).
TABLE 7.
ASSOCIATION BETWEEN COMMUNITY-ACQUIRED PNEUMONIA REQUIRING HOSPITALIZATION AND HAPLOTYPES BASED ON FOUR POLYMORPHISMS AMONG WHITE AND THREE POLYMORPHISMS AMONG BLACK PARTICIPANTS
| Haplotypes
|
Frequency of Haplotypes
|
|||||||
|---|---|---|---|---|---|---|---|---|
| PAI4G,5G | PAI2846 | PAI4588 | PAI7343 | PAI12750 | Overall | CAP Participants | Participants without CAP | P Value |
| Whites* | ||||||||
| 5G | A | C | G | 10.7 | 6.5 | 11.2 | 0.01 | |
| 5G | G | C | A | 18.9 | 19.0 | 18.9 | 0.93 | |
| 5G | G | C | G | 9.7 | 9.2 | 9.7 | 0.77 | |
| 5G | G | T | A | 6.3 | 6.2 | 6.3 | 0.99 | |
| 4G | G | C | A | 46.4 | 53.4 | 45.7 | 0.004 | |
| 4G | G | T | A | 6.6 | 5.2 | 6.7 | 0.40 | |
| Blacks† | ||||||||
| 5G | G | G | 65.8 | 64.0 | 65.9 | 0.59 | ||
| 4G | G | G | 27.5 | 27.6 | 27.4 | 0.93 | ||
Definition of abbreviation: CAP = community-acquired pneumonia.
A total of 1,562 participants had genotype data for the four single nucleotide polymorphisms in whites and 1,116 participants had genotype data for the three single nucleotide polymorphisms in blacks.
Exact P value for marker trait association was 0.01.
Exact P value for marker trait association was 0.33.
Magnitude of Association between SNPs and Haplotypes with Phenotypes
In the Health ABC cohort, the PAI4G,5G alleles described only 1 and 0.3% of variation in the circulating PAI-1 concentrations measured in the healthy state in whites and blacks. Furthermore, the 4G/G/C/A and 5G/A/C/G haplotypes, associated with CAP susceptibility in whites, could explain only 1 and 0.1% of the variation in circulating PAI-1 concentrations measured in the healthy state. However, PAI4G,5G alleles described 20 and 22% of variation in PAI-1 expression after stimulation with LPS and PGN in healthy white volunteers. Finally, 43% of CAP cases among white subjects in the Health ABC cohort could be attributed to the 4G,5G alleles.
Sensitivity Analyses
We conducted sensitivity analyses to assess associations between the PAI4G,5G SNP and the haplotypes with CAP among whites after excluding CAP events before enrollment in the Health ABC study. One hundred and ninety-eight participants (6.4%) had at least one hospitalization for CAP after enrollment in the Health ABC study: 126 were whites and 72 were blacks. Again, a codominant effect for the presence of a 4G allele at the PAI4G,5G site was seen in white participants. The OR for the combined 4G/4G and 4G/5G genotypes versus the 5G/5G genotype was similar to that when including all known cases in the analysis. Results did not reach statistical significance in univariate analysis (relative risk [RR], 1.6; 95% CI, 0.97–2.7; P = 0.069) but were significant in multivariable analysis (RR, 1.8; 95% CI, 1.03–3.2; P = 0.04). Haplotype analysis showed that the 4G/G/C/A haplotype was associated with lower odds of CAP susceptibility (P = 0.007), whereas the 5G/A/C/G haplotype approached, but did not reach, statistical significance (P = 0.06) (Table E3A).
DISCUSSION
Consistent with our hypothesis, we demonstrated an association between genotypes associated with higher circulating PAI-1 concentrations at PAI4G,5G, PAI2846, and PAI7343 sites and increased odds of CAP requiring hospitalization in well-functioning elderly whites. Only the previously functional PAI4G,5G genotype was associated with higher PAI expression during whole blood stimulation. Half of white subjects had the hypersecretor 4G allele, and most had the 4G/G/C/A haplotype at PAI4G,5G/PAI2846/PAI4588/PAI7343 SNPs, which was associated with increased CAP susceptibility. Of the subjects with the hyposecretor 5G allele, only 11% had the 5G/A/C/G haplotype, which was associated with lower CAP susceptibility. These associations were independent of demographic characteristics and comorbid conditions. In contrast, among blacks, the PAI4G,5G polymorphism was also associated with higher circulating PAI-1 concentrations, but neither this polymorphism nor any of the haplotypes were associated with CAP susceptibility.
To our knowledge, this is the first report describing the role of PAI-1 in pneumonia susceptibility in humans. In contrast to a previous study that examined PAI-1 gene–deficient mice (27), our results suggest that genetic variation associated with increased PAI-1 expression increases susceptibility. The differences in PAI-1 concentrations between the groups were small and the PAI4G,5G genotypes explained only 1% of variation in the circulating PAI-1 concentrations. Genotyping additional SNPs did not explain additional variability in circulating PAI-1 concentrations or alter the OR of CAP susceptibility compared with the PAI4G,5G SNP alone in whites. Our results are consistent with recent studies in which the 4G,5G alleles and more comprehensive genotyping techniques, such as high-density SNP analysis of the PAI-1 gene (28, 29), explained only a small fraction of variation in circulating PAI-1 concentrations. Furthermore, similar results have been observed when comprehensive methods were used to ascertain contribution of genetic variation to circulating concentrations of other inflammatory markers, including C-reactive protein (30). In contrast to the association between these alleles and PAI-1 concentrations measured in a healthy state, 20% of variation in PAI-1 expression could be attributed to the 4G,5G allele in whole blood stimulation assay. These results are consistent with circulating concentrations for the 4G,5G alleles observed during an acute infection (14). In this study, PAI-1 concentrations were twofold higher in subjects with the 4G/4G genotype compared with the 5G/5G genotype. Therefore, the PAI 4G,5G SNP may contribute to greater variability in PAI-1 expression on exposure to infection compared with a healthy state.
We speculate that increased PAI-1 expression among participants with the 4G allele may dampen the proinflammatory activity of uPA (3, 31–36). Our results also suggest that increased PAI-1 concentrations in the elderly could be a marker of high-risk PAI-1 genotypes and haplotypes and increased risk of CAP. Therefore, our findings have important implications in the frail elderly who have up-regulated markers of inflammation and coagulation and are at higher risk for infection (37). Furthermore, our results suggest that, although increased PAI-1 expression is associated with chronic health conditions that increase risk of CAP, such as diabetes (38), PAI-1 expression during host response to infection may independently influence CAP susceptibility in the elderly.
An important strength of our study is the cohort design compared with earlier case-control designs (39, 40). Designing studies to ascertain the role of genetic variants in susceptibility to infection poses several challenges. First, an ideal study design would be a prospective data collection with lifetime follow-up of a large cohort to identify cases and control subjects. Because this is often impractical, we used a cohort of 70- to 79-year-old participants because the likelihood of developing CAP increases exponentially in this age group (41). We also included events in the 5 years preceding enrollment in addition to the 7 years of follow-up in the Health ABC study to better classify cases and noncases. Although we cannot exclude CAP in our control subjects before this period, the incidence is low in individuals younger than 65 years. Second, we had to use two definitions to identify CAP: the ICD-9 diagnoses using HCFA data during the initial 5 years, and a more precise method during the ensuing 7 years. Although ICD-9 diagnosis is sensitive in detecting CAP events, it lacks specificity (42). Our sensitivity analysis demonstrates that ORs for the reported associations remained unchanged after excluding events before Health ABC enrollment, but some of these associations did not meet statistical significance due to reduced sample size. Third, a potential problem of including cases occurring before the assembly of the cohort is selection bias from a variety of potential sources, including differential mortality. Results can be biased if genotype determines the probability that individuals would be included in the study at the outset. PAI-1 genotypes were not associated with mortality after CAP in whites, and the frequency of the 4G,5G genotypes was similar across different age ranges in our cohort (Table E4A). Therefore, selection bias is possible, but unlikely to affect our results. An additional strength of our study is its generalizability to other well-functioning populations in the United States. The cohort was randomly chosen from two urban areas in the United States, and the incidence of CAP in our cohort is similar to other population-based studies (41, 43). The genotype and haplotype frequencies are consistent with previous studies in blacks and whites, including recently published high-density SNP map analysis of the PAI-1 gene (28, 29, 44, 45).
The lack of association of the 4G,5G genotype and CAP susceptibility among blacks and the results of our haplotype analysis in whites suggests that additional polymorphisms within the PAI-1 gene may regulate PAI-1 expression. We tried to identify these additional genetic variants while choosing candidate SNPs. Our approach was to identify functional or potentially functional SNPs with an allele frequency of greater than 10%. However, none of the nonsynonymous SNPs in our study are likely to be functional. The PAI2846 SNP was associated with both PAI-1 concentrations and susceptibility to CAP, and the 5G/A/C/G haplotype was associated with lower odds of CAP susceptibility in whites. However, this SNP was neither associated with PAI-1 expression after whole blood stimulation in whites nor associated with PAI-1 concentrations or CAP susceptibility in blacks in our study. It was also not associated with PAI-1 concentrations among whites in a previous report (29). Another potential explanation for the lack of association between haplotypes and CAP susceptibility among blacks in our study could be differences in haplotype frequencies. Our findings corroborate previous reports demonstrating that blacks have fewer common haplotypes compared with whites (46) and rare haplotypes may be more important in CAP susceptibility among blacks. Our power calculations suggest that our study could detect modest effects (OR ⩾ 2), as seen with the 4G allele in whites. Our study was not powered to detect ORs less than 2, especially for uncommon haplotypes that may play an important role (47). Therefore, associations between less frequent haplotypes and CAP susceptibility among blacks could not be ascertained in the present study. The population-attributable risk in detecting smaller effects of gene variants, especially for rare alleles, is presently not clear (48).
Our study has limitations. First, we used candidate SNPs from the SeattleSNPs website. Using SNPs from the HapMap project or more comprehensive methods to identify tag SNPs, including high-density SNP analysis, may yield different results. Our study was designed in 2002, before initiation of the HapMap project and therefore these SNPs were not included in our study. Second, it is possible that the reported association is due to linkage with other polymorphisms on chromosome 7q. Replication of our results is other population-based cohorts should be conducted to confirm our results.
In conclusion, PAI-1 genotypes associated with increased circulating PAI-1 concentrations are associated with increased susceptibility to CAP in well-functioning elderly men and women. Our results suggest that genetic variants in addition to the commonly described 4G,5G functional polymorphism may regulate PAI-1 expression and CAP susceptibility.
Supplementary Material
Supported by the National Institute on Aging (N01-AG-6-2101, N01-AG-6-2103, N01-AG-6-2106), National Heart, Lung, and Blood Institute (R01HL74104), National Institute of General Medical Sciences (R01 GM61992), and an internal Seed grant from the Department of Critical Care Medicine at the University of Pittsburgh.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200605-644OC on August 29, 2007
Conflict of Interest Statement: S.Y. has no financial relationship with a commercial entity that has an interest in the subject of this manuscript. D.C.A. has no financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.D. has no financial relationship with a commercial entity that has an interest in the subject of this manuscript. A.B.N. has no financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.A.K. has no financial relationship with a commercial entity that has an interest in the subject of this manuscript. R.L. has no financial relationship with a commercial entity that has an interest in the subject of this manuscript. R.E.F. has no financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.Z. has no financial relationship with a commercial entity that has an interest in the subject of this manuscript. S.B.K. has no financial relationship with a commercial entity that has an interest in the subject of this manuscript. T.B.H. has no financial relationship with a commercial entity that has an interest in the subject of this manuscript. M.G. has no financial relationship with a commercial entity that has an interest in the subject of this manuscript. K.Y. has no financial relationship with a commercial entity that has an interest in the subject of this manuscript. R.G.W. has received investigator-initiated research grants from Eli Lilly & Co. and Chiron, Inc., for genetic association studies of coagulation and inflammatory genes.
References
- 1.Horrevoets AJ. Plasminogen activator inhibitor 1 (PAI-1): in vitro activities and clinical relevance. Br J Haematol 2004;125:12–23. [DOI] [PubMed] [Google Scholar]
- 2.Abraham E. Coagulation abnormalities in acute lung injury and sepsis. Am J Respir Cell Mol Biol 2000;22:401–404. [DOI] [PubMed] [Google Scholar]
- 3.Gyetko MR, Chen GH, McDonald RA, Goodman R, Huffnagle GB, Wilkinson CC, Fuller JA, Toews GB. Urokinase is required for the pulmonary inflammatory response to Cryptococcus neoformans: a murine transgenic model. J Clin Invest 1996;97:1818–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi G, Schultz MJ, van Till JW, Bresser P, Van Der Zee JS, Boermeester MA, Levi M, van der Poll T. Disturbed alveolar fibrin turnover during pneumonia is restricted to the site of infection. Eur Respir J 2004;24:786–789. [DOI] [PubMed] [Google Scholar]
- 5.Prabhakaran P, Ware LB, White KE, Cross MT, Matthay MA, Olman MA. Elevated levels of plasminogen activator inhibitor-1 in pulmonary edema fluid are associated with mortality in acute lung injury. Am J Physiol Lung Cell Mol Physiol 2003;285:L20–L28. [DOI] [PubMed] [Google Scholar]
- 6.El Solh AA, Bhora M, Pineda L, Aquilina A, Abbetessa L, Berbary E. Alveolar plasminogen activator inhibitor-1 predicts ARDS in aspiration pneumonitis. Intensive Care Med 2006;32:110–115. [DOI] [PubMed] [Google Scholar]
- 7.Sawdey MS, Loskutoff DJ. Regulation of murine type 1 plasminogen activator inhibitor gene expression in vivo: tissue specificity and induction by lipopolysaccharide, tumor necrosis factor-alpha, and transforming growth factor-beta. J Clin Invest 1991;88:1346–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aillaud MF, Pignol F, Alessi MC, Harle JR, Escande M, Mongin M, Juhan-Vague I. Increase in plasma concentration of plasminogen activator inhibitor, fibrinogen, von Willebrand factor, factor VIII:C and in erythrocyte sedimentation rate with age. Thromb Haemost 1986;55:330–332. [PubMed] [Google Scholar]
- 9.Hashimoto Y, Kobayashi A, Yamazaki N, Sugawara Y, Takada Y, Takada A. Relationship between age and plasma t-PA, PA-inhibitor, and PA activity. Thromb Res 1987;46:625–633. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto K, Shimokawa T, Yi H, Isobe K, Kojima T, Loskutoff DJ, Saito H. Aging accelerates endotoxin-induced thrombosis: increased responses of plasminogen activator inhibitor-1 and lipopolysaccharide signaling with aging. Am J Pathol 2002;161:1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson SJ, Wiman B, Hamsten A, Green F, Humphries S, Henney AM. The two allele sequences of a common polymorphism in the promoter of the plasminogen activator inhibitor-1 (PAI-1) gene respond differently to interleukin-1 in HepG2 cells. J Biol Chem 1993;268:10739–10745. [PubMed] [Google Scholar]
- 12.Eriksson P, Kallin B, van't Hooft FM, Bavenholm P, Hamsten A. Allele-specific increase in basal transcription of the plasminogen-activator inhibitor 1 gene is associated with myocardial infarction. Proc Natl Acad Sci USA 1995;92:1851–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westendorp RG, Hottenga JJ, Slagboom PE. Variation in plasminogen-activator-inhibitor-1 gene and risk of meningococcal septic shock. Lancet 1999;354:561–563. [DOI] [PubMed] [Google Scholar]
- 14.Hermans PW, Hibberd ML, Booy R, Daramola O, Hazelzet JA, de Groot R, Levin M. 4G/5G promoter polymorphism in the plasminogen-activator-inhibitor-1 gene and outcome of meningococcal disease. Meningococcal Research Group. Lancet 1999;354:556–560. [DOI] [PubMed] [Google Scholar]
- 15.Kim KK, Flaherty KR, Long Q, Hattori N, Sisson TH, Colby TV, Travis WD, Martinez FJ, Murray S, Simon RH. A plasminogen activator inhibitor-1 promoter polymorphism and idiopathic interstitial pneumonia. Mol Med 2003;9:52–56. [PMC free article] [PubMed] [Google Scholar]
- 16.Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, Hauser SL, Smith MW, O'Brien SJ, Altshuler D, et al. Methods for high-density admixture mapping of disease genes. Am J Hum Genet 2004;74:979–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med 1990;9:811–818. [DOI] [PubMed] [Google Scholar]
- 18.Sebastiani P, Lazarus R, Weiss ST, Kunkel LM, Kohane IS, Ramoni MF. Minimal haplotype tagging. Proc Natl Acad Sci USA 2003;100:9900–9905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao JH, Curtis D, Sham PC. Model-free analysis and permutation tests for allelic associations. Hum Hered 2000;50:133–139. [DOI] [PubMed] [Google Scholar]
- 20.Yende S, Tuomanen EI, Wunderink RG, Kanaya A, Newman AB, Harris T, de Rekeneire N, Kritchevsky SB. Preinfection systemic inflammatory markers and risk of hospitalization due to pneumonia. Am J Respir Crit Care Med 2005;172:1440–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jokinen C, Heiskanen L, Juvonen H, Kallinen S, Karkola K, Korppi M, Kurki S, Ronnberg PR, Seppa A, Soimakallio S. Incidence of community-acquired pneumonia in the population of four municipalities in eastern Finland. Am J Epidemiol 1993;137:977–988. [DOI] [PubMed] [Google Scholar]
- 22.LaCroix AZ, Lipson S, Miles TP, White L. Prospective study of pneumonia hospitalizations and mortality of US older people: the role of chronic conditions, health behaviors, and nutritional status. Public Health Rep 1989;104:350–360. [PMC free article] [PubMed] [Google Scholar]
- 23.Baik I, Curhan GC, Rimm EB, Bendich A, Willett WC, Fawzi WW. A prospective study of age and lifestyle factors in relation to community-acquired pneumonia in US men and women. Arch Intern Med 2000;160:3082–3088. [DOI] [PubMed] [Google Scholar]
- 24.Koivula I, Sten M, Makela PH. Risk factors for pneumonia in the elderly. Am J Med 1994;96:313–320. [DOI] [PubMed] [Google Scholar]
- 25.Zaykin DV, Westfall PH, Young SS, Karnoub MA, Wagner MJ, Ehm MG. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum Hered 2002;53:79–91. [DOI] [PubMed] [Google Scholar]
- 26.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 2003;19:149–150. [DOI] [PubMed] [Google Scholar]
- 27.Rijneveld AW, Florquin S, Bresser P, Levi M, De Waard V, Lijnen R, Van Der Zee JS, Speelman P, Carmeliet P, van der Poll T. Plasminogen activator inhibitor type-1 deficiency does not influence the outcome of murine pneumococcal pneumonia. Blood 2003;102:934–939. [DOI] [PubMed] [Google Scholar]
- 28.Festa A, D'Agostino R Jr, Rich SS, Jenny NS, Tracy RP, Haffner SM. Promoter (4G/5G) plasminogen activator inhibitor-1 genotype and plasminogen activator inhibitor-1 levels in blacks, Hispanics, and non-Hispanic whites: the Insulin Resistance Atherosclerosis Study. Circulation 2003;107:2422–2427. [DOI] [PubMed] [Google Scholar]
- 29.Kathiresan S, Gabriel SB, Yang Q, Lochner AL, Larson MG, Levy D, Tofler GH, Hirschhorn JN, O'Donnell CJ. Comprehensive survey of common genetic variation at the plasminogen activator inhibitor-1 locus and relations to circulating plasminogen activator inhibitor-1 levels. Circulation 2005;112:1728–1735. [DOI] [PubMed] [Google Scholar]
- 30.Carlson CS, Aldred SF, Lee PK, Tracy RP, Schwartz SM, Rieder M, Liu K, Williams OD, Iribarren C, Lewis EC, et al. Polymorphisms within the C-reactive protein (CRP) promoter region are associated with plasma CRP levels. Am J Hum Genet 2005;77:64–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gyetko MR, Todd RF III, Wilkinson CC, Sitrin RG. The urokinase receptor is required for human monocyte chemotaxis in vitro. J Clin Invest 1994;93:1380–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sitrin RG, Shollenberger SB, Strieter RM, Gyetko MR. Endogenously produced urokinase amplifies tumor necrosis factor-alpha secretion by THP-1 mononuclear phagocytes. J Leukoc Biol 1996;59:302–311. [DOI] [PubMed] [Google Scholar]
- 33.Beck JM, Preston AM, Gyetko MR. Urokinase-type plasminogen activator in inflammatory cell recruitment and host defense against Pneumocystis carinii in mice. Infect Immun 1999;67:879–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gyetko MR, Sud S, Chen GH, Fuller JA, Chensue SW, Toews GB. Urokinase-type plasminogen activator is required for the generation of a type 1 immune response to pulmonary Cryptococcus neoformans infection. J Immunol 2002;168:801–809. [DOI] [PubMed] [Google Scholar]
- 35.Gyetko MR, Sud S, Chensue SW. Urokinase-deficient mice fail to generate a type 2 immune response following schistosomal antigen challenge. Infect Immun 2004;72:461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gyetko MR, Aizenberg D, Mayo-Bond L. Urokinase-deficient and urokinase receptor-deficient mice have impaired neutrophil antimicrobial activation in vitro. J Leukoc Biol 2004;76:648–656. [DOI] [PubMed] [Google Scholar]
- 37.Fried LP, Hadley EC, Walston JD, Newman A, Guralnik JM, Studenski S, Harris TB, Ershler WB, Ferrucci L. From bedside to bench: research agenda for frailty. Sci Aging Knowledge Environ 2005;2005:e24. [DOI] [PubMed] [Google Scholar]
- 38.Kanaya AM, Wassel Fyr C, Vittinghoff E, Harris TB, Park SW, Goodpaster BH, Tylavsky F, Cummings SR. Adipocytokines and incident diabetes mellitus in older adults: the independent effect of plasminogen activator inhibitor 1. Arch Intern Med 2006;166:350–356. [DOI] [PubMed] [Google Scholar]
- 39.Gallagher PM, Lowe G, Fitzgerald T, Bella A, Greene CM, McElvaney NG, O'Neill SJ. Association of IL-10 polymorphism with severity of illness in community acquired pneumonia. Thorax 2003;58:154–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waterer GW, Quasney MW, Cantor RM, Wunderink RG. Septic shock and respiratory failure in community-acquired pneumonia have different TNF polymorphism associations. Am J Respir Crit Care Med 2001;163:1599–1604. [DOI] [PubMed] [Google Scholar]
- 41.Fry AM, Shay DK, Holman RC, Curns AT, Anderson LJ. Trends in hospitalizations for pneumonia among persons aged 65 years or older in the United States, 1988–2002. JAMA 2005;294:2712–2719. [DOI] [PubMed] [Google Scholar]
- 42.Guevara RE, Butler JC, Marston BJ, Plouffe JF, File TM Jr, Breiman RF. Accuracy of ICD-9-CM codes in detecting community-acquired pneumococcal pneumonia for incidence and vaccine efficacy studies. Am J Epidemiol 1999;149:282–289. [DOI] [PubMed] [Google Scholar]
- 43.O'Meara ES, White M, Siscovick DS, Lyles MF, Kuller LH. Hospitalization for pneumonia in the Cardiovascular Health Study: incidence, mortality, and influence on longer-term survival. J Am Geriatr Soc 2005;53:1108–1116. [DOI] [PubMed] [Google Scholar]
- 44.Bensen JT, Hsu FC, Brown WM, Sutton BS, Norris JM, Tracy RP, Jenny NS, Saad MF, Haffner S, Bowden DW, et al. Association analysis of the plasminogen activator inhibitor-1 4G/5G polymorphism in Hispanics and African Americans: the IRAS family study. Hum Hered 2004;57:128–137. [DOI] [PubMed] [Google Scholar]
- 45.Hooper WC, Lally C, Austin H, Renshaw M, Dilley A, Wenger NK, Phillips DJ, Whitsett C, Rawlins P, Evatt BL. The role of the t-PA I/D and PAI-1 4G/5G polymorphisms in African-American adults with a diagnosis of myocardial infarction or venous thromboembolism. Thromb Res 2000;99:223–230. [DOI] [PubMed] [Google Scholar]
- 46.Crawford DC, Carlson CS, Rieder MJ, Carrington DP, Yi Q, Smith JD, Eberle MA, Kruglyak L, Nickerson DA. Haplotype diversity across 100 candidate genes for inflammation, lipid metabolism, and blood pressure regulation in two populations. Am J Hum Genet 2004;74:610–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu PY, Zhang YY, Lu Y, Long JR, Shen H, Zhao LJ, Xu FH, Xiao P, Xiong DH, Liu YJ, et al. A survey of haplotype variants at several disease candidate genes: the importance of rare variants for complex diseases. J Med Genet 2005;42:221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Risch NJ. Searching for genetic determinants in the new millennium. Nature 2000;405:847–856. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.