Abstract
The European medicinal leech is one of vanishingly few animal species with direct application in modern medicine. In addition to the therapeutic potential held by many protease inhibitors purified from leech saliva, and notwithstanding the historical association with quackery, Hirudo medicinalis has been approved by the United States Food and Drug Administration as a prescription medical device. Accurate annotation of bioactive compounds relies on precise species determination. Interpretations of developmental and neurophysiological characteristics also presuppose uniformity within a model species used in laboratory settings. Here, we show, with mitochondrial sequences and nuclear microsatellites, that there are at least three species of European medicinal leech, and that leeches marketed as H. medicinalis are actually Hirudo verbana. Beyond the obvious need for reconsideration of decades of biomedical research on this widely used model organism, these findings impact regulatory statutes and raise concerns for the conservation status of European medicinal leeches.
Keywords: Annelida, anticoagulants, biodiversity, conservation biology, DNA barcoding, neurobiology
1. Introduction
Medicinal leeches have been facilitators of phlebotomy since the time of Galen with their use for balancing humours peaking during the post-Napoleonic era when tens of millions were employed across Europe (Moquin-Tandon 1846; Harding & Moore 1927; Elliott & Tullett 1992). At that time, the annual leech harvest was sufficiently intense to stimulate some of the earliest legislative efforts at biological conservation (Elliott & Tullett 1992). Long since repudiated as suitable devices for the treatment of conditions ranging from obesity to pneumonitis (Louis 1828), these much maligned invertebrates are again seeing wide use as biomedical tools. European medicinal leeches returned to the modern clinical toolbox in the context of flap and replantation surgery (Derganc & Zdravic 1960; Foucher et al. 1981). Specifically, with artery-only replantation (e.g. of digits), free tissue transfer surgery (e.g. skin flaps) and cases of venous obstruction, the post-operative use of leeches to ameliorate congestion improves prognosis and tissue survival in a cost-effective and reliable manner that has yet to be supplanted by mechanical or pharmaceutical alternatives (Foucher et al. 1981; Chepeha et al. 2002; Graf et al. 2006). Their potential as living apothecaries has stimulated the isolation of scores of bioactive compounds including important anticoagulants, antistasins and other protease inhibitors (Sollner et al. 1994; Baskova & Zavalova 2001; Salzet 2002). Leeches also have been favoured model organisms in developmental genetics and in neurobiology in light of their large, easily manipulated neurons, complex behaviours and integrated locomotion (Kristan et al. 2005). More recently, they have become a compelling research model for enteric symbioses (Graf et al. 2006). In addition to stimulating lucrative leech farming industries, an enviable status was achieved in 2004 with United States Food and Drug Administration (US FDA) approval to market the European medicinal leech, Hirudo medicinalis, as a prescription medical device (Rados 2004)—provided that accurate labelling and branding regulations were followed.
Hirudo medicinalis, described by Linnaeus in 1758, has long been considered the sole European medicinal leech (Sawyer 1986). Yet, by 1827, at least five additional species were recognized (Moquin-Tandon 1827), the varieties of which even Darwin (1859) noted ‘cannot be kept together’. The lack of clear internal anatomical differences has been an impediment to accurately delimiting leech species and taxonomic descriptions often were predicated on subtle variations in colour pattern later considered too variable to be reliable (Moquin-Tandon 1846). In the context of identifying wild and commercially marketed medicinal leeches, we provide DNA barcoding data and, independent of the barcoding locus, nuclear microsatellite data regarding multiple species of Hirudo in comparison with commercially available leeches.
2. Material and methods
Wild European medicinal leeches were collected from France, Italy, Slovenia, Croatia, Ukraine and Macedonia (table 1 and see Trontelj et al. 2004). These included 13 wild H. medicinalis and 13 wild Hirudo verbana distinguished on the basis of colour patterns (figure 1). Commercially available leeches were obtained from four suppliers (Leeches USA Ltd, Westbury, NY, USA; Biopharm Leeches supplied through Carolina Biological Supply Company, Burlington, NC, USA; Ward's Natural Science, Rochester, NY, USA; and Biopharm Leeches (UK), Hendy, Carmarthenshire, UK) as well as from the laboratories of Joerg Graf (University of Connecticut, Storrs, CT, USA) and Leathem Mehaffey (Vassar College, Poughkeepsie, NY, USA) wherein leeches are used as model organisms. Microsatellite libraries were enriched and screened using a protocol (Budinoff et al. 2004) modified from Dr Travis Glenn's group (Savannah River Ecology Laboratory, University of Georgia, Aiken, SC, USA). Twenty-nine clones contained microsatellite regions with flanking sequences appropriate for primer design. Primers were designed using MacVector v. 7.0 (Accelrys, San Diego, CA, USA). An M13 forward primer sequence was added to the 5′ end of each forward primer, following the protocol of Boutin-Ganache et al. (2001). Amplification reactions for microsatellite analyses were performed with genomic DNA from 13 specimens of H. medicinalis and 13 specimens of wild H. verbana using equal quantities of each microsatellite primer pair and a 6-FAM fluorescently labelled M13 forward primer. Reactions consisted of 1 μl genomic DNA, 0.5 μl of each 10 μM primer, 2.5 μl 10× PCR Buffer II (Amersham Pharmacia Biotech, Piscataway, NJ, USA), 2 μl of 10 μM dNTP mixture, 0.13 μl AmpliTaq Gold (Amersham Biosciences) and 16.5 μl H2O. Reactions were run at 94°C for 10 min, followed by 40 cycles of 94°C for 30 s, primer-specific annealing temperature (table 2) for 30 s and 72°C for 45 s. Reactions ended with a 72°C extension for 7 min.
Table 1.
Allelic profiles for European medicinal leeches.
| sample | locality | morphospecies | HvA10 | HvH07 | Hm8 | Hm10 | Hm12 | Hv351 | HvT379 | Hm1 | Hm2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| allele2 | allele1 | allele2 | allele1 | allele2 | allele1 | allele2 | allele1 | allele2 | allele1 | allele2 | allele1 | allele2 | allele1 | allele2 | allele1 | allele2 | allele1 | |||
| HM04 | Paimpont, France | H. medicinalis | — | — | 234 | 329 | 209 | 148 | 133 | 104 | 158 | |||||||||
| HR20 | Hrast, SE Slovenia | H. medicinalis | — | — | 231 | 329 | 209 | 150 | 133 | 104 | 158 | |||||||||
| HR21 | Hrast, SE Slovenia | H. medicinalis | — | — | 231 | 329 | 205 | 209 | 150 | 133 | 104 | 158 | ||||||||
| HR22 | Hrast, SE Slovenia | H. medicinalis | — | — | 231 | 329 | 209 | 150 | 133 | 104 | 107 | 158 | ||||||||
| X2 | Kharkiv, E Ukraine | H. medicinalis | — | — | 231 | 329 | 209 | 148 | 150 | 133 | 104 | 158 | ||||||||
| KO66 | Kocevje, S Slovenia | H. medicinalis | — | — | 231 | 329 | 209 | 150 | 168 | 133 | 104 | 158 | ||||||||
| KO67 | Kocevje, S Slovenia | H. medicinalis | — | — | 231 | 329 | 209 | 150 | 168 | 133 | 104 | 158 | ||||||||
| X3 | Leipzig, E Germany | H. medicinalis | — | — | 237 | 329 | 197 | 136 | 148 | 133 | 104 | 158 | ||||||||
| PO56 | Podvinci, NE Slovenia | H. medicinalis | — | — | 231 | 329 | 205 | 217 | 146 | 148 | 133 | 104 | 155 | |||||||
| PO58 | Podvinci, NE Slovenia | H. medicinalis | — | — | 231 | 329 | 205 | 148 | 150 | 133 | 104 | 158 | ||||||||
| PO59 | Podvinci, NE Slovenia | H. medicinalis | — | — | 231 | 234 | 329 | 205 | 209 | 146 | 168 | 133 | 104 | 158 | ||||||
| PO60 | Podvinci, NE Slovenia | H. medicinalis | — | — | 231 | 329 | 209 | 148 | 150 | 133 | 104 | 158 | ||||||||
| PO61 | Podvinci, NE Slovenia | H. medicinalis | — | — | 231 | 329 | 209 | 150 | 168 | 133 | 104 | 158 | ||||||||
| HM 97 | Paimpont, France | H. medicinalis | — | — | 234 | 329 | 209 | 146 | 168 | 133 | 110 | 158 | ||||||||
| C1 | S Italy | H. verbana | 321 | 348 | 174 | 177 | — | 316 | 319 | — | 140 | 136 | 138 | 92 | 95 | 158 | ||||
| KR40 | Gabrovica, SW Slovenia | H. verbana | 309 | 171 | — | 316 | 319 | — | 153 | 136 | 92 | 155 | ||||||||
| KR41 | Gabrovica, SW Slovenia | H. verbana | 309 | 171 | — | 316 | 319 | — | 188 | 136 | 92 | 155 | ||||||||
| PA54 | Island Pag, Croatia | H. verbana | 306 | 348 | 174 | 177 | — | 323 | — | 162 | 172 | 136 | 95 | 158 | ||||||
| PA55 | Island Pag, Croatia | H. verbana | 306 | 174 | — | 323 | — | 162 | 136 | 95 | 155 | 158 | ||||||||
| OH31 | Lake Ohrid, Macedonia | H. verbana | 309 | 174 | — | 319 | — | 166 | 136 | 92 | 101 | 155 | ||||||||
| OH32 | Lake Ohrid, Macedonia | H. verbana | 306 | 174 | — | 319 | — | 155 | 190 | 133 | 136 | 92 | 155 | |||||||
| OH33 | Lake Ohrid, Macedonia | H. verbana | 309 | 174 | 177 | — | 313 | — | 166 | 133 | 136 | 92 | 101 | 161 | ||||||
| OH34 | Lake Ohrid, Macedonia | H. verbana | 309 | 174 | 177 | — | 316 | — | 142 | 166 | 133 | 136 | 92 | 155 | 161 | |||||
| X1 | Lecce, S Italy | H. verbana | 306 | 174 | — | 313 | — | 140 | 166 | 136 | 92 | 95 | 158 | |||||||
| KA46 | Povir, SW Slovenia | H. verbana | 309 | 171 | 180 | — | 319 | — | 180 | 136 | 92 | 158 | ||||||||
| KA47 | Povir, SW Slovenia | H. verbana | 306 | 180 | — | 319 | — | 146 | 136 | 92 | 158 | |||||||||
| KA49 | Povir, SW Slovenia | H. verbana | 309 | 171 | 180 | — | 316 | 319 | — | 180 | 136 | 92 | 98 | 155 | ||||||
| Graf | Commercial Graf Lab | 306 | 177 | — | 316 | 329 | — | 170 | 172 | 136 | 92 | 161 | ||||||||
| Carolina | Commercial C. Biological | 306 | 183 | — | 316 | — | 170 | 138 | 92 | 155 | 161 | |||||||||
| HM LM 2 | Commercial Mehaffey Lab | 306 | 171 | 177 | — | 316 | — | 172 | 176 | 136 | 92 | 98 | 155 | 164 | ||||||
| HM UK | Commercial Biopharm UK | 306 | 183 | — | 313 | — | 170 | 138 | 92 | 164 | ||||||||||
| HM LM 1 | Commercial Mehaffey Lab | 306 | 171 | 177 | — | 313 | — | 170 | 180 | 136 | 92 | 155 | ||||||||
| Leeches USA | Commercial | 309 | 177 | 265 | 316 | 252 | 283 | 180 | 194 | 136 | 92 | — | ||||||||
| Leeches USA | Commercial | 306 | 183 | 274 | 316 | 223 | 248 | 172 | 184 | 139 | 92 | 161 | ||||||||
| Leeches USA | Commercial | 306 | 183 | 274 | 282 | 316 | 248 | 252 | 172 | 136 | 139 | 92 | 95 | — | ||||||
| Leeches USA | Commercial | 306 | 177 | 183 | 262 | 271 | 316 | 248 | 252 | 186 | 192 | 139 | 95 | 161 | ||||||
Figure 1.
Distinctive colour patterns for two species of European medicinal leech. (a) Hirudo medicinalis Linnaeus 1758. (b) Hirudo verbana Carena 1820.
Table 2.
Microsatellite loci, primers and annealing temperatures.
| locus | forward primera | reverse primer | annealing T (°C) |
|---|---|---|---|
| HvA10 | CACGACGTTGTAAAACGACGAAAAGGTCAAAGGTCACAACGAC | TTGCCCTGGATGCCATACAG | 50 |
| HvH07 | CACGACGTTGTAAAACGACCTCGTAAGAAATGGAAACTG | GATGTTGAAACCCGAAGC | 55 |
| Hm8 | CACGACGTTGTAAAACGACTGGATGATAATGCGTCGTCAGTC | TCTTTGGTTCACAGGGCTTAGTTG | 55 |
| Hm10 | CACGACGTTGTAAAACGACCTCTCCACAGCCTTATGGTGTTG | AGGATGAGGTGAAGCGGGGATG | 55 |
| Hm12 | CACGACGTTGTAAAACGACGCACCAAGCCAGAATAAGGTTAG | TCTCCACTTTCACTCCCGCTC | 50 |
| Hv351 | CACGACGTTGTAAAACGACCGCAATCTGCCTGAAAAAGTAAAC | CGAATGTGTTGGATGCCACTAAC | 50 |
| HvT379 | CACGACGTTGTAAAACGACATTGGGAACAAGAACAAGCC | CAGAAGGTGGAGAAGTGGTTG | 50 |
| Hm1 | CACGACGTTGTAAAACGACTCAGGCGACATCCTCTTCATCG | ATGGCTACCACTGCGTTGTTG | 50 |
| Hm2 | CACGACGTTGTAAAACGACAGGAACTCATCATTTATCTCCAGCC | ACGGAAACCACGGTCTCCAAGAG | 50 |
| HvD7 | CACGACGTTGTAAAACGACTCTACTTCCAGAACTCGTG | GCAGCCTTTACGGAGCGG | 50 |
The first 19 nt of each forward primer corresponds to a fluorescently labelled M13 primer used in the reaction.
Twenty primer sets were eliminated from consideration in light of mismatches between observed and expected amplification product sizes, inconsistent or weak amplification, hypervariability and lack of variability. Nine primers sets (table 2) were employed for this study for which all 26 wild leeches and 10 commercially obtained leeches were scored with GeneMapper (Chatterji & Pachter 2006). Microsatellite profiles were subjected to analyses of population structure with STRUCTURE (Pritchard et al. 2000) and analysis of molecular variance (AMOVA) with Arlequin (Schneider et al. 1996).
Each leech included in the microsatellite analyses was also subjected to DNA Barcoding protocols. The universal primers (Folmer et al. 1994), LCO1490, 5′ GGTCAACAAATCATAAAGATATTGG 3′ and HCO2198, 5′ TAAACTTCAGGGTGACCAAAAAATCA 3′, were used to amplify cox1. All amplifications used Ready-To-Go PCR Beads (Amersham Pharmacia Biotech, Piscataway, NJ), 0.5 μl of each 10 μM primer, 1 μl DNA template and 23 μl RNase-free H2O run for 35 cycles of 94°C (45 s), 46°C (30 s) and 68°C (45 s). Amplification products were purified with the AMPure system PCR Cleaning protocol (Agencourt, Beverly, MA, USA) and were sequenced in both directions with the same primers. Each sequencing reaction, including 1 μl BigDye (Applied Biosystems, Perkin–Elmer Corporation), 1 μl of 1 μM primer (single primer for each direction) and 3 μl of DNA template, ran for 35 cycles of 96°C (30 s), 50°C (30 s) and 60°C (4 min). Sequences were purified by following the CleanSEQ protocol (Agencourt, Beverly, MA, USA). Products were re-suspended in 6 μl of formamide and electrophoresed in an ABI 3730 sequencer (Applied Biosystems, Foster City, CA, USA). Sequences of complimentary strands were edited and reconciled using CodonCode Aligner (CodonCode, Dedham, MA, USA). Aligned cox1 sequences (GenBank Accession Members—AF116029, AY425449-52, AY763148-55, AY786458, EF446680–EF446713) were subjected to parsimony analysis, 100 bootstrapping replicates as well as Kishino–Hasegawa (KH) and Templeton tests in PAUP* (Swofford 2003). Branch lengths were calculated using a general time-reversible model and gamma-distributed rate parameter as specified by ModelTest (Posada & Crandall 1998).
3. Results
Microsatellite profiles (table 1) clearly distinguished between wild H. medicinalis and H. verbana. Four microsatellite loci showed clear evidence of species-level segregation: two loci, Hm8 and Hm12, would not amplify from wild H. verbana. Two other loci, HvA10 and HvH07, would not amplify from wild H. medicinalis. Three of the five loci that amplified from all wild European medicinal leeches, Hm2, Hm10, and HvT397, showed no allelic variation in wild H. medicinalis, yet were variable for wild H. verbana. The remaining two amplifiable loci with variability for all wild leech isolates, Hm1 and Hv351, nonetheless had no alleles in common between the two species. In light of these clear differences, it is not surprising that analyses of population structure assigned wild-caught individuals to either H. medicinalis or H. verbana with more than 98.5% confidence in a manner precisely concordant with dorsal colour patterns evident in figure 1. AMOVA demonstrated that differences between these two species accounted for nearly half of all of the genetic variability evidenced in the microsatellite loci (FCT=0.481; p<0.004). Inverted nesting of the AMOVA (i.e., presuming phenotypic plasticity in two colour variants of but a single widespread species) implied that more than 90% of allelic variation was accounted for by this obvious morphological difference and with no remaining variation explainable by geography (FCT=−0.37; p<0.97).
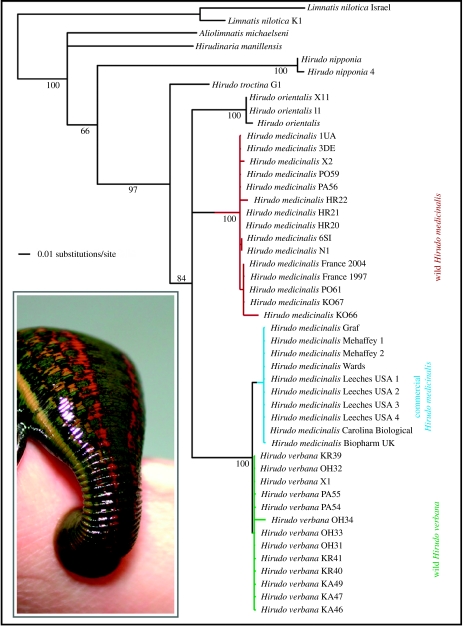
In addition to definitively distinguishing the two wild species of Hirudo, our microsatellite loci also inferred, for all commercially obtained leeches, an unambiguous ancestry (p<0.006) with H. verbana notwithstanding their having been marketed as H. medicinalis. Though the commercially available leeches, unlike wild H. verbana, amplified for loci Hm8 and HM12, none of the alleles found were shared with wild H. medicinalis. Lest this slightly different microsatellite profile be perceived as ambiguous, forcing an analysis of population structure to consider the possibility of three distinct groups split the wild H. verbana roughly into northwestern and southeastern groups and confidently (p<0.01) placed the commercially marketed leeches among these two. DNA barcoding was consistent with all of the foregoing in depicting distinct monophyletic groups for H. medicinalis and H. verbana each with independent histories and averaging 8.5% sequence divergence between the two species relative to less than 2% sequence variation within either species (figure 2). Commercially labelled H. medicinalis leeches were monophyletic (non-parametric bootstrap=100) with wild H. verbana (uncorrected p distance averaged 1.81%) and distinct from wild H. medicinalis (uncorrected p distance averaged 9.03%). Trees constraining commercially available leeches to be monophyletic with wild H. medicinalis were significantly suboptimal in KH and Templeton tests (p=0.0019 and 0.0020, respectively).
Figure 2.
DNA barcoding distinguishes European medicinal leech species. Multiple equally parsimonious trees varied on resolution within each of the wild or commercial medicinal leech clades, and on whether Hirudo orientalis is sister to H. medicinalis or to the other European medicinal leeches. Clade support values are non-parametric bootstrap proportions. Mean genetic distances: 8.55% between wild H. medicinalis and H. verbana; 1.81% between leeches commercially marketed Hirudo medicinalis and wild H. verbana; 9.03% between leeches commercially marketed Hirudo medicinalis and wild H. medicinalis; 8.60% between H. orientalis and the other wild Hirudo species. Branch lengths are based on a GTR +G model of nucleotide substitution. A commercially obtained Hirudo medicinalis (i.e. H. verbana) is shown feeding (inset).
4. Discussion
The foregoing confirms recent work pointing to more than one distinct species of European medicinal leech (Nesemann & Neubert 1999; Trontelj et al. 2004; Trontelj & Utevsky 2005) and a suspicion that widely used laboratory model organisms are incorrectly identified (Kutschera 2006). Whether the erroneous marketing of H. verbana as H. medicinalis might be relevant to efficacy is not thoroughly determined but seems doubtful. Nevertheless, US FDA regulations currently do not expressedly permit the use of H. verbana, and specifically proscribe the mislabelling of a device. The implications of our findings are more far reaching. Already invertebrate neurobiological and developmental biology communities are grappling with the recognition that apparent phenotypic plasticity in some leeches actually belies species heterogeneity (Bely & Weisblat 2006). A detailed EST project, supposedly for H. medicinalis, is well underway at the French National Sequencing Centre, Genoscope. There are more than 300 entries in public genome databases and nearly 700 entries in PubMed with H. medicinalis as the primary taxonomic key for work in neurobiology, developmental genetics and even enteric symbiosis models (Graf et al. 2006). Nearly all of these concern research using commercially available leeches as model organisms. Indeed, more than 300 scholarly articles concerning leeches indicate just such a commercial source. Given our results, molecular characterization of laboratory strains of apparent H. medicinalis is now warranted. To our knowledge, there are no isogenic strains of H. medicinalis, though many undoubtedly exist for H. verbana which is undoubtedly the object of genomic and peptidome studies (Baskova et al. 2004; Wang et al. 2005; Yanes et al. 2005).
At least 115 bioactive compounds have been isolated from medicinal leeches mostly being putative anticoagulants, antistasins and other protease inhibitors (Sollner et al. 1994; Baskova & Zavalova 2001; Salzet 2002). Antistasins, like other coagulation factor Xa inhibitors, have antimetastatic properties (Tuszinsky et al. 1987). Amino acid sequences from these short peptides are substantially different between closely related species (Kim & Kang 1998) and even among paralogues of this serine protease family within a species (Moser et al. 1998). Hirudin and calin, heretofore presumed to be from H. medicinalis, inhibit coagulation by interfering with thrombin or platelet aggregation, respectively (Markwardt 1955; Deckmyn et al. 1995). A thorough investigation of the various species of European medicinal leech should reveal yet more of these powerful peptides and could further enrich the suite of available therapeutic agents and biomedical research tools.
Medicinal leeches, once abundant across Europe, suffered a precipitous decline in the nineteenth century. A century of harvesting tens of millions of leeches each year, the advent of intensive farming and widespread draining of European wetlands together led to dramatic shortages, unregulated smuggling and some of the first European legislative efforts in biological conservation in the early 1900s (Harding & Moore 1927; Elliott & Tullett 1992). In UK, it is still an offence to injure, possess and sell medicinal leeches or damage their natural habitat (Elliott & Tullett 1992). Hirudo medicinalis is afforded threatened species status under IUCN, is regulated by CITES, the Berne Convention and the European Union Habitat Directive (Elliott & Tullett 1992; Trontelj & Utevsky 2005). Such regulations necessarily are species specific and H. verbana has no legal protection whatsoever. Pending a more detailed evaluation of the conservation status of these historically compelling and medically valuable invertebrate animals, we urge the extension of all proscriptions equally to H. verbana and to the newly discovered H. orientalis (Utevsky & Trontelj 2005).
Acknowledgments
We thank Matjaz Bedjanic, Elizabeth Borda, Pierre Deleporte, Dario Ferreri, Otto Friesen, Joerg Graf, Klemen Koselj, Leathem Mehaffey and Anna Phillips for providing specimens of wild caught and laboratory model organism leeches. Andrei Utevsky provided technical assistance for photography. This work was supported by grants from the US National Science Foundation (DEB 0119329 and DBI 0353817), the Richard Lounsbery Foundation, the Slovenian Research Agency and a grant from INTAS (YSF 01/2-0062).
References
- Baskova I.P, Zavalova L.L. Proteinase inhibitors from the medicinal leech Hirudo medicinalis. Biochemistry Mosc. 2001;66:703–717. doi: 10.1023/a:1010223325313. doi:10.1023/A:1010223325313 [DOI] [PubMed] [Google Scholar]
- Baskova I.P, Zavalova L.L, Basanova A.V, Moshkovskii S.A, Zgoda V.G. Protein profiling of the medicinal leech salivary gland secretion by proteomic analytical methods. Biochemistry Mosc. 2004;69:770–775. doi: 10.1023/b:biry.0000040202.21965.2a. doi:10.1023/B:BIRY.0000040202.21965.2a [DOI] [PubMed] [Google Scholar]
- Bely A.E, Weisblat D.A. Lessons from leeches: a call for DNA barcoding in the lab. Evol. Dev. 2006;8:491–501. doi: 10.1111/j.1525-142X.2006.00122.x. doi:10.1111/j.1525-142X.2006.00122.x [DOI] [PubMed] [Google Scholar]
- Boutin-Ganache I, Raposo M, Raymond M, Deschepper C.F. M13-tailed primers improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. Biotechniques. 2001;31:24–27. [PubMed] [Google Scholar]
- Budinoff R.B, Siddall A.M, Siddall M.E. Twelve variable microsatellite loci for the North American medicinal leech, Macrobdella decora. Mol. Ecol. Notes. 2004;4:491–493. doi:10.1111/j.1471-8286.2004.00723.x [Google Scholar]
- Chatterji S, Pachter L. GeneMapper: reference based gene annotation. Genome Biol. 2006;7:R29. doi: 10.1186/gb-2006-7-4-r29. doi:10.1186/gb-2006-7-4-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepeha D.B, Nussenbaum B, Bradford C.R, Teknos T.N. Leech therapy for patients with surgically unsalvageable venous obstruction after revascularized free tissue transfer. Arch. Otolaryngol. Head Neck Surg. 2002;128:960–965. doi: 10.1001/archotol.128.8.960. [DOI] [PubMed] [Google Scholar]
- Darwin C. John Murray; London, UK: 1859. On the origin of species. [Google Scholar]
- Deckmyn H, Stassen J.M, Vreys I, Van Houtte E, Sawyer R.T, Vermylen J. Calin from Hirudo medicinalis, an inhibitor of platelet adhesion to collagen, prevents platelet-rich thrombosis in hamsters. Blood. 1995;85:712–719. [PubMed] [Google Scholar]
- Derganc M, Zdravic F. Venous congestion of flaps treated by application of leeches. Br. J. Plast. Surg. 1960;13:187–192. doi: 10.1016/s0007-1226(60)80036-7. doi:10.1016/S0007-1226(60)80036-7 [DOI] [PubMed] [Google Scholar]
- Elliott J.M, Tullett P.A. The medicinal leech. Biologist. 1992;39:153–158. [Google Scholar]
- Folmer O, Black M, Hoen W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metozoan invertebrates. Mol. Mar. Biol. Biotech. 1994;3:294–299. [PubMed] [Google Scholar]
- Foucher G, Henderson H.R, Maneau M, Merle M, Braun F.M. Distal digital replantation: one of the best indications for microsurgery. Int. J. Microsurg. 1981;3:263–270. [Google Scholar]
- Graf J, Kikuchi Y, Rio R.V.M. Leeches and their microbiota: naturally simple symbiosis models. Trends Microbiol. 2006;14:365–371. doi: 10.1016/j.tim.2006.06.009. doi:10.1016/j.tim.2006.06.009 [DOI] [PubMed] [Google Scholar]
- Harding W.A, Moore J.P. Taylor and Francis; London, UK: 1927. The fauna of British India: Hirudinea. [Google Scholar]
- Kim D.R, Kang K.W. Amino acid sequence of piguamerin, an antistasin-type protease inhibitor from the blood sucking leech Hirudo nipponia. Eur. J. Biochem. 1998;254:692–697. doi: 10.1046/j.1432-1327.1998.2540692.x. doi:10.1046/j.1432-1327.1998.2540692.x [DOI] [PubMed] [Google Scholar]
- Kristan W.B, Calabrese R.L, Friesen W.O. Neuronal basis of leech behaviors. Prog. Neurobiol. 2005;76:279–327. doi: 10.1016/j.pneurobio.2005.09.004. doi:10.1016/j.pneurobio.2005.09.004 [DOI] [PubMed] [Google Scholar]
- Kutschera U. The infamous blood suckers from Lacus Verbanus. Lauterbornia. 2006;56:1–4. [Google Scholar]
- Louis P.C.A. Recherche sur les effets de la saignée dans plusieurs maladies inflammatoires. Archives Générales de Médecine. 1828;18:321–336. [Google Scholar]
- Markwardt F. Untersuchungen fiber Hirudin. Natur-wissenschaften. 1955;42:537–538. [Google Scholar]
- Moquin-Tandon A. Maison de Commerce; Montpellier, France: 1827. Monographie de la Famille des Hirudinées. [Google Scholar]
- Moquin-Tandon A. Ballière; Paris, France: 1846. Monographie de la Famille des Hirudinées. [Google Scholar]
- Moser M, Auerswald E, Mentele R, Eckerskorn C, Fritz H, Fink E. Bdellastasin, a serine protease inhibitor of the antistasin family from the medical leech Hirudo medicinalis—primary structure, expression in yeast, and characterisation of native and recombinant inhibitor. Eur. J. Biochem. 1998;253:212–220. doi: 10.1046/j.1432-1327.1998.2530212.x. doi:10.1046/j.1432-1327.1998.2530212.x [DOI] [PubMed] [Google Scholar]
- Nesemann, H. & Neubert, E. 1999 Annelida: Clitellata: Branchiobdellida, Acanthobdellea, Hirudinea, In Süßwasserfauna von Mitteleuropa Heidelberg, Germany: Spektrum Akad Verl.
- Posada D, Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Pritchard J.K, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rados C. Beyond bloodletting: FDA gives leeches a medical makeover. FDA Consum. 2004;38:9. [PubMed] [Google Scholar]
- Salzet M. Leech thrombin inhibitors. Curr. Pharm. Des. 2002;8:493–503. doi: 10.2174/1381612023395664. doi:10.2174/1381612023395664 [DOI] [PubMed] [Google Scholar]
- Sawyer R.T. Clarendon Press; Oxford, UK: 1986. Leech biology and behaviour. [Google Scholar]
- Schneider, S., Kueffe, J.-M., Roessli, D. & Excoffier, L. 1996 Arlequin, a software for population genetic data analysis Geneva, Switzerland: University of Geneva.
- Sollner C, Mentele R, Eckerskorn C, Fritz H, Sommerhoff C.P. Isolation and characterization of hirustasin, an antistasin-type serine-proteinase inhibitor from the medical leech Hirudo medicinalis. Eur. J. Biochem. 1994;219:937–943. doi: 10.1111/j.1432-1033.1994.tb18575.x. doi:10.1111/j.1432-1033.1994.tb18575.x [DOI] [PubMed] [Google Scholar]
- Swofford, D. L. 2003 PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sunderland, MA: Sinauer Associates.
- Trontelj P, Utevsky S.Y. Celebrity with a neglected taxonomy: molecular systematics of the medicinal leech (genus Hirudo) Mol. Phylogenet. Evol. 2005;34:616–624. doi: 10.1016/j.ympev.2004.10.012. doi:10.1016/j.ympev.2004.10.012 [DOI] [PubMed] [Google Scholar]
- Trontelj P, Sotler M, Verovnik R. Genetic differentiation between two species of the medicinal leech, Hirudo medicinalis and the neglected H. verbana, based on random-amplified polymorphic DNA. Parasitol. Res. 2004;94:118–124. doi: 10.1007/s00436-004-1181-x. [DOI] [PubMed] [Google Scholar]
- Tuszinsky G.P, Gasic T.B, Gasic G.J. Isolation and characterization of antistasin: an inhibitor of metastasis and coagulation. J. Biol. Chem. 1987;262:9718–9723. [PubMed] [Google Scholar]
- Utevsky S.Y, Trontelj P. A new species of the medicinal leech (Oligochaeta, Hirudinida, Hirudo) from Transcaucasia and an identification key for the genus Hirudo. Parasitol. Res. 2005;98:61–66. doi: 10.1007/s00436-005-0017-7. doi:10.1007/s00436-005-0017-7 [DOI] [PubMed] [Google Scholar]
- Wang W.-Z, Emes R.D, Christoffers K, Verrall J, Blackshaw S.E. Hirudo medicinalis: a platform for investigating genes in neural repair. Cell. Mol. Neurobiol. 2005;25:427–440. doi: 10.1007/s10571-005-3151-y. doi:10.1007/s10571-005-3151-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanes O, Villanueva J, Querol E, Aviles F.X. Functional screening of serine protease inhibitors in the medical leech Hirudo medicinalis monitored by intensity fading MALDI-TOF MS. Mol. Cell. Proteomics. 2005;4:1602–1613. doi: 10.1074/mcp.M500145-MCP200. doi:10.1074/mcp.M500145-MCP200 [DOI] [PubMed] [Google Scholar]