Abstract
During the past decade, there has been a renewed interest in using P. aeruginosa as a model system for biofilm development and pathogenesis. Since the biofilm matrix represents a critical interface between the bacterium and the host or its environment, considerable effort has been expended to acquire a more complete understanding of the matrix composition. Here, we focus on recent developments regarding the roles of alginate, Psl, and Pel polysaccharides in the biofilm matrix.
Introduction
The bacterium Pseudomonas aeruginosa is a remarkably adept opportunist. If provided a breach in local or systemic immunity, the organism can cause severe, life-threatening infections. These infections persist despite aggressive antimicrobial therapy and a robust inflammatory response. Perhaps the most widely recognized P. aeruginosa persistent infection occurs in individuals with the genetic disease cystic fibrosis (CF).
The persistence of P. aeruginosa during CF infections has been linked, in part, to its ability to form biofilms [1,2]. Biofilms, which are defined as communities of microorganisms that are attached to a surface, play a significant role in infectious disease [3]. Bacteria within biofilms are usually embedded within a matrix, which can consist of protein, polysaccharide, and nucleic acid. The matrix provides a critical role in the biofilm resistance phenotype. A complete understanding of the organization and composition of the P. aeruginosa biofilm matrix may assist in the development of novel therapeutics aimed at disrupting biofilms, which will translate into improved clearance of these infections. Here, we focus on the polysaccharide components of the extracellular polymeric substance (EPS) of P. aeruginosa biofilms.
Alginate
The CF lung is initially colonized with nonmucoid P. aeruginosa strains, but with time, mucoid variants emerge and become the predominant lung pathogen [4]. The mucoid phenotype is due to the overproduction of alginate, a capsular polysaccharide virulence factor that confers a selective advantage for P. aeruginosa in the CF airway. Alginate is a high molecular weight, acetylated polymer composed of non-repetitive monomers of β-1,4 linked L-guluronic and D-mannuronic acids. Alginate-producing variants arise in vivo most frequently due to mutations in the negative regulator mucA [5]. There is a distinct correlation between the appearance of mucoid P. aeruginosa and a worsening clinical prognosis for CF patients [4]. Infection of the CF lung causes inflammatory cells to be recruited to the site of infection, where they release reactive oxygen species and cause extensive tissue damage [4,6]. Alginate appears to protect P. aeruginosa from the consequences of this inflammation, since it scavenges free radicals released by activated macrophages in vitro and appears to provide protection from phagocytic clearance [4,6]. Although antibodies to alginate are found in the sera of chronically infected CF patients, these antibodies fail to mediate opsonic killing of P. aeruginosa in vitro [7]. Likely each of these properties contributes to the ability of mucoid P. aeruginosa to persist and establish chronic infections in the CF lung. Since aspects regarding the genetics, pathogenesis, and biochemistry of alginate have been reviewed elsewhere [4,8,9], we focus attention on observations regarding the role of alginate in biofilms of mucoid P. aeruginosa.
Results from several independent laboratories have shown that overproduction of acetylated alginate leads to significant architectural and morphological changes in the biofilm [10–13]. This translates into increased resistance to antimicrobials [10] or IFN-gamma-mediated killing by cells of the innate immune system [14]. In support of this, Alkawash and colleagues showed that degradation of mucoid P. aeruginosa biofilms by alginate lyase led to enhanced killing by gentamicin [15]. Surprisingly however, studies from two of these groups and our work showed that alginate synthesis is not required for biofilm development [11,12,16]. In these studies, alginate proficient and deficient P. aeruginosa formed morphologically similar biofilms. However, overproduction of alginate, which occurs primarily as a result of mucA mutations, clearly affects resistance properties of the biofilm.
Psl
The above-mentioned studies were performed in alginate overproducing strains (i.e. mucA mutants). During the past decade, there has been a renewed interest in using P. aeruginosa as a model system for biofilm development and pathogenesis. Most of these studies have been performed with nonmucoid (i.e. mucA+) P. aeruginosa strains such as PAO1 or PA14, which produce little to no detectable alginate in vitro. Furthermore, when the alginate genes were disrupted in PAO1 and PA14, these strains were fully capable of forming biofilms. The biofilms formed by these mutants retained what appeared to be polysaccharide matrix material [16]. This suggested that one or more polysaccharides independent of alginate might be essential for biofilm development in nonmucoid P. aeruginosa strains. Several groups initiated studies to identify alternative polysaccharide-encoding genes, and two loci were discovered. The first, pel (Fig. 1), was found to be involved in pellicle formation in strain PA14 [17]. The role of pel in the biofilm matrix will be discussed below. The second polysaccharide locus designated psl (polysaccharide synthesis locus, Fig. 1), is an operon composed of 15 genes encoding the Psl biosynthetic machinery. The psl operon was shown to be essential for biofilm formation in strains PAO1 and ZK2870 [18–21]. In these studies, inactivation of the psl gene cluster led to a significant defect in cell-surface and cell-cell interactions. Psl is also required for adherence to mucin-coated surfaces and airway epithelial cells; biotic surfaces that are clearly relevant to CF [20]. A study in 2006 utilizing an inducible psl construct found that in addition to being required for cell-surface and cell-cell interactions, psl is also needed for maintenance of the biofilm structure post-attachment. This led the authors to conclude that Psl functions as a scaffold, holding biofilm cells together in the matrix [20].
Fig. 1.
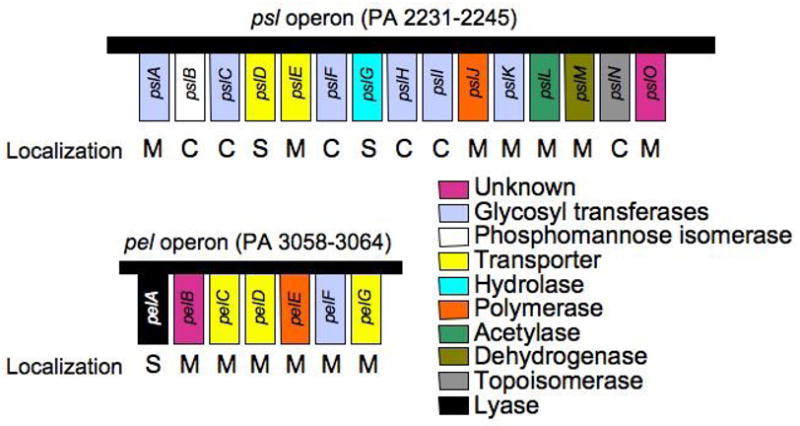
Structure of psl and pel operons. Putative functions and localization of Psl and Pel enzymes are shown (M-membrane, C-cytoplasm, S-secreted).
Carbohydrate and lectin staining analyses indicate that Psl is a mannose- and galactose-rich polysaccharide, however the precise Psl structure has not been elucidated [19,21,22]. This is an area requiring future research. Overhage and Campisano et al. demonstrated that pslA and pslD, which encode a putative UDP-glucose lipid carrier and exporter, respectively, are essential for biofilm formation in strain PAO1 [23,24]. They also demonstrated that while psl is constitutively expressed in planktonic cells, its expression is localized to the center of developing biofilm microcolonies [24]. This implies that Psl has a role in biofilm differentiation. Our lectin staining studies show Psl is equally distributed in undifferentiated, flat multiple-layer biofilms. However, mature microcolonies reveal peripheral staining of Psl with minimal staining of matrix in the center of the microcolonies. Instead, this region has numerous motile cells representing a biofilm at a developmental stage just prior to dispersion (L. Ma and D.J. Wozniak, unpublished data).
Pel
P. aeruginosa is able to form biofilms not only on mucosal and other solid surfaces but also at the air-liquid interface of standing cultures. The genetic basis for these structures, known as pellicles, was elucidated by screening a PA14 transposon library for pellicle-deficient mutants. This revealed a seven-gene operon, named pel (Fig. 1), which is necessary for maintenance of biofilm structure in strain PA14 [17]. The authors hypothesized that pel is involved in the production of an extracellular matrix material. To determine the nature of this pel-associated matrix material, mutants lacking one or more pel genes were evaluated for biofilm initiation, colony morphology, and mature biofilm integrity. While biofilm initiation per se was not significantly affected in PA14 pel mutants, the colony morphology was affected as well as the ability of these cells to bind Congo red [17]. Carbohydrate and linkage analyses provide evidence that pel encodes a glucose-rich matrix polysaccharide polymer, which does not appear to be cellulose [17,19]. As with Psl, the Pel structure is unknown and further biochemical analyses of Pel polysaccharide is necessary.
In a similar study using a non-piliated PAK strain, a transposon screen for nonadherent mutants generated several mutations that mapped to the pel locus [25]. The authors suggested that any role for pel in attachment might not be observed because type IV pili may compensate for a lack of pel during attachment [25]. As predicted, biofilm initiation was significantly reduced in non-piliated PAK pel mutants. Clearly the roles of pel and type IV pili in the initial attachment process will need to be delineated.
Genes pelA-G (Fig. 1) are highly conserved in other P. aeruginosa strains, including the common laboratory strain PAO1 [17]. The Gram-negative plant pathogen Ralstonia solanacearum contains a homologous gene cluster that, when mutated, resulted in a biofilm-defective phenotype similar to that observed in P. aeruginosa pel mutants [25]. Putative functions in polysaccharide processing have been assigned to most of the pel genes, (Fig. 1) and [17,19,25,26].
Thus, the pel locus produces a glucose-rich matrix polysaccharide that is essential for pellicle formation and biofilm structure in P. aeruginosa strains PA14 and PAK. The pel-encoded polysaccharide is biochemically and genetically distinct from Psl. To date, no immunological or lectin reagents are available to probe Pel expression or localization in developing P. aeruginosa biofilms.
Regulation of Psl and Pel polysaccharide
Currently, little is known regarding the regulation of Psl and Pel but several seemingly disparate findings have begun to shed some light on this issue. D’Argenio et al. performed transposon mutagenesis of strain PAO1 and isolated a number of colonies that exhibited a “wrinkly” colony phenotype [27]. Genetic analysis of these mutants revealed that mutations in the wspF gene lead to the hyperaggregative phenotype. The wsp locus was first described in P. fluorescens and appears to encode a chemosensory system involved in the wrinkly spreader phenotype [28]. Here, WspF controls the methylation state of WspA, which subsequently controls activation of the response regulator WspR. If wspF is inactivated, WspA is hypermethylated and WspR is constitutively active.
In a later study, Kirisits et al. reported similar hyperaggregative and hyperadherent PAO1-derived colony variants isolated from biofilm reactors [29]. Transcriptional profiling of these variants showed increased psl and pel expression, when compared with the parental PAO1 strain. Disruption of the psl operon in the variant reversed the hyperaggregative and hyperadherent phenotype but colonies still retained a wrinkled phenotype, presumably due to Pel overexpression. Therefore, the authors conclude that psl, in addition to pel and perhaps other components are expressed at a higher level and responsible for the hyperaggregative and hyperadherent variant phenotype [29]. Similar conclusions were drawn by Friedman and Kolter when analyzing the autoaggregative P. aeruginosa variant ZK2870 [19]. In support of this, over-expression of Psl via a pBAD-derived promoter system is sufficient to convert P. aeruginosa to a phenotype that resembles the above-mentioned autoaggregative variants [20].
It is reasonable to assume that the wrinkled colonies in the aforementioned D’Argenio study [27] also overexpress psl and pel and that this overexpression is caused by activation of WspR. In fact, a later study by Hickman et al. showed an increase in psl and pel expression in a wspF mutant [30]. This study further evaluated the effect of the Wsp system by investigating the role of WspR as a diguanylate cyclase. Proteins with this activity generate cyclic-di-GMP, a small signaling molecule involved in many cellular processes [31]. The levels of cellular c-di-GMP appear to correlate with the biofilm forming ability of P. aeruginosa. When WspR was constitutively activated, as seen in a wspF mutant, c-di-GMP levels were high and biofilm formation was greater than in the wild type strain. As seen in the above-mentioned autoaggregative variants, a transcriptional profile of wspF mutant bacteria revealed elevated levels of psl and pel transcription, when compared with the parental strain. This study also elegantly illustrated that when c-di-GMP was degraded, biofilm formation decreased substantially. This strengthens the relationship between psl and pel expression, the Wsp chemosensory system, and cellular levels of c-di-GMP.
Another regulatory system controlling psl and pel expression is the GacS/GacA/rsmZ system. In a study of two-component systems in strain PAK, a mutation in retS, encoding a hybrid sensor kinase/response regulator, elevated psl and pel expression resulting in enhanced biofilm formation [32]. A second round of transposon mutagenesis in the retS strain revealed that mutations in the GacS/GacA/rsmZ regulatory pathway reversed the retS phenotype. GacS and GacA are a sensor-regulator two-component pair and rsmZ is a small regulatory RNA that represses the activity of the posttranscriptional RNA binding protein RsmA. The model for how this system functions is as follows [33]: signals that activate RetS repress expression of rsmZ whereas signals that activate GacS induce rsmZ expression. When rsmZ levels are high, RsmA is inactive and this results in increased psl and pel expression. The opposite is true: low levels of rsmZ favor repression of pel and pel.. More recent work identified LadS, encoding a hybrid sensor kinase, which also modulates rsmZ levels [34] and therefore indirectly affects psl and pel expression.
Conclusions and perspectives
Since discovery of the Psl and Pel polysaccharides four years ago, much progress has been made in our understanding of the P. aeruginosa biofilm matrix. This has forced us to reconsider the notion that all P. aeruginosa biofilms are composed of alginate. We also must consider that the relationship between biofilm formation by nonmucoid strains and those in which conversion to mucoidy has occurred does not appear to be a change in the alginate levels in the biofilm matrix, but a fundamental change in its carbohydrate constituents. In the context of CF infections, the discovery of alternative polysaccharides that contribute to biofilm formation in nonmucoid strains, which are believed to be the first to colonize, implies that during initial infection, biofilm formation may precede the switch to mucoidy. Clearly the potential role of Psl and Pel in P. aeruginosa pathogenesis is an area needing further investigation.
Future studies aimed at an understanding of individual psl and pel gene functions and regulation of their expression will provide more information about the selective advantage of individual P. aeruginosa polysaccharides. Additionally, the potential overlapping or redundant functions of the Pel and Psl polysaccharides of nonmucoid P. aeruginosa is an area in need of study. A seminal question is whether individual cells have the capacity to produce more than one polysaccharide simultaneously. Do mucoid biofilms also contain Pel and/or Psl? Collectively, the regulation studies illustrate that psl and pel appear to be coordinately regulated and that c-di-GMP levels play a critical role in this control. The challenge in the coming years will be to discern how the Wsp and RsmA/rsmZ pathways are integrated to control the acute vs. biofilm lifestyle of P. aeruginosa. Finally, the role of polysaccharide-independent components, such as extracellular DNA, LPS, membrane vesicles, lectins, and cup fimbriae must be placed in context with emerging data regarding alginate, Psl, and Pel polysaccharides.
Since Psl and Pel polysaccharides are expressed on planktonic P. aeruginosa cells, play a critical role in biofilm development, and are conserved among P. aeruginosa strains examined, these polysaccharides clearly represent attractive targets for agents aimed at disrupting the matrix. As chronic infections due to mucoid P. aeruginosa are almost impossible to eradicate, targeting Psl and Pel may allow us to prevent initial infection or enhance the clearance of an existing nonmucoid infection. Once a better understanding of the chemical nature of the polysaccharide matrix and its link to colonization and pathogenesis of P. aeruginosa develops, this may be feasible.
Acknowledgments
Cystic Fibrosis Foundation Grant WOZNIA06P0 and Public Health Service grant AI061396 (D.J.W) supported this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
• of special interest
•• of outstanding interest
- •1.Lam J, Chan R, Lam K, Costerton JRW. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect Immun. 1980;28:546–556. doi: 10.1128/iai.28.2.546-556.1980. Microscopic evidence for P. aeruginosa biofilms in CF patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •2.Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. Biochemical and microscopic evidence for P. aeruginosa biofilms in CF patients. [DOI] [PubMed] [Google Scholar]
- 3.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- ••4.Govan JRW, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. A bit dated, but an outstanding and thorough review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••5.Martin DW, Schurr MJ, Mudd MH, Govan JRW, Holloway BW, Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci USA. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. First to show the molecular mechanism underlying P. aeruginosa mucoid conversion in CF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pier GB. Pseudomonas aeruginosa: a key problem in cystic fibrosis. ASM News. 1998;6:339–347. [Google Scholar]
- 7.Pier GB, Boyer D, Preston M, Coleman FT, Llosa N, Mueschenborn-Koglin S, Theilacker C, Goldenberg H, Uchin J, Priebe GP, et al. Human monoclonal antibodies to Pseudomonas aeruginosa alginate that protect against infection by both mucoid and nonmucoid strains. J Immunol. 2004;173:5671–5678. doi: 10.4049/jimmunol.173.9.5671. [DOI] [PubMed] [Google Scholar]
- 8.Gacesa P. Bacterial alginate biosynthesis--recent progress and future prospects. Microbiology. 1998;144:1133–1143. doi: 10.1099/00221287-144-5-1133. [DOI] [PubMed] [Google Scholar]
- 9.Ramsey DM, Wozniak DJ. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol Microbiol. 2005;56:309–322. doi: 10.1111/j.1365-2958.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- 10.Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, Parsek MR. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol. 2001;183:5395–5401. doi: 10.1128/JB.183.18.5395-5401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •11.Nivens DE, Ohman DE, Williams J, Franklin MJ. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J Bacteriol. 2001;183:1047–1057. doi: 10.1128/JB.183.3.1047-1057.2001. First to show alginate is not essential for P. aeruginosa biofilm development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stapper AP, Narasimhan G, Ohman DE, Barakat J, Hentzer M, Molin S, Kharazmi A, Hoiby N, Mathee K. Alginate production affects Pseudomonas aeruginosa biofilm development and architecture, but is not essential for biofilm formation. J Med Microbiol. 2004;53:679–690. doi: 10.1099/jmm.0.45539-0. [DOI] [PubMed] [Google Scholar]
- 13.Tielen P, Strathmann M, Jaeger KE, Flemming HC, Wingender J. Alginate acetylation influences initial surface colozniation by mucoid Pseudomonas aeruginosa. Microbiol Res. 2005;160:165–176. doi: 10.1016/j.micres.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Leid JG, Willson CJ, Shirtliff ME, Hassett DJ, Parsek MR, Jeffers AK. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-{gamma}-mediated macrophage killing. J Immunol. 2005;175:7512–7518. doi: 10.4049/jimmunol.175.11.7512. [DOI] [PubMed] [Google Scholar]
- 15.Alkawash MA, Soothill JS, Schiller NL. Alginate lyase enhances antibiotic killing of mucoid Pseudomonas aeruginosa in biofilms. APMIS. 2006;114:131–138. doi: 10.1111/j.1600-0463.2006.apm_356.x. [DOI] [PubMed] [Google Scholar]
- •16.Wozniak DJ, Wyckoff TJO, Starkey M, Keyser R, Azadi P, O’Toole GA, Parsek MR. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci U S A. 2003;100:7907–7912. doi: 10.1073/pnas.1231792100. First to provide evidence for polysaccharides other than alginate as matrix material in P. aeruginosa biofilms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol. 2004;51:675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- 18.Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol. 2004;186:4466–4475. doi: 10.1128/JB.186.14.4466-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •19.Friedman L, Kolter R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aerguinosa biofilm matrix. J Bacteriol. 2004;186:4457–4465. doi: 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma L-Y, Jackson K, Landry RM, Parsek MR, Wozniak DJ. Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol. 2006;188:8213–8221. doi: 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsukawa M, Greenberg EP. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J Bacteriol. 2004;186:4449–4456. doi: 10.1128/JB.186.14.4449-4456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma L, Lu H, Sprinkle A, Parsek MR, Wozniak D. Pseudomonas aeruginosa Psl is a galactose- and mannose-rich exopolysaccharide. J Bacteriol. 2007 doi: 10.1128/JB.00620-07. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campisano A, Schroeder C, Schemionek M, Overhage J, Rehm BHA. PslD is a secreted protein required for biofilm formation by Pseudomonas aeruginosa. Appl Environ Microbiol. 2006;72:3066–3068. doi: 10.1128/AEM.72.4.3066-3068.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Overhage J, Schemionek M, Webb JS, Rehm BHA. Expression of the psl operon in Pseudomonas aeruginosa PAO1 biofilms: PslA performs an essential function in biofilm formation. Appl Environ Microbiol. 2005;71:4407–4413. doi: 10.1128/AEM.71.8.4407-4413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vasseur P, Vallet-Gely I, Soscia C, Genin S, Filloux A. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology. 2005;151:985–997. doi: 10.1099/mic.0.27410-0. [DOI] [PubMed] [Google Scholar]
- 26.Vasseur P, Soscia C, Voulhoux R, Filloux A. PelC is a Pseudomonas aeruginosa outer membrane lipoprotein of the OMA family of proteins involved in exopolysaccharide transport. Biochimie. 2007;89:903–915. doi: 10.1016/j.biochi.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 27.D’Argenio DA, Calfee MW, Rainey PB, Pesci EC. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J Bacteriol. 2002;184:6481–6489. doi: 10.1128/JB.184.23.6481-6489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spiers AJ, Kahn SG, Bohannon J, Travisano M, Rainey PB. Adaptive divergence in experimental populations of Pseudomonas fluorescens. I Genetic and phenotypic bases of wrinkly spreader fitness. Genetics. 2002;161:33–46. doi: 10.1093/genetics/161.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirisits MJ, Prost L, Starkey M, Parsek MR. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2005;71:4809–4821. doi: 10.1128/AEM.71.8.4809-4821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••30.Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci USA. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. A seminal paper in the field. Clearly linked c-di-GMP and the wsp signal transduction system with P. aeruginosa biofilm formation as well as polysaccharide gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annual Review of Genetics. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- ••32.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. A highly significant and stimulating study that defined a molecular basis for the inverse control of genes associated with acute and chronic P. aeruginosa infection. [DOI] [PubMed] [Google Scholar]
- 33.Yahr TL, Greenberg EP. The genetic basis for the commitment to chronic versus acute infection in Pseudomonas aeruginosa. Mol Cell. 2004;16:497–498. doi: 10.1016/j.molcel.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 34.Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci USA. 2006;103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]


