Abstract
Primary biliary cirrhosis (PBC) is a cholestatic disease associated with autoimmune phenomena and alterations in both biliary bicarbonate excretion and expression of the bicarbonate carrier AE2. The bile acid ursodeoxycholic acid (UCDA) is currently used in treatment of cholestatic liver diseases and is the treatment of choice in PBC; however, a subset of PBC patients respond poorly to UDCA monotherapy. In these patients, a combination of UDCA and glucocorticoid therapy appears to be beneficial. To address the mechanism of this benefit, we analyzed the effects of UDCA and dexamethasone on AE2 gene expression in human liver cells from hepatocyte and cholangiocyte lineages. The combination of UDCA and dexamethasone, but not UDCA or dexamethasone alone, increased the expression of liver-enriched alternative mRNA isoforms AE2b1 and AE2b2 and enhanced AE2 activity. Similar effects were obtained after replacing UDCA with UDCA conjugates. In in vitro and in vivo reporter assays, we found that a UDCA/dexamethasone combination upregulated human AE2 alternate overlapping promoter sequences from which AE2b1 and AE2b2 are expressed. In chromatin immunoprecipitation assays, we demonstrated that combination UCDA/dexamethasone treatment induced p300-related interactions between HNF1 and glucocorticoid receptor on the AE2 alternate promoter. Our data provide a potential molecular explanation for the beneficial effects of the combination of UDCA and glucocorticoids in PBC patients with inadequate response to UDCA monotherapy.
Introduction
Ursodeoxycholic acid (UDCA) is currently being used for the treatment of cholestatic liver diseases (1–3). In contrast to classical immunosuppressors, UDCA is particularly useful in primary biliary cirrhosis (PBC), a chronic cholestatic disease of unknown pathogenesis that is associated with characteristic autoimmune phenomena (4–9). When adequate UDCA therapy — i.e., daily administration of high doses of this hydrophilic bile acid — is initiated at early stages (I–II) of the disease, about 60% of PBC patients normalize their biochemical parameters and reach a survival comparable to that in the general population (8, 9). There are reports suggesting that the remaining patients who exhibit a suboptimal response to UDCA alone may experience some benefit when UDCA therapy is combined with glucocorticoids (10–15).
The precise molecular mechanism by which UDCA is beneficial for the treatment of intrahepatic cholestasis has not yet been fully elucidated (1–3). Among other effects, UDCA administration is known to result in bicarbonate-rich choleresis (16, 17), although this bile acid has been reported to exert modulatory effects in the particular BDL rat model, in which obstructive cholestasis is associated with bile ductular cell hyperplasia and increased secretin-stimulated ductal secretory activity (18). In any case, PET technology in untreated PBC patients (and other cholestatic patients) indicates that they have impaired biliary bicarbonate excretion in response to secretin and that UDCA may restore the secretin response in those patients with PBC (19). On the other hand, PBC patients have diminished expression of anion exchanger 2 (AE2) protein (also known as SLC4A2) in the liver, and UDCA treatment appears to be associated with improved AE2 expression (20). Recently, AE2 has been demonstrated to mediate secretin-stimulated biliary excretion of bicarbonate in the normal rat (21). AE2 is a Na+-independent anion exchanger that exerts electroneutral exchange of Cl– and HCO3– across the plasma membrane, usually through HCO3– efflux and Cl– influx, and is thus also involved in the regulation of the intracellular pH (pHi) (22, 23).
There are 3 N-terminal variants of the human AE2: the full-length isoform AE2a and the alternative isoforms, AE2b1 and AE2b2 (24). AE2a mRNA is transcribed from the upstream AE2 promoter (seemingly in a constitutive manner) and occurs in most human tissues, including the liver. On the other hand, expression of alternative isoforms is driven from AE2b2/AE2b1 overlapping promoter sequences within intron 2 of the AE2 gene. In humans, this alternative expression happens in a more tissue-restricted fashion, mainly in liver and kidney (24). Recent experiments have shown that alternative AE2b1 and AE2b2 protein isoforms are rather active and are able to exchange anions at rates even higher than that of the complete AE2a isoform (25). The expression of alternative isoforms and the regulation of their AE2b2/AE2b1 overlapping promoter in the liver therefore deserves careful attention. A binding site for the liver-enriched hepatocyte nuclear factor 1α (HNF1α) and/or HNF1β was localized near upstream to alternative exon 1b2 at position –113b2 (or –654b1) in the overlapping promoter sequence and was found to be responsible for approximately 70% of the baseline alternative transcriptional activity (26). HNF1α has been reported to participate cooperatively with the glucocorticoid receptor (GR, also known as NR3C1) in the transcriptional activation of several liver-specific genes (27–29). A good example of such an interplay is the response of the rat phenylalanine hydroxylase gene (Pah) to hormone stimulation, which involves binding of GR and HNF1α to a glucocorticoid response element (GRE) core sequence (GREcore) and an HNF1 site in the Pah promoter, as well as interactions of GR and HNF1α via coactivators (29). Additional reports suggest that HNF1α (and also HNF1β) can recruit coregulatory factors such as cAMP response element–binding protein–binding protein/E1A-binding protein (300 kDa) (CBP/p300) (30–34), which may in turn interact with GR (35). Notably, the AE2b2/AE2b1 promoter shows 2 potential GREcore motifs between exons 1b2 and 1b1 (at positions –327b1 and –245b1) (24), which might also be relevant for the transcriptional activity of this region through interplay(s) with HNF1α and/or HNF1β.
UDCA, a hydrophilic dihydroxy bile acid that possesses structural similarities to steroids, has been shown to interact with GR in vitro (36–38). Here we investigated whether UDCA, alone or in combination with glucocorticoids, could upregulate the alternative expression of the AE2 gene in the liver. We then analyzed the possible mechanisms involved.
Results
Treatment with UDCA together with dexamethasone leads to upregulated alternative expression of the AE2 gene in liver cells.
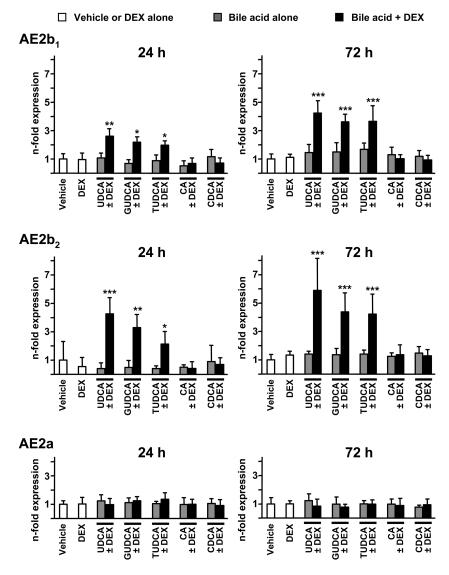
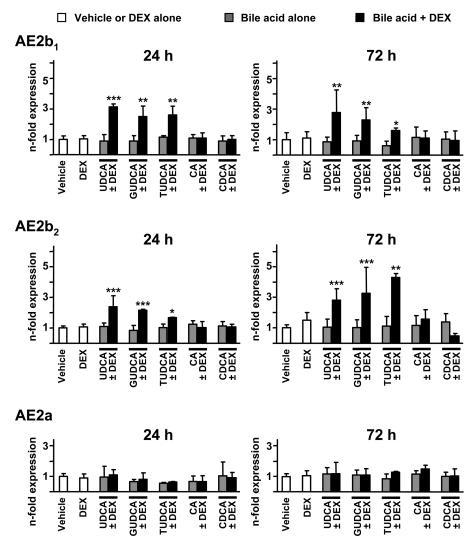
We first analyzed the effects of UDCA and dexamethasone on the expression of AE2 mRNA isoforms in human liver cells from both cholangiocyte and hepatocyte lineages. As shown in Figure 1, combined administration of UDCA and dexamethasone (100 μM each) to cultured normal human cholangiocytes (NHCs) for 24 hours resulted in increased expression of mRNAs for alternative variants AE2b1 and AE2b2. Increased alternative expression in biliary cells was even more pronounced after 72 hours of combined treatment (Figure 1). No significant effects, however, were observed when UDCA or dexamethasone was administered alone (Figure 1). Similarly to NHC cells, hepatocyte lineage PLC/PRF/5 cells also showed increased levels of mRNAs for AE2b1 and AE2b2 after 24 and 72 hours of treatment with the combination of UDCA and dexamethasone, but not with any of these compounds alone (Figure 2). In these hepatocytic cells, the increased alternative expression appeared to be more rapid than in cholangiocytes, reaching statistical significance at 6 hours with the combined treatment (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI33156DS1), while alternative upregulation in cholangiocytes became in turn rather robust after longer treatment, i.e., after 72 hours (Figure 1). When unconjugated UDCA was replaced by equimolar concentration of glycine- or taurine-conjugated UDCA (GUDCA or TUDCA, respectively), the combination effects on AE2 alternative expression remained in both cell types (Figures 1 and 2). These combination effects were specific for these hydrophilic dihydroxy bile acids, as combination of dexamethasone with equimolar concentrations of cholic acid (CA) or chenodeoxycholic acid (CDCA) had no effect on AE2 alternative expression (Figures 1 and 2). On the other hand, none of the combined treatments resulted in changes in the expression levels of the full-length AE2a mRNA in either of the 2 liver cell types (Figures 1 and 2).
Figure 1. The combination of UDCA and dexamethasone upregulates AE2 alternative expression in NHCs.
The mRNA levels for AE2b1, AE2b2, and AE2a isoforms were determined by real-time PCR in cultured NHCs treated for 24 or 72 hours with either dexamethasone (DEX) and/or UDCA, GUDCA, TUDCA, CA, or CDCA (100 μM each) or with just vehicle. Values (normalized for GAPDH mRNA) are given as fold expression relative to values in the respective controls with just vehicle. Data are mean ± SD; n = 6 (except for GUDCA, TUDCA, CA, and CDCA — with and without DEX — for which n = 3 each). *P < 0.05, **P < 0.01, ***P < 0.001 versus vehicle control.
Figure 2. UDCA together with dexamethasone upregulates AE2 alternative expression in hepatocyte lineage PLC/PRF/5 cells.
The mRNA levels for AE2b1, AE2b2, and AE2a isoforms were determined by real-time PCR in PLC/PRF/5 cells treated for 24 or 72 hours with either dexamethasone and/or UDCA, GUDCA, TUDCA, CA, or CDCA (100 μM each) or with just vehicle. Values (normalized for GAPDH mRNA) are given as fold expression relative to values in the respective controls with just vehicle. Data are mean ± SD; n = 6 (except for GUDCA, TUDCA, CA, and CDCA — with and without DEX — for which n = 3 each). *P < 0.05, **P < 0.01, ***P < 0.001 versus vehicle control.
The combination of UDCA and dexamethasone increases the AE2 anion exchange activity in liver cells.
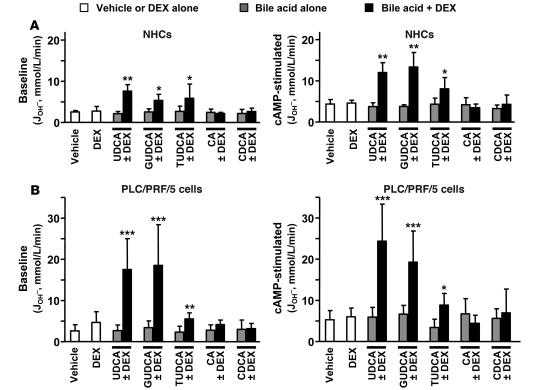
The aforementioned effect of UDCA plus dexamethasone combination on the alternative transcription from the AE2 gene in NHCs and PLC/PRF/5 cells was associated with physiological changes in the Cl–/HCO3– anion exchange (AE) activity displayed by these cells. Both baseline and cAMP-stimulated AE activities were markedly enhanced in the 2 cell lines after 24 hours with the combinations of dexamethasone plus UDCA — either unconjugated or conjugated — compared with controls in the absence of dexamethasone and/or these hydrophilic bile acids (Figure 3). Again, the role of these hydrophilic dihydroxy bile acids in the increased AE activity when combined with dexamethasone was specific, as no effects were observed when using combinations with either CA or CDCA (Figure 3).
Figure 3. The combination of UDCA and dexamethasone increases AE activities in hepatobiliary cells.
Cl–/HCO3– AE activity was determined by microfluorimetry in NHCs (A) and PLC/PRF/5 cells (B) following treatments for 24 hours with either dexamethasone and/or UDCA, GUDCA, TUDCA, CA, or CDCA (100 μM each) or with just vehicle. Treated cells were analyzed for AE activities, both baseline (left) and in the presence of a cAMP-stimulation mixture (right). The AE activity was estimated as the rate of pHi recovery from propionate-induced intracellular alkalinization. Rates of pHi change were measured as δpHi/δt from the tangent to the experimental plot; transmembrane acid fluxes (or equivalent transmembrane base fluxes, JOH– [mmol/l/min]) were calculated as βtot × δpHi/δt, where βtot is the total intracellular buffering power in the presence of CO2/HCO3–. Data are mean ± SD; n = 9 each. *P < 0.05, **P < 0.01, ***P < 0.001 versus vehicle control.
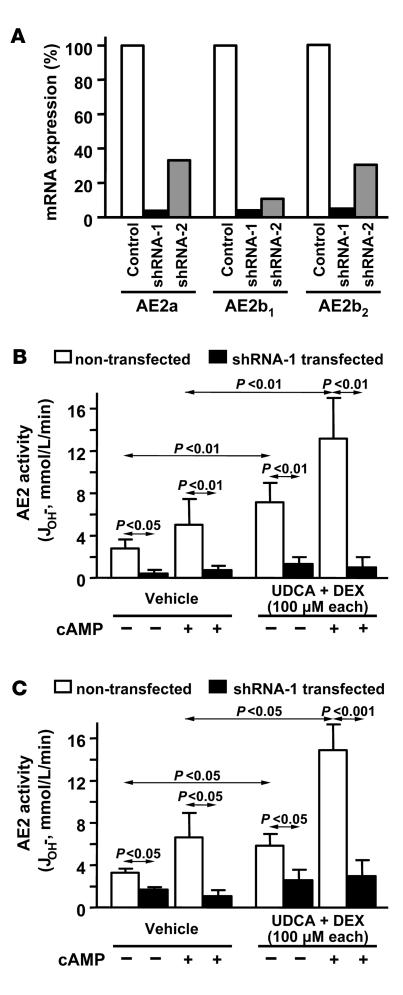
Previous data from knockdown experiments in normal rat cholangiocytes have indicated that the AE activity displayed by these cells is mediated by AE2 protein (21). We therefore inferred that the AE activity displayed by NHCs and PLC/PRF/5 cells is also mediated by AE2 and that the increased AE activity after UDCA plus dexamethasone treatment is most probably due to the enhanced expression of AE2b1 and AE2b2 mRNAs observed in these cells. To assess this possibility, we performed AE2-knockdown experiments in both human liver cell types through cotransfection with silencing pSUPER plasmid constructs — either shRNA-1 or shRNA-2 (described in ref. 21) — together with a normalizing GFP expression vector. PLC/PRF/5 cells could be transfected very efficiently (Supplemental Figure 2), and real-time PCR quantitation showed decreased AE2 mRNA levels with both shRNA plasmid constructs, the silencing effect being more pronounced with shRNA-1 (Figure 4A). We therefore used shRNA-1 for efficient silencing and further microfluorimetric experiments to determine whether AE2 was the carrier mediating the AE activity in PLC/PRF/5 cells as well as in NHCs. After 48 hours of cotransfection, PLC/PRF/5 cells were incubated with and without the combination of UDCA and dexamethasone, and 24 hours later, baseline and cAMP-stimulated AE activities were determined. As shown in Figure 4B, AE activities were markedly reduced under all circumstances in shRNA-1–transfected cells compared with nontransfected cells. These data indicate that AE2 indeed mediates the AE activities we detected in hepatocyte lineage PLC/PRF/5 cells. It may therefore be assumed that the increased AE activity observed in PLC/PRF/5 cells after combined treatment with UDCA plus dexamethasone (Figure 3) is directly related to the enhanced alternative expression of AE2b1 and AE2b2 mRNAs observed in these cells (Figure 2).
Figure 4. AE2 silencing experiments indicate that AE activities displayed by hepatobiliary cells are mediated by AE2.
(A) AE2 silencing was assessed in PLC/PRF/5 cells by quantifying the mRNA levels for AE2a, AE2b1, and AE2b2 isoforms 3 days after transient transfection with pSUPER plasmid constructs (either shRNA-1 silencing construct, shRNA-2 silencing construct, or pSUPER empty vector as control, each together with GFP expression vector). AE2 mRNA values were normalized with GAPDH mRNA levels and corrected for the transfection efficiency with GFP mRNA levels. Values are given as percentage of mRNA levels relative to the control. (B) Baseline and cAMP-stimulated AE activities were determined by microfluorimetry in PLC/PRF/5 cells transfected with shRNA-1 silencing construct (together with GFP expression vector) and treated for 24 hours either with UDCA plus dexamethasone or with just vehicle. (C) Baseline and cAMP-stimulated AE activities were determined by microfluorimetry in NHCs transfected with shRNA-1 silencing construct (together with GFP expression vector) and treated for 24 hours either with UDCA plus dexamethasone or with just vehicle. In both B and C, cotransfected cells could be differentiated from nontransfected cells in the same field of the microscope for their appropriate analysis. Data are mean ± SD; n = 6 each.
Transfection efficiency in cultured primary cholangiocytes was much lower than that in PLC/PRF/5 cells, as indicated by the scarce green fluorescence signals displayed by the NHCs cotransfected with the normalizing GFP expression vector (Supplemental Figure 2). Although such a low transfection efficiency, with most of the cells not being transfected, precluded an appropriate assessment of mRNA silencing in NHC total RNA, GFP-based discrimination of the few transfected cells and subsequent microfluorimetric analysis of their AE activities showed a significant reduction in those transfected cells (putatively silenced for AE2 expression), compared with nontransfected cells (Figure 4C).
Combined treatment with UDCA and dexamethasone enhances the transcriptional activity of the AE2 alternate promoter.
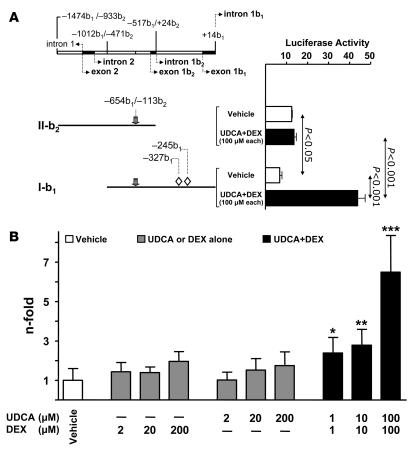
We then carried out in vitro and in vivo luciferase reporter gene assays to analyze the effects of UDCA and dexamethasone on the human AE2 alternate promoter regions. For in vitro experiments, we used PLC/PRF/5 cells because of their adequate transfection efficiency, which is much higher than that of cultured primary cholangiocytes (Supplemental Figure 2). Thus, PLC/PRF/5 cells were transiently transfected with either: (a) construct I-b1 which contains 1 kb of the AE2b2/AE2b1 overlapping promoter sequence proceeding upstream from exon 1b1; or (b) construct II-b2, which contains a 0.9-kb sequence upstream to exon 1b2 and lacks the promoter region downstream to exon 1b2 (and therefore does not contain the 2 putative GREcore motifs located between exons 1b2 and 1b1, at positions –327b1 and –245b1; Figure 5). Dual luciferase analysis of transfected PLC/PRF/5 cells showed that II-b2 promoter displayed higher baseline transcriptional activity than I-b1 (~2-fold; P < 0.05; Figure 6A), and this is in line with previous findings in HepG2 cells (24). But only PLC/PRF/5 cells with the I-b1 construct (containing the AE2b2/AE2b1 overlapping promoter region with the 2 GREcore motifs) displayed a significant response to the administration of UDCA and dexamethasone (100 μM each), with a 6.5-fold increase in the baseline luciferase activity (Figure 6A). The response of the I-b1 promoter region to the UDCA plus dexamethasone treatment was dose dependent, as shown with equimolar combinations ranging from 1 μM to 100 μM each (Figure 6B).
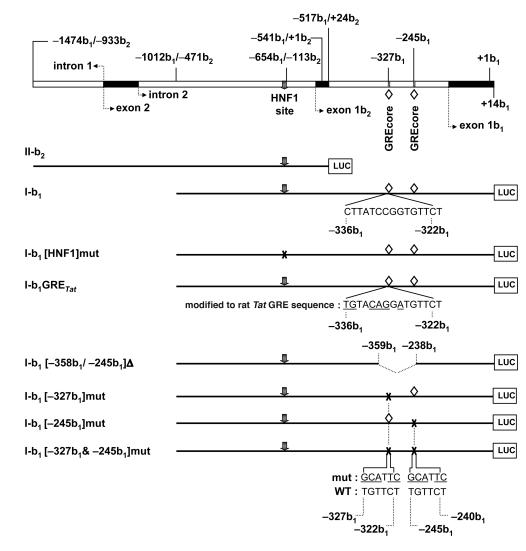
Figure 5. Luciferase constructs with sequences of the human AE2 alternate promoter.
The upper diagram shows AE2 alternate promoter regions from the distal part of intron 1 to the end of exon 1b1. Negative numbers followed by b1 or b2 indicate nucleotide positions upstream to respective +1 positions (i.e., A in ATG start codons) in exons 1b1 or 1b2, respectively. HNF1 site (gray arrow) and 2 GREcore motifs (open rhombuses) are also shown. Luciferase constructs II-b2 and I-b1 contain partial sequences of AE2 alternate overlapping promoter. Construct I-b1[HNF1]mut has the HNF1 site mutated (indicated by X instead of gray arrow). In construct I-b1GRETat, 6 bases (underlined) had been replaced to form the complete GRE site of the rat Tat promoter. I-b1[–358b1/–239b1]Δ is a construct internally deleted for the –358b1/–239b1 region and thus lacking the 2 GREcore motifs. Constructs I-b1[–327b1]mut, I-b1[–245b1]mut, and I-b1[–327b1&–245b1]mut have abrogating mutations in GREcore motifs (indicated by underlined nucleotides above wild-type motif sequences).
Figure 6. In vitro, the combination of UDCA and dexamethasone upregulates the transcriptional activity of AE2 alternate promoter regions in transfected PLC/PRF/5 cells.
(A) Diagram of AE2 promoter regions (gray arrows indicate the HNF1 site; open rhombuses represent GREcore motifs) and luciferase constructs II-b2 and I-b1, with luciferase activities of PLC/PRF/5 cells transiently transfected (0.1 μg of respective construct) and treated for 24 hours either with UDCA plus dexamethasone or with just vehicle. (B) Dose-dependent luciferase activities of PLC/PRF/5 cells transiently transfected with luciferase construct I-b1 (0.1 μg) and treated for 24 hours with different concentrations of UDCA and/or dexamethasone. Luciferase activities (normalized with Renilla activities) are given as fold activity relative to the activity of construct I-b1 in the absence of UDCA and DEX. Data are mean ± SD; n = 9 each. *P < 0.05, **P < 0.01, ***P < 0.001 versus vehicle control.
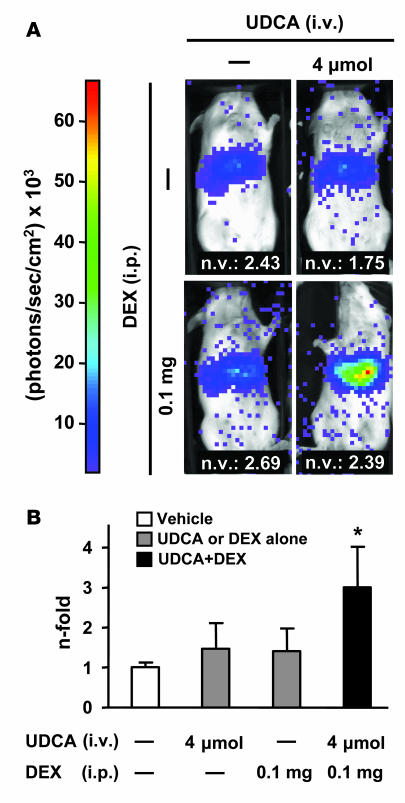
Experiments using hydrodynamic injection with promoter constructs in BALB/c mice and detection of the luciferase activity through in vivo imaging system (IVIS) technology provided parallel results. Mice injected with construct I-b1 responded to the combination of UDCA and dexamethasone with a significant increase in the luciferase activity at the abdominal area corresponding to the liver (Figure 7). On the other hand, the combination of UDCA and dexamethasone had no effect on the in vivo luciferase activity in mice injected with construct II-b2, which does not contain the GREcore motifs (data not shown).
Figure 7. In vivo, the combination of UDCA and dexamethasone upregulates the transcriptional activity of AE2 alternate promoter sequences in injected BALB/c mice.
(A) Seven days after their hydrodynamic injection with luciferase construct I-b1 (30 μg), BALB/c mice were treated with UDCA, dexamethasone, UDCA plus dexamethasone, or just vehicle. After 1 hour, luminescence in the abdominal area was determined by mouse bioluminescence as described in Methods. Typical mouse images corresponding to each treatment are shown. Normalizing values (n.v.) are quotients obtained for each animal after DNA quantifications (through real-time PCR) for the luciferase gene (as transfected gene) and the albumin gene (as endogenous gene) in the liver. (B) Abdominal luminescence in injected animals after the different treatments. Luminescence units were corrected with normalizing values. Normalized values are given as fold corrected luminescence units relative to those in nontreated mice. Data are mean ± SD; n = 4 each. *P < 0.05 versus vehicle control.
Role of specific regulatory elements in the stimulatory effect of UDCA and dexamethasone in the AE2 alternate promoter activity.
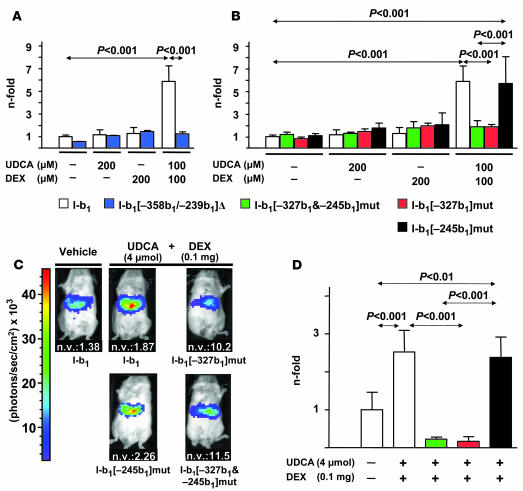
Experiments involving deletion and site-directed mutagenesis were further carried out to clarify the role of the GREcore motifs at positions –327b1 and –245b1 for UDCA plus dexamethasone effects. Deletion of the region –358b1/–239b1, which contains the GREcore motifs (construct I-b1[–358b1/–239b1]Δ), blocked the stimulatory effects of the combination of UDCA and dexamethasone on the luciferase activity in transiently transfected PLC/PRF/5 cells (Figure 8A). Likewise, site-directed mutagenesis of the 2 GREcore motifs (construct I-b1[–327b1&–245b1]mut) led to a similar blockade. Independent site-directed mutagenesis of each GREcore motif indicated that only changes in the motif at position –327b1 (construct I-b1[–327b1]mut) blocked the stimulatory effects of UDCA plus dexamethasone, whereas no blockade was observed after changing the GREcore at –245b1 (construct I-b1[–245b1]mut) compared with the wild-type construct I-b1 (Figure 8B). These in vitro observations with mutated constructs were confirmed in vivo through hydrodynamic injections in BALB/c mice and IVIS luciferase detection. Mice injected with either construct I-b1[–327b1&–245b1]mut or construct I-b1[–327b1]mut showed no response to the combination of UDCA and dexamethasone, while mice injected with construct I-b1[–245b1]mut did respond to the combined treatment, as did mice injected with the wild-type construct I-b1 (Figure 8, C and D).
Figure 8. In vitro and in vivo role of GREcore motifs for the effects of UDCA and dexamethasone on AE2 alternate promoter.
(A) Luciferase activity of PLC/PRF/5 cells transiently transfected either with wild-type construct I-b1 or with deleted construct I-b1[–358b1/–239b1]Δ (cf. Figure 5) and treated for 24 hours with UDCA and/or dexamethasone. (B) Luciferase activity in PLC/PRF/5 cells transfected either with construct I-b1 or with constructs mutated at the GREcore motif(s) (cf. Figure 5) and treated for 24 hours with UDCA and/or DEX. In both A and B, luciferase activities (normalized with Renilla activity) are given as fold activity relative to the activity in cells transfected with construct I-b1 in the absence of UDCA and DEX; data are mean ± SD; n = 9 each. (C) Representative images of abdominal luminescence in BALB/c mice injected with 30 μg of luciferase constructs mutated at the GREcore motif(s) (cf. Figure 5) and treated with UDCA, DEX, UDCA plus dexamethasone, or just vehicle, as described in Methods. Normalizing values are quotients obtained for each animal after DNA quantifications (through real-time PCR) for the luciferase gene (as transfected gene) and the albumin gene (as endogenous gene) in the liver. (D) Values of abdominal luminescence in mice injected and treated as in C. Luminescence units from each animal had been corrected with corresponding normalizing values and are shown as fold corrected luminescence units relative to those in mice injected with construct I-b1 and with just vehicle (i.e., with no UDCA or DEX administration). Data are mean ± SD; n = 4 each.
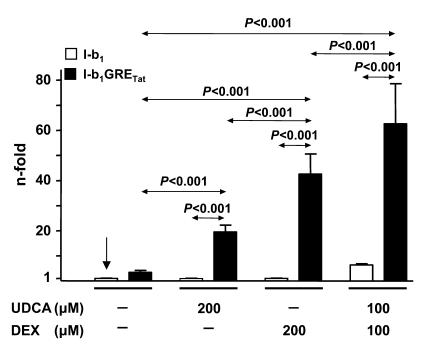
We decided to assess the steric suitability of the AE2 alternate promoter sequences that surround GREcore –327b1 for the GR-mediated response to UDCA and dexamethasone and converted the core motif into a complete GRE site. Mutation of 6 of 9 nucleotides immediately upstream to GREcore –327b1 resulted in the GRE sequence reported for the rat tyrosine aminotransferase (Tat) promoter (39, 40) (construct I-b1GRETat; Figure 5). This complete GRE sequence proved to be functional in the context of the AE2b2/AE2b1 promoter, with good responsiveness to administration of dexamethasone alone; i.e., the luciferase activity in PLC/PRF/5 cells transfected with construct I-b1GRETat was 13-fold higher than that in cells transfected with the wild-type construct I-b1 (Figure 9). Cells transfected with construct I-b1GRETat could also respond to UDCA alone, showing a 6-fold higher luciferase activity than cells with the wild-type construct I-b1 (Figure 9).
Figure 9. Mutations immediately upstream from the GREcore –327b1 site leading to a complete GRE site resulted in full responsiveness of AE2 alternate promoter to either UDCA or dexamethasone alone.
Luciferase activities of PLC/PRF/5 cells transiently transfected with construct I-b1 or I-b1GRETat — the latter having the complete sequence of the GRE site in the rat Tat promoter, as depicted in Figure 5 — and treated for 24 hours with UDCA and/or dexamethasone. Values of luciferase activity (normalized with Renilla values) are given as fold activity relative to the activity in cells transfected with construct I-b1 in the absence of UDCA and DEX (indicated by a vertical arrow). Data are mean ± SD; n = 9 each.
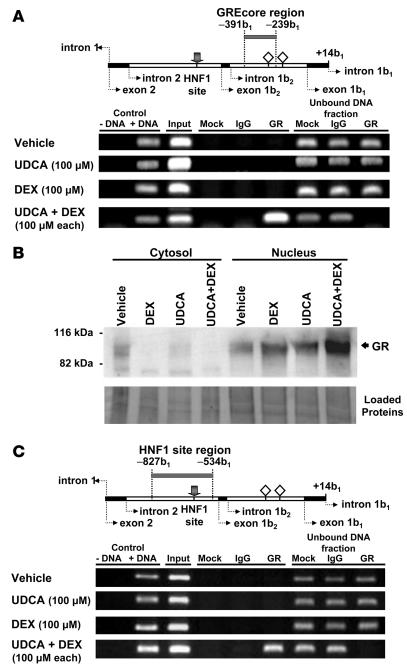
We then explored the occurrence of interaction between the wild-type GREcore –327b1 site and GR and carried out ChIP assays against GR in nuclear extracts from nontransfected PLC/PRF/5 cells exposed to UDCA and/or dexamethasone. As shown in Figure 10A, PCR amplification of the region –391b1/–239b1 (which contains the GREcore –327b1) was obtained when using immunoprecipitates from cells treated with the combination of UDCA and dexamethasone but not those from cells treated with UCDA or dexamethasone alone. On the other hand, compartmentalization of GR via its immunodetection in cytosolic and nuclear fractions showed that each treatment (UDCA plus dexamethasone, UDCA alone, and dexamethasone alone) resulted in translocation of GR to the nucleus (Figure 10B), suggesting that UDCA plus dexamethasone–dependent interaction between GR and the GREcore element at position –327b1 has further requirements in addition to GR translocation.
Figure 10. Interaction of GR with the GREcore site at position –327b1 in the AE2 alternate promoter depends on simultaneous treatment of PLC/PRF/5 cells with UDCA and dexamethasone.
(A) ChIP assays against human GR from extracts of PLC/PRF/5 cells treated for 24 hours with UDCA, dexamethasone, UDCA plus dexamethasone, or just vehicle. In the upper diagram, the gray bar refers to the amplicon –391b1/–239b1 (which includes GREcore –327b1) expected in the PCR amplifications shown below. The following templates were used: DNA negative (just water as negative control), DNA positive (human genomic DNA as positive PCR control), input (start material), mock (no antibody), IgG (rabbit total IgG) — the latter 2 as negative immunoprecipitation controls — and GR immunoprecipitate. Protein A/G–agarose–unbound DNA fractions were also used as control templates. (B) Immunoblots of cytosolic and nuclear extracts from PLC/PRF/5 cells treated as described above, using an antibody against human GR. Control of loaded proteins shown below resulted from ponceau staining of the transferred membrane. (C) In the upper diagram, the gray bar refers to the amplicon –827b1/–534b1 (which includes the HNF1 element). Amplifications shown below were obtained with the same templates as in A. In upper diagrams of A and C, gray arrows indicate the HNF1 site; open rhombuses represent GREcore motifs.
As previously mentioned, AE2b2/AE2b1 alternate promoter possesses a functional HNF1 element near upstream to alternative exon 1b2, at position –113b2 (or –654b1), which is responsible for approximately 70% of the baseline promoter activity (26). To assess a possible interaction of GR with a nuclear factor bound to this HNF1 site (i.e., with HNF1α and/or HNF1β), we used GR immunoprecipitates for PCR amplification of the region –827b1/–534b1, which contains the HNF1 site. Amplification of this HNF1 site region was obtained only after combined treatment with UDCA and dexamethasone (Figure 10C), which indicates that either direct or indirect interaction of GR with HNF1α and/or HNF1β may occur on AE2b2/AE2b1 alternate promoter when these compounds are administered together.
Role of HNF1α in the stimulation of AE2 alternate promoter by UDCA plus dexamethasone in hepatocyte lineage cells.
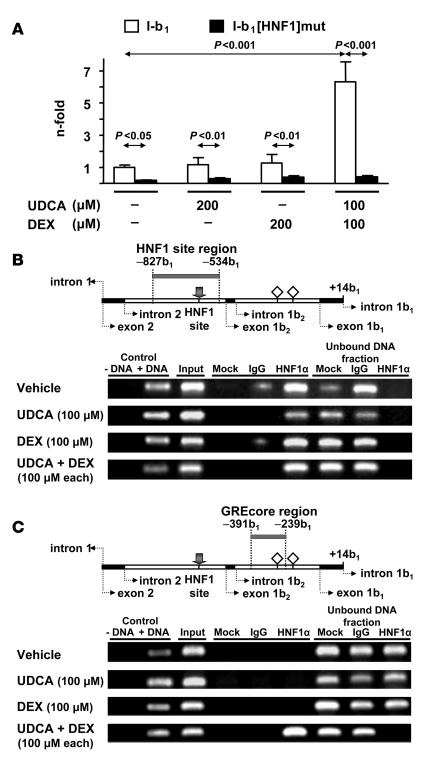
Dual luciferase assay of PLC/PRF/5 cells transiently transfected with construct I-b1 mutated at the HNF1 site (construct I-b1[HNF1]mut; Figure 5) indicated that abrogation of the HNF1 element results in low promoter activity, with no response to the combination of UDCA and dexamethasone or to either of these compounds alone (Figure 11A). The fact that baseline transcriptional activity of construct I-b1[HNF1]mut was also markedly attenuated compared with that of the wild-type construct (4.5-fold; Figure 11A) suggested that an HNF1-bound transcription factor forms part of the baseline transcription machinery of AE2 alternate promoter in intact liver cells. ChIP assays against HNF1α in nuclear extracts from PLC/PRF/5 cells (with and without treatments) further supported such a view. PCR amplification of the region –827b1/–534b1 (with the HNF1 site at position –654b1) using HNF1α immunoprecipitates as template thus yielded a band of the expected size under all circumstances (Figure 11B), which indicates that interaction of HNF1α with its site may also occur at baseline. By contrast, PCR amplification of the region –391b1/–239b1 (where GREcore –327b1 is located), using HNF1α immunoprecipitates as template, yielded the corresponding band only after combined administration of UDCA and dexamethasone (Figure 11C). These data and aforementioned ChIP findings with GR immunoprecipitates (Figure 10C) indicate that while HNF1α may interact at baseline with its site in AE2 alternate promoter, interaction of HNF1α with GR appears to be strictly dependent on the simultaneous administration of UDCA and dexamethasone.
Figure 11. Role of the HNF1α and its interaction with the HNF1 element for the effects on AE2 alternate promoter following simultaneous treatment of PLC/PRF/5 cells with UDCA and dexamethasone.
(A) Luciferase activities of PLC/PRF/5 cells transiently transfected with wild-type construct I-b1 or with HNF1-mutated construct I-b1[HNF1]mut and treated for 24 hours with UDCA and/or dexamethasone. Values of luciferase activity (normalized with Renilla values) are given as fold activity relative to the activity in cells transfected with construct I-b1 treated with just vehicle (i.e., in the absence of UDCA and DEX). Data are mean ± SD; n = 9 each. (B) ChIP assays with HNF1α immunoprecipitates from extracts of PLC/PRF/5 cells treated as described in Figure 10, in which the region that includes the HNF1 element was amplified (amplicon –827b1/–534b1; see the gray bar in the upper diagram). Positive and negative controls were equivalent to those in Figure 10A, but IgG for the negative immunoprecipitation control was goat total IgG. (C) ChIP assays with HNF1α immunoprecipitates in which the region that includes GREcore –327b1 (amplicon –391b1/–239b1 indicated by a gray bar in the upper diagram) was amplified using the same templates as in B. In upper diagrams of B and C, gray arrows indicate the HNF1 site; open rhombuses represent GREcore motifs.
In contrast to the aforesaid results with HNF1α immunoprecipitates, ChIP assays against HNF1β in nuclear extracts from nontransfected PLC/PRF/5 cells showed negligible amplification bands when the HNF1β immunoprecipitates were used as templates (Supplemental Figure 3), suggesting that HNF1β plays no major role in AE2 alternate transcription in these nontransfected hepatocyte lineage cells.
Involvement of p300 in the UDCA plus dexamethasone–dependent interaction of GR with HNF1α on AE2 alternate promoter.
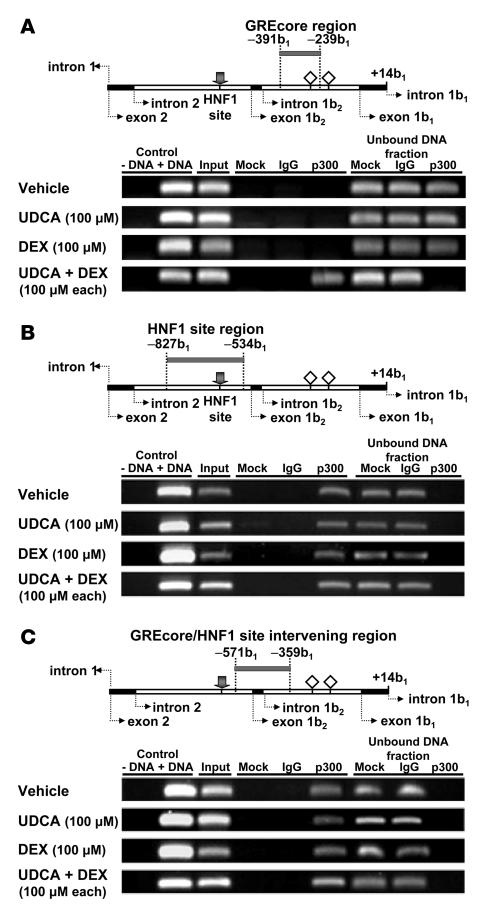
On the basis of previous reports concerning the interaction of GR and HNF1α with coregulatory factors CBP/p300 (30–33, 35) in liver-expressed genes, we carried out additional ChIP assays using an antibody against p300 and searched for a possible linker role of p300 in the UDCA plus dexamethasone–dependent interaction of GR with HNF1α. As shown in Figure 12, A and B, PCR amplifications using p300 immunoprecipitates as template could only detect the GREcore –327b1 region (amplicon –391b1/–239b1) after combined treatment with UDCA and dexamethasone, while the HNF1 site region (amplicon –827b1/–534b1) was detected under all circumstances, i.e., with UDCA alone, dexamethasone alone, UDCA plus dexamethasone, and just vehicle of 50% ethanol. Accordingly, p300 seems to interact with HNF1α regardless of stimulation and could therefore participate in baseline transcription from AE2 alternate promoter. On the other hand, p300 could interact with GR when UDCA and dexamethasone are administered together, suggesting an involvement of p300 in UDCA/dexamethasone-dependent upregulation of AE2 alternate promoter. Because CBP and p300 usually function together as coregulatory factors that interact with DNA-bound transcription factors and other coactivators to form a large acetyltransferase complex, it is possible that CBP/p300 is recruited between HNF1α and GR once GR interacts with GREcore –327b1 following UDCA plus dexamethasone treatment. PCR amplification of the intervening region –571b1/–359b1 between the HNF1 site (at position –654b1 or –113b2) and GREcore –327b1 in the AE2b2/AE2b1 overlapping promoter provides further support for such a view. Thus, while a band of the expected size was regularly obtained, the intensity of the amplified band appeared to be greater after the combined treatment with UDCA and dexamethasone (Figure 12C).
Figure 12. The coregulatory factor p300 may function as a scaffold protein for the interaction of GR and HNF1α on the AE2 alternate promoter in PLC/PRF/5 cells following simultaneous treatment with UDCA and dexamethasone.
(A) ChIP assays with p300 immunoprecipitates from extracts of PLC/PRF/5 cells treated as described in Figure 10, in which the region that includes GREcore –327b1 (amplicon –391b1/–239b1) was amplified. Positive and negative controls were equivalent to those in Figure 10A, but IgG for the negative immunoprecipitation control was mouse total IgG. (B) ChIP assays with p300 immunoprecipitates in which the region that includes the HNF1 element (amplicon –827b1/–534b1) was amplified using the same templates as in A. (C) ChIP assays with p300 immunoprecipitates in which the intervening region between HNF1 and GREcore –327b1 elements (amplicon –571b1/–359b1) was amplified using the same templates as in A. Gray bars in the upper diagrams refer to the corresponding amplicons; gray arrows indicate the HNF1 site; open rhombuses represent GREcore motifs.
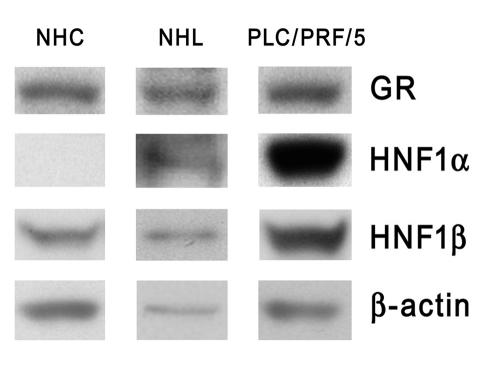
HNF1α is absent in normal human cholangiocytes, and its role might be replaced by HNF1β.
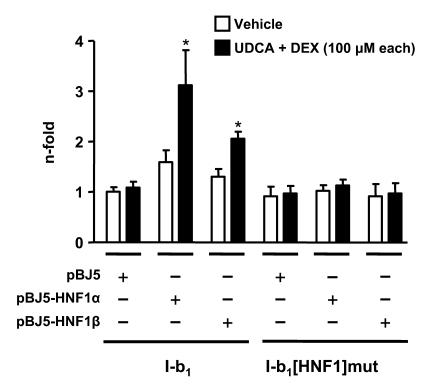
The parallel responses of cholangiocyte and hepatocyte lineage cells to UDCA plus dexamethasone combination (Figures 1–3) supported the idea that the mechanism described above for PLC/PRF/5 cells was also applicable to NHCs. Immunoblot analysis indicated that the 2 cell lineages express GR at similar levels. However, while HNF1α was readily detectable in PLC/PRF/5 cells (as well as in normal human liver extract), it was absent in NHCs, which expressed only the HNF1β isoform (Figure 13). We therefore tried to assess the capability of HNF1β to transactivate the AE2 alternate promoter compared with HNF1α and carried out transactivation experiments in PC3 cells, a cell line that is virtually devoid of HNF1 isoforms (26). Dual luciferase assays showed that PC3 cells indeed became responsive to UDCA plus dexamethasone combination when cotransfected with the luciferase construct I-b1 together with either expression vectors for HNF1α or HNF1β (Figure 14). Cotransfection of any of these HNF1 expression vectors with construct I-b1[HNF1]mut (which is mutated at the HNF1 site; Figure 5), however, resulted in no responses to the combination therapy (Figure 14). These findings indicate that HNF1β is able to specifically transactivate the AE2b2/AE2b1 promoter in a fashion similar to HNF1α.
Figure 13. Western blot analysis using whole-cell extracts from NHCs and PLC/PRF/5 cells.
Total lysate from normal human liver (NHL) (shown in between) was used for a comparison with a normal expression pattern of these proteins in whole liver tissue. Electrophoresed proteins were electrotransferred and probed with antibodies against GR, HNF1α, and HNF1β. Loading control was carried out with an antibody against β-actin.
Figure 14. Non-liver PC3 cells transfected with a luciferase construct with the AE2b2/AE2b1 overlapping promoter sequence become responsive to the combination of UDCA and dexamethasone when cotransfected with expression vectors for either HNF1α or HNF1β.
PC3 cells transfected with construct I-b1 or HNF1 site mutated construct I-b1[HNF1]mut were each cotransfected with either pBJ5-HNF1α, pBJ5-HNF1β, or empty pBJ5 expression vectors and then treated for 24 hours with UDCA and dexamethasone or with just vehicle. Luciferase activities (normalized with Renilla activities) are given as fold activity relative to the activity of construct I-b1 in the absence of UDCA and DEX. Data are mean ± SD; n = 9 each. *P < 0.001 versus vehicle control.
To ascertain possible nuclear interactions involving HNF1β through ChIP assays, cultured NHCs proved unsuitable for this demanding procedure, as the required high number of cells (and continuous cell growth) resulted in a less-differentiated phenotype of these primary cells. We therefore turned to the easily transfectable PLC/PRF/5 cells. ChIP assays against HNF1β in nuclear extracts from PLC/PRF/5 cells transfected with the HNF1β expression vector and further treated with UDCA and/or dexamethasone suggested the occurrence of HNF1β/GR/p300 interactions, as supported by the amplification of expected bands when using the HNF1β immunoprecipitates as templates (Supplemental Figure 4). These results closely resemble those from ChIP assays against HNF1α in nuclear extracts from nontransfected PLC/PRF/5 cells treated with UDCA plus dexamethasone (Figure 11). On the other hand, they are in contrast to the virtual absence of amplified bands when using HNF1β immunoprecipitates from these same nontransfected PLC/PRF/5 cells ( Supplemental Figure 3). Together, our results provide evidence supporting the hypothesis that HNF1β is able to replace HNF1α for interactions with GR — possibly through p300 — on the AE2b2/AE2b1 overlapping promoter after treatment with UDCA plus dexamethasone combination.
Discussion
In recent years, several potential mechanisms have been proposed to explain the beneficial effects of UDCA in various cholestatic conditions (1–3). Among chronic cholestatic disorders, PBC is the most representative disease in which treatment with UDCA is recommended. Daily administration of high doses of UDCA (13–15 mg/kg body wt) has been reported to ameliorate the natural course of the disease (8, 9). Moreover, UDCA has been shown to restore the deficient secretin-dependent biliary bicarbonate excretion in PBC patients (19). Also in PBC, UDCA appeared to improve the diminished liver expression of AE2, which is the anion carrier involved in biliary bicarbonate excretion (20, 21, 41). But almost 40% of PBC patients show incomplete clinical response to UDCA alone, and they are at high risk of severe disease (42). Preliminary trials have provided evidence for the combination of UDCA with glucocorticoids being promising for this subset of PBC patients, as it appears to relieve their cholestasis and liver histological alterations (10–15). To investigate the mechanisms involved in this benefit, we focused on the effects of such a combined therapy in the liver expression of AE2 gene and carried out both in vitro and in vivo experiments. Our findings can be summarized as follows: (a) In human liver cells of both cholangiocyte lineage (NHCs) and hepatocyte lineage (PLC/PRF/5 cells), combination of UDCA and dexamethasone, but not UDCA or dexamethasone alone, markedly upregulated the expression of alternative AE2b1 and AE2b2 mRNA isoforms, with no changes in the levels of AE2a mRNA. (b) Upregulated alternative expression after UDCA/dexamethasone combined treatment was associated with increased AE activities in the 2 liver cell types. (c) UDCA/dexamethasone-dependent upregulation of AE2 alternate promoter was confirmed using luciferase promoter constructs both in vitro and in vivo (transient transfections/dual luciferase assays in PLC/PRF/5 cells and hydrodynamic injections/IVIS luciferase detection in mice). (d) Deletion and site-directed mutagenesis of putative consensus sequences identified a GREcore element directly involved in UDCA/dexamethasone-dependent upregulation. (e) Luciferase assays using a promoter construct with mutated HNF1 site revealed that this element is also involved in UDCA plus dexamethasone effects. And (f) ChIP assays showed that UDCA plus dexamethasone treatment results in interactions between GR and HNF1 isoforms, which possibly involve p300 as well. Together, our findings indicate that the combination of UDCA and glucocorticoids may stimulate AE2-mediated hepatobiliary excretion of bicarbonate through GR/HNF1/p300-dependent upregulation of AE2 alternative expression in liver cells.
Our findings in the initial in vitro experiments with cultured human liver cells from the 2 hepatobiliary cell lineages clearly showed that upregulated AE2 alternative expression following simultaneous administration of UDCA and dexamethasone is associated with increased AE activities (both baseline and cAMP-stimulated). AE2-knockdown experiments confirmed that AE activities displayed by these cells are mediated by AE2 (Figure 4, B and C), which is consistent with earlier data in normal rat cholangiocytes (21). It might therefore be assumed that UDCA/dexamethasone-dependent increases in the AE activity currently observed in our liver cell lineages are related to the enhanced expression of mRNAs for alternative isoforms AE2b1 and AE2b2. It is possible that modulated expression of these 2 highly active AE2 isoforms (25) by UDCA and glucocorticoids may play an important role in the canalicular generation and ductal modification of the bile flow under pathophysiological conditions. During UDCA therapy administered daily to cholestatic patients, and also due to the permanent enterohepatic circulation, the biliary tree becomes highly enriched with this hydrophilic bile acid, which is secreted at the hepatocyte canaliculi after its conjugation with taurine or glycine. Noticeably, the effects observed in our primary human cholangiocytes (and also in hepatocyte lineage PLC/PRF/5 cells) with the combination of UDCA together with dexamethasone remained when using conjugated UDCA (either GUDCA or TUDCA) instead of unconjugated UDCA. On the other hand, the observed effects are specific for these hydrophilic bile acids, as they did not occur when UDCA was replaced by either CA or CDCA in the combination with dexamethasone. Primary bile acids CA and CDCA are less hydrophilic than UDCA because they both have 7-α hydroxylation instead of 7-β hydroxylation (in fact, CDCA is just the 7-α epimer of UDCA).
Both in vitro and in vivo experiments with luciferase constructs indicated that the combination of UDCA and dexamethasone is able to increase the transcriptional activity from AE2b2/AE2b1 overlapping promoter sequences, but only when the region between exons 1b2 and 1b1 is included (Figures 6 and 7). This region has no complete GRE motif, but rather 2 putative GREcore motifs close to each other at positions –327b1 and –245b1 (24). Experiments of deletion and site-directed mutagenesis showed that GREcore –327b1 is the relevant site for the response to the combined treatment with UDCA and dexamethasone (Figure 8).
Baseline GR is known to be present in the cytoplasm as an inactive complex with chaperones (i.e., hsp90 and hsp70), and upon its interaction with glucocorticoids, GR translocates to the nucleus. Once translocated to the nucleus, GR may exert transactivation of genes containing a functional, complete GRE site in their promoters (43–45). But there are genes in which just half of a GRE site, i.e., a GREcore, appears to be involved in glucocorticoid signaling (44). It is possible that GR may need further assistance in addition to its interaction with glucocorticoids for appropriate binding to GREcore sites. In fact, our data suggest that UDCA could provide such assistance to GR for its interaction with GREcore –327b1 in the AE2 alternate promoter and further upregulation. Certainly UDCA has structural similarities to steroids, and administration of UDCA has been previously reported to result in translocation of GR to the nucleus, although the precise interaction of this receptor with UDCA is not yet fully elucidated (36–38). In PLC/PRF/5 cells, we found that UDCA administration indeed resulted in translocation of GR to the nucleus, similar to what was observed following administration of dexamethasone (Figure 10B). We tested whether the lack of effect of any of these compounds on the AE2 alternate promoter was related to a lack of the complete GRE site, or whether it was instead dependent on the structural architecture of AE2 alternate promoter sequences surrounding our GREcore –327b1. As shown in Figure 9, conversion of the core sequence into a complete GRE site in the luciferase reporter–linked AE2b2/AE2b1 promoter construct rendered the construct responsive to the administration of either dexamethasone or UDCA alone. This effect suggests that the promoter sequence surrounding our GREcore –327b1 is not a hindrance for GR to bind its target sequence, but rather that GR is able to acquire a proper steric position on the AE2 alternate promoter for GR/DNA interaction. In the case of the wild-type GREcore –327b1 in AE2 alternate promoter, it is possible that correct GR positioning, GR/GREcore-site binding, and gene transactivation require that GR interact with both UDCA and dexamethasone simultaneously. Our findings with ChIP assays against GR from PLC/PRF/5 nuclear extracts support this view, as they reveal that GR indeed binds to the GREcore site in the AE2 alternate promoter when cells have been treated with the combination of UDCA and dexamethasone, but not when treated with dexamethasone or UDCA alone (Figure 10A), in spite of the fact that each treatment elicits GR translocation (Figure 10B).
In addition to GR, we found that other nuclear factors may be involved in UDCA/dexamethasone-mediated upregulation of AE2 alternate promoter in the liver. An HNF1 site accounting for approximately 70% of the baseline AE2 alternate promoter activity was previously localized 326 bp upstream to the GREcore –327b1 (26). Our present results using a luciferase AE2 promoter construct with mutated HNF1 site for transient transfection of PLC/PRF/5 cells indicate that this site is essential for the response to the combination of UDCA and dexamethasone. Interestingly, ChIP assays against HNF1α and HNF1β in nuclear extracts from nontransfected PLC/PRF/5 cells revealed that HNF1α rather than HNF1β is binding to the HNF1 site and that this occurs regardless of treatment (Figure 11; see also Supplemental Figure 3 for the negligible amplification of bands in HNF1β immunoprecipitates from these nontransfected hepatocytic cells). However, interaction of HNF1α with GR was only observed in PLC/PRF/5 cells that had been treated with UDCA plus dexamethasone combination (Figures 10 and 11). GR and HNF1α have been reported to participate cooperatively in the transcriptional activation of liver-specific genes (27–29). For instance, upregulation of the Pah gene following stimulation with glucocorticoids — or simply cAMP — was found to involve binding of GR and HNF1α to their respective GREcore and HNF1 motifs in the promoter as well as cooperative interplay between GR and HNF1α by unknown mechanisms (29). None of these cases, however, seemed to require UDCA together with dexamethasone for such an interplay.
Additionally, we obtained evidence of involvement of the general coregulatory factor p300 acetyltransferase in the UDCA plus dexamethasone–dependent interaction between HNF1α and GR on AE2 alternate promoter in nontransfected PLC/PRF/5 cells. It is currently known that p300, together with its closely related factor CBP, is present in most cell types (46), where it may interact with a large number of DNA-bound nuclear receptors and nuclear factors to form a large acetyltransferase complex — a useful database for the CBP/p300 interactome is currently available (“CBP & p300 interactome database”; found at http://www.stjude.org/brindle) — and participate in second messenger–regulated gene expression (47–50). Curiously, high doses of UDCA (500 μM) have recently been reported to diminish histone acetylation in several cell lines from colon cancer (51), but this effect appears unrelated to what we have observed in our study, and a direct effect of UDCA on the CBP/p300 complex and interacting nuclear proteins remains to be elucidated. Although the detailed mechanism of action of p300 for AE2 alternate promoter deserves deeper studies, our present data from ChIP assays against p300 in nuclear extracts from nontransfected PLC/PRF/5 cells suggest that this coregulator may interact with DNA-bound HNF1α on the AE2 alternate promoter regardless of any treatment (Figure 12, B and C). HNF1α/p300 interaction has been consistently demonstrated to be involved in liver upregulation of several genes (30, 32, 33). On the other hand, p300 has been described to interact with GR (35, 52). Our ChIP assays against p300 indicate that this interaction between p300 and GR may also occur on the AE2 alternate promoter, but only when cells are treated with the combination of UDCA and dexamethasone (Figure 12A).
Together, these findings suggest that GR coupled to UDCA and dexamethasone can display concerted interactions with GREcore –327b1, CBP/p300, and HNF1α to upregulate AE2 alternate transcription and enhance Cl–/HCO3– exchange activity in hepatocyte lineage PLC/PRF/5 cells. Upregulated expression of AE2b2 transcript appears to be particularly robust compared with that of AE2b1 transcript, and it is possible that exon 1b2 and flanking regions in the AE2b2/AE2b1 overlapping promoter become embraced by the steric disposition of HNF1 site and GREcore –327b1 element and their interacting transcription and/or coregulatory factors. The interactions proposed for the hepatocyte lineage PLC/PRF/5 cells may be similarly postulated for the effects of UDCA plus dexamethasone in upregulating AE2 alternative expression and enhancing Cl–/HCO3– exchange activity in cholangiocyte lineage NHCs. Human cholangiocytes have no expression of HNF1α but do express HNF1β instead, and we have shown that this isoform may replace HNF1α for the proposed interactions. In fact, similar to HNF1α, HNF1β has also been shown to be able to recruit coregulatory factors including CBP/p300 (34). Moreover, we found that human cholangiocytes do express GR — like rat cholangiocytes (53) and many other cell types.
In summary, our findings in liver cells give evidence for a relevant role of the alternative expression from AE2 gene in the physiology and pathophysiology of the hepatobiliary tract. Moreover, they reveal the potentiality of naturally occurring compounds with therapeutic properties such as UDCA and glucocorticoids to modulate the alternative expression. In the case of human intrahepatic cholestatic diseases, activation of this mechanism may result in improved bicarbonate-rich choleresis. This provides rational support for the use of combined therapy with UDCA and glucocorticoids in PBC patients with poor response to UDCA alone.
Methods
Measurement of AE activity in human liver cells.
NHCs, isolated from normal liver explants and cultured as described previously (54), were used after no more than 6 passages. Hepatocyte lineage PLC/PRF/5 cells were grown in DMEM with glutamax, supplemented with 10% FCS and penicillin/streptomycin. To determine AE activity, both cell types were seeded on sterile 12-mm glass coverslips. When cells reached confluence, they were treated for 24 hours with 100 μM dexamethasone and/or 100 μM UDCA (or either TUDCA or CDCA [both from Sigma-Aldrich], GUDCA [Calbiochem], or CA [Fluka]) in 0.5% ethanol or with just vehicle. Baseline and cAMP-stimulated AE activities were assessed by microfluorimetry as described previously (21).
Isolation of total RNA and real-time RT-PCR.
Total RNA was extracted from both NHCs and PLC/PRF/5 cells after 6, 24, or 72 hours of treatment (100 μM UDCA, 100 μM dexamethasone, equimolar combination of both, or vehicle [0.5% ethanol]) with TRI Reagent (Sigma-Aldrich). Additional experiments in which UDCA was replaced by either GUDCA, TUDCA, CA, or CDCA (100 μM each) were also carried out. Reverse transcription was followed by quantitative real-time PCR on resultant cDNA in an iCycler iQ5 Apparatus (Bio-Rad Laboratories) using specific primers for AE2a, AE2b1, and AE2b2 isoforms and for the normalizing control GAPDH, as described previously (24).
Recombinant plasmids with promoter sequences.
Constructs I-b1 and II-b2 were as previously reported (24) (see Figure 5). Construct I-b1[HNF1]mut was obtained from construct I-b1, in which the wild-type HNF1 motif was replaced with the mutated motif in construct –286b2/+24b2mut (26) by using flanking Bsu36I restriction sites. Internally deleted construct I-b1[–358b1/–239b1]Δ with no GREcore sites was obtained through PCR amplifications in 2 steps. First, upstream and downstream fragments with a 14-bp overlapping sequence were amplified from construct I-b1 with the following primers: forward 5′-CCTACCCTGACTTTGCATGA-3′ and tagged reverse 5′-CAGCAGCCTGGCTTCTCCCCTCTGTGA-3′ for fragment –1,012b1/–359b1 and tagged forward 5′-AAGCCAGGCTGCTGGCACCGCTATGGA-3′ and reverse 5′-ACCAACAGTACCGGAATGC-3′ for fragment –238b1/+9b1 (tagged sequences underlined). In a second step, purified overlapping fragments were used for a new amplification with flanking nontagged primers, and resultant amplicon was inserted in SmaI-linearized pGL3-basic (Promega). Mutagenesis to obtain constructs I-b1[–327b1]mut, I-b1[–245b1]mut, and I-b1[–327b1&–245b1]mut with abrogated GREcore motif(s) (TGTTCT→GCATTC) and construct I-b1GRETat with a complete GRE motif (39, 40) (Figure 5) was performed using a Site-Directed Mutagenesis Kit (Stratagene) on construct I-b1 as template and with specific mutagenic 38- to 44-mer oligonucleotides designed according to the manufacturer’s instructions. The integrity of all inserts was confirmed by sequence analysis.
Transient transfection of hepatocyte lineage PLC/PRF/5 cells.
Transient transfections and assessment for luciferase activities with the Dual-Luciferase Reporter Assay System (Promega) were carried out as described previously (26) with minor modifications. Briefly, PLC/PRF/5 cells were seeded on 24-well culture plates and transfected for 16 hours with 0.5 μl Lipofectamine 2000 (Invitrogen) and 1 μg of DNA (900 ng of carrier DNA [vector pGL3-basic] with 1 ng of the internal standard pRL-SV40 construct, both from Promega, and 100 ng of the corresponding promoter construct). Then cells were treated for 24 hours with UDCA and/or dexamethasone at different concentrations (from 1 to 200 μM) and assessed for dual luciferase activities. For the transfection of PLC/PRF/5 cells with the HNF1β expression vector, details are given in Supplemental Figure 4. For transactivation assays with PC3 cells, pBJ5 expression vectors (pBJ5-HNF1α, pBJ5-HNF1β, or the empty vector pBJ5 [all kindly provided by G.R. Crabtree, Stanford University, Stanford, California, USA; ref. 55]; 200 ng each, supplemented with 800 ng of pGL3-basic and 1 ng of pRL-SV40 construct) were added to the cotransfection reaction with the luciferase reporter–linked AE2b2/AE2b1 promoter constructs and treated as described above for PLC/PRF/5 cells (Figure 14).
AE2 gene silencing.
For AE2-knockdown experiments in hepatocyte lineage PLC/PRF/5 cells, we used the previously described plasmid constructs shRNA-1 and shRNA-2 in vector pSUPER (21), which express shRNA molecules under the H1 promoter. PLC/PRF/5 cells were transiently cotransfected using 4 μl of Lipofectamine 2000 with 2 μg of either shRNA construct and 1 ng of GFP expression vector. Three days after cotransfection, the levels of AE2a, AE2b1, and AE2b2 mRNAs were determined by real-time quantitative PCR. AE2 mRNA values were normalized with GAPDH mRNA values as described previously (24). The GAPDH values used were previously corrected for transfection efficiencies using GFP mRNA values (obtained with GFP primers 5′-ATGCCACCTACGGCAAGCTGA-3′ and 5′-GTCCTTGAAGAAGATGGTGC-3′), whole calculations being according to the formula AE2 values/(GAPDH values/GFP values). Silencing of AE activity was determined by microfluorimetry as described previously (21), after discriminating transfected cells by their GFP fluorescence. For AE2 silencing in NHCs, the low transfection efficiency in these primary cells (Supplemental Figure 2) did not allow for appropriate assessments at the mRNA level, but silencing of the AE activity could be determined in the few transfected NHCs, in a manner similar to that described for PLC/PRF/5 cells.
In vivo transfection and treatments of animals.
Aliquots of 2.5 ml of 0.9% NaCl with 30 μg of different AE2 promoter luciferase constructs were injected into 6-week-old female BALB/c mice for hydrodynamics-based in vivo transfection (56, 57). Seven days after transfection, we determined the baseline luciferase activity and treated the animals as follows: controls were given i.v. injection of 50 μl UDCA solvent (0.06 mmol/l NaOH; 0.9% NaCl) and i.p. injection of 50 μl dexamethasone solvent (0.9% NaCl/ethanol mixture; 1:1, vol/vol); for treatments with UDCA and/or dexamethasone, these compounds (4 μmol UDCA and/or 0.1 mg dexamethasone) were added to the respective solvents and injected as for the controls. Anesthetized animals were injected i.p. with 100 μl of 30 mg/ml luciferin (Xenogen) 1 hour after treatments, and luciferase activity was measured 10 minutes afterward using an IVIS Imaging System (Xenogen). Images were processed with Living Image Software version 2.20.1, also from Xenogen. Then animals were sacrificed for liver collection and DNA extraction using a QIAamp DNA Mini Kit (QIAGEN). To normalize the values of luciferase activity, we carried out relative estimation of the copy numbers of luciferase gene (transfection control) and albumin gene (endogenous gene control) by using real-time PCR with 3 μl of DNA samples as template and gene-specific primers. Thus, whole calculations were according to the formula luminescence values/normalizing values, the latter being the ratio of luciferase gene values to albumin gene values. All animal experiments were approved by the Animal Care Committee of the University of Navarra.
Western blot assays.
PLC/PRF/5 cells treated for 24 hours with 100 μM UDCA and/or 100 μM dexamethasone were lysed in 10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, 0.6% NP-40, and Complete Protease Inhibitor (CPI; Roche Diagnostics), followed by 30 seconds of centrifugation (16,000 g at 4°C). Once supernatants (cytosolic fractions) were collected, remaining pellets were washed with lysis buffer (without NP-40) and dissolved in 20 mM HEPES pH 7.9, 420 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 10% glycerol, and CPI, through vigorous shaking (20 minutes). After 10 minutes of centrifugation (16,000 g at 4°C), the resulting supernatants were used as nuclear extracts. Proteins (20 μg) from both cytosolic fractions and nuclear extracts were subjected to SDS-PAGE, blotted to nitrocellulose membranes, and probed with a rabbit antibody against human GR (Santa Cruz Biotechnology Inc.). We also carried out immunodetections in whole extracts from NHCs and PLC/PRF/5 cells. Thus, 20 μg of whole cell extracts were resolved in SDS-PAGE gels, transferred to nitrocellulose, and probed with antibodies against human GR, HNF1α (both from Santa Cruz Biotechnology Inc.), HNF1β (Everest Biotech), and β-actin (Sigma-Aldrich). Immunoblot visualization was performed with ECL reagent (PerkinElmer LAS) and ImageQuant Capture (GE Healthcare).
ChIP assays.
We proceeded as previously described (58) with minor modifications. Briefly, hepatocyte lineage PLC/PRF/5 cells treated with UDCA and/or dexamethasone were fixed with 1% formaldehyde (10 minutes at room temperature), washed twice with ice-cold PBS, and lysed in 5 mM Tris-HCl pH 8.0, 85 mM KCl, 0.5% NP-40, and CPI. Nuclei were pelleted at 3,000 g and resuspended in 50 mM Tris-HCl pH 8.1, 10 mM EDTA, 1% SDS, and CPI. Chromatin was sheared into approximately 500-bp fragments using a Bandelin sonicator (6 cycles at 15 seconds each, at 30% maximum power) followed by centrifugation to pellet debris. Aliquots with supernatant proteins (150 μg in 500 μl of 20 mM HEPES, pH 7.9, 0.5 mM DTT, 20% glycerol, and CPI) were incubated for 15 min at 4°C with 50 μl of protein A/G–agarose beads, BSA, and total IgG from mouse, goat, or rabbit — depending on the host species for immunoprecipitating antibodies (each total IgG was from Santa Cruz Biotechnology Inc.). Immunoprecipitations were carried out overnight at 4°C with the following antibodies (2 μg): rabbit anti-human GR, goat anti-human HNF1α, goat anti-human HNF1β (sc-8992 X, sc-6547 X, sc-7411 X, respectively; all from Santa Cruz Biotechnology Inc.) or mouse anti-human p300 (BD Biosciences — Pharmingen), as well as species-specific total IgG for the negative controls. Immunoprecipitates were collected and washed twice with 2 mM EDTA and 50 mM Tris-HCl pH 8.0 and then 4 times with 100 mM Tris-HCl pH 8.0, 500 mM LiCl, 1% NP-40, and 1% deoxycholic acid. Protein-DNA complexes were then eluted from protein A/G–agarose beads by 2 washings with a freshly prepared solution (50 mM NaHCO3, 1% SDS), with brief shaking and further centrifugation (1,000 g, 5 minutes). Collected supernatants were incubated overnight with RNase A (10 μg/ml) at 67°C. Prior to PCR, immunoprecipitates and corresponding input chromatin were digested with proteinase K and extracted with phenol-chloroform as described previously (58). Amplicons –391b1/–239b1 (which includes GREcore site at –327b1) and –827b1/–534b1 (with the HNF1 element) were obtained by using the following primers: forward 5′-ACAGGAGCCTTCCTCACAGA-3′ and reverse 5′-CAGAACAAGGAAGAGCGCGGGGAGGGCT-3′ for the former and forward 5′-TAGAGGCGCAGGTCAAGTCT-3′ and reverse 5′-AAGTCCATGGCCGGCAGGGGTGAAC-3′ for the latter. Finally, to amplify a promoter region between HNF1 element and GREcore site at –327b1 (stretch from –571b1 and –359b1), we used 5′-CCCTCCTTCTCAGGTTCACCCCTGCC-3′ and 5′-CTGGCTTCTCCCCTCTGTGA-3′ as forward and reverse primers, respectively. PCR products electrophoresed in agarose gels with ethidium bromide were assessed in a GelDoc XR Analyzer (Bio-Rad).
Statistics.
Data were analyzed by nonparametric tests (Kruskall-Wallis test followed by Mann-Whitney U test adjusted with Bonferroni correction) and expressed as mean ± SD. Two-tailed P values less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We are very grateful to E. Sáez and S. Sarvide for excellent technical assistance. We would also like to thank I. Uriarte and S. Melero for their help. Supported through the Spanish “UTE for CIMA project” and by grants from the Spanish Ministry of Education and Science (SAF2005-02468 and SAF2006-07818) and the Carlos III Institute of Health (FIS-01/0777, C03/02, and CIBERehd).
Footnotes
Nonstandard abbreviations used: AE, anion exchange; AE2, anion exchanger 2; CA, cholic acid; CBP, cAMP response element–binding protein–binding protein; CDCA, chenodeoxycholic acid; GR, glucocorticoid receptor; GRE, glucocorticoid response element; GREcore, GRE core sequence; GUDCA, glycine-conjugated UDCA; HNF1α, hepatocyte nuclear factor 1α; IVIS, in vivo imaging system; NHC, normal human cholangiocyte; p300, E1A-binding protein (300 kDa); Pah, rat phenylalanine hydroxylase gene; PBC, primary biliary cirrhosis; Tat, rat tyrosine aminotransferase gene; TUDCA, taurine-conjugated UDCA; UDCA, ursodeoxycholic acid.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 118:695–709 (2008). doi:10.1172/JCI33156
References
- 1.Beuers U. Drug insight: mechanisms and sites of action of ursodeoxycholic acid in cholestasis. Nat. Clin. Pract. Gastroenterol. Hepatol. 2006;3:318–328. doi: 10.1038/ncpgasthep0521. [DOI] [PubMed] [Google Scholar]
- 2.Paumgartner G., Beuers U. Ursodeoxycholic acid in cholestatic liver disease: mechanisms of action and therapeutic use revisited. Hepatology. 2002;36:525–531. doi: 10.1053/jhep.2002.36088. [DOI] [PubMed] [Google Scholar]
- 3.Lazaridis K.N., Gores G.J., Lindor K.D. Ursodeoxycholic acid ‘mechanisms of action and clinical use in hepatobiliary disorders’. J. Hepatol. 2001;35:134–146. doi: 10.1016/s0168-8278(01)00092-7. [DOI] [PubMed] [Google Scholar]
- 4.Poupon R., et al. Is ursodeoxycholic acid an effective treatment for primary biliary cirrhosis? Lancet. 1987;1:834–836. doi: 10.1016/s0140-6736(87)91610-2. [DOI] [PubMed] [Google Scholar]
- 5.Heathcote E.J., et al. The Canadian Multicenter Double-blind Randomized Controlled Trial of ursodeoxycholic acid in primary biliary cirrhosis. Hepatology. 1994;19:1149–1156. [PubMed] [Google Scholar]
- 6.Lindor K.D., et al. Ursodeoxycholic acid in the treatment of primary biliary cirrhosis. Gastroenterology. 1994;106:1284–1290. doi: 10.1016/0016-5085(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 7.Angulo P., et al. Long-term ursodeoxycholic acid delays histological progression in primary biliary cirrhosis. Hepatology. 1999;29:644–647. doi: 10.1002/hep.510290301. [DOI] [PubMed] [Google Scholar]
- 8.Corpechot C., et al. The effect of ursodeoxycholic acid therapy on the natural course of primary biliary cirrhosis. Gastroenterology. 2005;128:297–303. doi: 10.1053/j.gastro.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 9.Pares A., Caballeria L., Rodes J. Excellent long-term survival in patients with primary biliary cirrhosis and biochemical response to ursodeoxycholic acid. Gastroenterology. 2006;130:715–720. doi: 10.1053/j.gastro.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 10.Leuschner M., et al. Ursodeoxycholic acid and prednisolone versus ursodeoxycholic acid and placebo in the treatment of early stages of primary biliary cirrhosis. J. Hepatol. 1996;25:49–57. doi: 10.1016/s0168-8278(96)80327-8. [DOI] [PubMed] [Google Scholar]
- 11.Leuschner M., et al. Oral budesonide and ursodeoxycholic acid for treatment of primary biliary cirrhosis: results of a prospective double-blind trial. Gastroenterology. 1999;117:918–925. doi: 10.1016/s0016-5085(99)70351-3. [DOI] [PubMed] [Google Scholar]
- 12.Holtmeier J., Leuschner U. Medical treatment of primary biliary cirrhosis and primary sclerosing cholangitis. Digestion. 2001;64:137–150. doi: 10.1159/000048854. [DOI] [PubMed] [Google Scholar]
- 13.Angulo P., et al. Oral budesonide in the treatment of patients with primary biliary cirrhosis with a suboptimal response to ursodeoxycholic acid. Hepatology. 2000;31:318–323. doi: 10.1002/hep.510310209. [DOI] [PubMed] [Google Scholar]
- 14.Rautiainen H., et al. Budesonide combined with UDCA to improve liver histology in primary biliary cirrhosis: a three-year randomized trial. Hepatology. 2005;41:747–752. doi: 10.1002/hep.20646. [DOI] [PubMed] [Google Scholar]
- 15.Sola S., Amaral J.D., Aranha M.M., Steer C.J., Rodrigues C.M. Modulation of hepatocyte apoptosis: cross-talk between bile acids and nuclear steroid receptors. Curr. Med. Chem. 2006;13:3039–3051. doi: 10.2174/092986706778521823. [DOI] [PubMed] [Google Scholar]
- 16.Knyrim K., Vakil N., Pfab R., Classen M. The effects of intraduodenal bile acid administration on biliary secretion of ionized calcium and carbonate in man. Hepatology. 1989;10:134–142. doi: 10.1002/hep.1840100203. [DOI] [PubMed] [Google Scholar]
- 17.Perez Barriocanal F., Marin J.J., Dumont M., Erlinger S. Influence of backward perfusion on ursodeoxycholate-induced choleresis in isolated in situ rat liver. J. Hepatol. 1990;11:165–171. doi: 10.1016/0168-8278(90)90108-4. [DOI] [PubMed] [Google Scholar]
- 18.Alpini G., et al. Ursodeoxycholate and tauroursodeoxycholate inhibit cholangiocyte growth and secretion of BDL rats through activation of PKC alpha. Hepatology. 2002;35:1041–1052. doi: 10.1053/jhep.2002.32712. [DOI] [PubMed] [Google Scholar]
- 19.Prieto J., et al. Assessment of biliary bicarbonate secretion in humans by positron emission tomography. Gastroenterology. 1999;117:167–172. doi: 10.1016/s0016-5085(99)70564-0. [DOI] [PubMed] [Google Scholar]
- 20.Medina J.F., Martinez A., Vazquez J.J., Prieto J. Decreased anion exchanger 2 immunoreactivity in the liver of patients with primary biliary cirrhosis. Hepatology. 1997;25:12–17. doi: 10.1002/hep.510250104. [DOI] [PubMed] [Google Scholar]
- 21.Banales J.M., et al. Bicarbonate-rich choleresis induced by secretin in normal rat is taurocholate-dependent and involves AE2 anion exchanger. Hepatology. 2006;43:266–275. doi: 10.1002/hep.21042. [DOI] [PubMed] [Google Scholar]
- 22.Alper S.L. Molecular physiology of SLC4 anion exchangers. Exp. Physiol. 2006;91:153–161. doi: 10.1113/expphysiol.2005.031765. [DOI] [PubMed] [Google Scholar]
- 23.Romero M.F., Fulton C.M., Boron W.F. The SLC4 family of HCO3– transporters. . Pflugers Arch. 2004;447:495–509. doi: 10.1007/s00424-003-1180-2. [DOI] [PubMed] [Google Scholar]
- 24.Medina J.F., Lecanda J., Acin A., Ciesielczyk P., Prieto J. Tissue-specific N-terminal isoforms from overlapping alternate promoters of the human AE2 anion exchanger gene. Biochem. Biophys. Res. Commun. 2000;267:228–235. doi: 10.1006/bbrc.1999.1951. [DOI] [PubMed] [Google Scholar]
- 25.Kurschat C.E., et al. Alkaline-shifted pHo sensitivity of AE2c1-mediated anion exchange reveals novel regulatory determinants in the AE2 N-terminal cytoplasmic domain. J. Biol. Chem. 2006;281:1885–1896. doi: 10.1074/jbc.M509734200. [DOI] [PubMed] [Google Scholar]
- 26.Malumbres R., et al. HNF1α upregulates the human AE2 anion exchanger gene (SLC4A2) from an alternate promoter. . Biochem. Biophys. Res. Commun. 2003;311:233–240. doi: 10.1016/j.bbrc.2003.09.200. [DOI] [PubMed] [Google Scholar]
- 27.Suh D.S., Rechler M.M. Hepatocyte nuclear factor 1 and the glucocorticoid receptor synergistically activate transcription of the rat insulin-like growth factor binding protein-1 gene. Mol. Endocrinol. 1997;11:1822–1831. doi: 10.1210/mend.11.12.0021. [DOI] [PubMed] [Google Scholar]
- 28.Lin B., Morris D.W., Chou J.Y. Hepatocyte nuclear factor 1α is an accessory factor required for activation of glucose-6-phosphatase gene transcription by glucocorticoids. DNA Cell Biol. 1998;17:967–974. doi: 10.1089/dna.1998.17.967. [DOI] [PubMed] [Google Scholar]
- 29.Bristeau A., Catherin A.M., Weiss M.C., Faust D.M. Hormone response of rodent phenylalanine hydroxylase requires HNF1 and the glucocorticoid receptor. Biochem. Biophys. Res. Commun. 2001;287:852–858. doi: 10.1006/bbrc.2001.5673. [DOI] [PubMed] [Google Scholar]
- 30.Soutoglou E., Papafotiou G., Katrakili N., Talianidis I. Transcriptional activation by hepatocyte nuclear factor-1 requires synergism between multiple coactivator proteins. J. Biol. Chem. 2000;275:12515–12520. doi: 10.1074/jbc.275.17.12515. [DOI] [PubMed] [Google Scholar]
- 31.Soutoglou E., et al. Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity. EMBO J. 2001;20:1984–1992. doi: 10.1093/emboj/20.8.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ban N., et al. Hepatocyte nuclear factor-1α recruits the transcriptional co-activator p300 on the GLUT2 gene promoter. Diabetes. 2002;51:1409–1418. doi: 10.2337/diabetes.51.5.1409. [DOI] [PubMed] [Google Scholar]
- 33.Dohda T., et al. Transcriptional coactivators CBP and p300 cooperatively enhance HNF1α–mediated expression of the albumin gene in hepatocytes. J. Biochem. (Tokyo) 2004;136:313–319. doi: 10.1093/jb/mvh123. [DOI] [PubMed] [Google Scholar]
- 34.Barbacci E., et al. HNF1β/TCF2 mutations impair transactivation potential through altered co-regulator recruitment. Hum. Mol. Genet. 2004;13:3139–3149. doi: 10.1093/hmg/ddh338. [DOI] [PubMed] [Google Scholar]
- 35.Almlof T., Wallberg A.E., Gustafsson J.A., Wright A.P. Role of important hydrophobic amino acids in the interaction between the glucocorticoid receptor tau 1-core activation domain and target factors. Biochemistry. 1998;37:9586–9594. doi: 10.1021/bi973029x. [DOI] [PubMed] [Google Scholar]
- 36.Miura T., et al. Functional modulation of the glucocorticoid receptor and suppression of NF-κB-dependent transcription by ursodeoxycholic acid. . J. Biol. Chem. 2001;276:47371–47378. doi: 10.1074/jbc.M107098200. [DOI] [PubMed] [Google Scholar]
- 37.Sola S., et al. Nuclear translocation of UDCA by the glucocorticoid receptor is required to reduce TGF-β1-induced apoptosis in rat hepatocytes. Hepatology. 2005;42:925–934. doi: 10.1002/hep.20870. [DOI] [PubMed] [Google Scholar]
- 38.Weitzel C., et al. Ursodeoxycholic acid induced activation of the glucocorticoid receptor in primary rat hepatocytes. Eur. J. Gastroenterol. Hepatol. 2005;17:169–177. doi: 10.1097/00042737-200502000-00007. [DOI] [PubMed] [Google Scholar]
- 39.Eloranta J.J., Jung D., Kullak-Ublick G.A. The human Na+-taurocholate cotransporting polypeptide gene is activated by glucocorticoid receptor and peroxisome proliferator-activated receptor-γ coactivator-1α, and suppressed by bile acids via a small heterodimer partner-dependent mechanism. . Mol. Endocrinol. 2006;20:65–79. doi: 10.1210/me.2005-0159. [DOI] [PubMed] [Google Scholar]
- 40.Truss M., Beato M. Steroid hormone receptors: interaction with deoxyribonucleic acid and transcription factors. Endocr. Rev. 1993;14:459–479. doi: 10.1210/edrv-14-4-459. [DOI] [PubMed] [Google Scholar]
- 41.Prieto J., Qian C., Garcia N., Diez J., Medina J.F. Abnormal expression of anion exchanger genes in primary biliary cirrhosis. Gastroenterology. 1993;105:572–578. doi: 10.1016/0016-5085(93)90735-u. [DOI] [PubMed] [Google Scholar]
- 42.Corpechot C., et al. Apolipoprotein E polymorphism, a marker of disease severity in primary biliary cirrhosis? J. Hepatol. 2001;35:324–328. doi: 10.1016/s0168-8278(01)00137-4. [DOI] [PubMed] [Google Scholar]
- 43.Beato M. Gene regulation by steroid hormones. Cell. 1989;56:335–344. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- 44.Schoneveld O.J., Gaemers I.C., Lamers W.H. Mechanisms of glucocorticoid signalling. Biochim. Biophys. Acta. 2004;1680:114–128. doi: 10.1016/j.bbaexp.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 45.Witchel S.F., DeFranco D.B. Mechanisms of disease: regulation of glucocorticoid and receptor levels — impact on the metabolic syndrome. Nat. Clin. Pract. Endocrinol. Metab. 2006;2:621–631. doi: 10.1038/ncpendmet0323. [DOI] [PubMed] [Google Scholar]
- 46.Yuan L.W., Giordano A. Acetyltransferase machinery conserved in p300/CBP-family proteins. Oncogene. 2002;21:2253–2260. doi: 10.1038/sj.onc.1205283. [DOI] [PubMed] [Google Scholar]
- 47.Collingwood T.N., Urnov F.D., Wolffe A.P. Nuclear receptors: coactivators, corepressors and chromatin remodeling in the control of transcription. J. Mol. Endocrinol. 1999;23:255–275. doi: 10.1677/jme.0.0230255. [DOI] [PubMed] [Google Scholar]
- 48.Vo N., Goodman R.H. CREB-binding protein and p300 in transcriptional regulation. . J. Biol. Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 49.Black J.C., Choi J.E., Lombardo S.R., Carey M. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol. Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 50.Guermah M., Palhan V.B., Tackett A.J., Chait B.T., Roeder R.G. Synergistic functions of SII and p300 in productive activator-dependent transcription of chromatin templates. Cell. 2006;125:275–286. doi: 10.1016/j.cell.2006.01.055. [DOI] [PubMed] [Google Scholar]
- 51.Akare S., et al. Ursodeoxycholic acid modulates histone acetylation and induces differentiation and senescence. Int. J. Cancer. 2006;119:2958–2969. doi: 10.1002/ijc.22231. [DOI] [PubMed] [Google Scholar]
- 52.Li Q., Su A., Chen J., Lefebvre Y.A., Hache R.J. Attenuation of glucocorticoid signaling through targeted degradation of p300 via the 26S proteasome pathway. Mol. Endocrinol. 2002;16:2819–2827. doi: 10.1210/me.2002-0154. [DOI] [PubMed] [Google Scholar]
- 53.Alvaro D., et al. Corticosteroids modulate the secretory processes of the rat intrahepatic biliary epithelium. Gastroenterology. 2002;122:1058–1069. doi: 10.1053/gast.2002.32374. [DOI] [PubMed] [Google Scholar]
- 54.Joplin R., et al. Human intrahepatic biliary epithelial cells proliferate in vitro in response to human hepatocyte growth factor. J. Clin. Invest. 1992;90:1284–1289. doi: 10.1172/JCI115992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Courtois G., Baumhueter S., Crabtree G.R. Purified hepatocyte nuclear factor 1 interacts with a family of hepatocyte-specific promoters. Proc. Natl. Acad. Sci. U. S. A. 1988;85:7937–7941. doi: 10.1073/pnas.85.21.7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu F., Song Y., Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 57.Zhang G., Budker V., Wolff J.A. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum. Gene Ther. 1999;10:1735–1737. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- 58.Johnson K.D., Bresnick E.H. Dissecting long-range transcriptional mechanisms by chromatin immunoprecipitation. Methods. 2002;26:27–36. doi: 10.1016/S1046-2023(02)00005-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.