Abstract
Transgenic mice expressing HCV core protein develop hepatic steatosis and hepatocellular carcinoma (HCC), but the mechanism underlying this process remains unclear. Because PPARα is a central regulator of triglyceride homeostasis and mediates hepatocarcinogenesis in rodents, we determined whether PPARα contributes to HCV core protein–induced diseases. We generated PPARα-homozygous, -heterozygous, and -null mice with liver-specific transgenic expression of the core protein gene (Ppara+/+:HCVcpTg, Ppara+/–:HCVcpTg, and Ppara–/–:HCVcpTg mice. Severe steatosis was unexpectedly observed only in Ppara+/+:HCVcpTg mice, which resulted from enhanced fatty acid uptake and decreased mitochondrial β-oxidation due to breakdown of mitochondrial outer membranes. Interestingly, HCC developed in approximately 35% of 24-month-old Ppara+/+:HCVcpTg mice, but tumors were not observed in the other genotypes. These phenomena were found to be closely associated with sustained PPARα activation. In Ppara+/–:HCVcpTg mice, PPARα activation and the related changes did not occur despite the presence of a functional Ppara allele. However, long-term treatment of these mice with clofibrate, a PPARα activator, induced HCC with mitochondrial abnormalities and hepatic steatosis. Thus, our results indicate that persistent activation of PPARα is essential for the pathogenesis of hepatic steatosis and HCC induced by HCV infection.
Introduction
HCV is one of the major causes of chronic hepatitis, whereas patients with persistent HCV infection have a high incidence of hepatocellular carcinoma (HCC) (1, 2). Occurrence of HCC associated with chronic HCV infection has increased over the past 2 decades (3–5), and chronic HCV infection is recognized as a serious debilitating disease. However, the mechanism in which chronic HCV infection mediates hepatocarcinogenesis remains unclear.
HCV core protein was shown to have oncogenic potential (6). To examine how HCV core protein participates in HCV-related hepatocarcinogenesis, transgenic mouse lines were established in which HCV core protein is expressed constitutively in liver at cellular levels similar to those found in chronic HCV-infected patients (7). These mice exhibited hepatic steatosis (7) and insulin resistance (8) as early as 3 months of age; on further aging, these symptoms worsened and hepatic adenomas developed in approximately 30% of mice between 16 and 18 months of age (9). Finally, HCC was found within hepatic adenomas in a classic “nodule-in-nodule” pathology (9). Interestingly, no hepatic inflammation or fibrosis was found in these mice throughout the course of HCC development (9), which suggested that the HCV core protein itself induces hepatic steatosis and HCC independently of hepatitis.
Several studies support the contention that hepatic steatosis promotes the development of HCC (10). Epidemiologic data have identified hepatic steatosis as a major accelerating factor of hepatocarcinogenesis in chronic HCV-infected patients (11). Moreover, increases in ROS production that can cause oxidative DNA damage, mitochondrial abnormalities, and accelerated hepatocyte proliferation were observed in the steatotic livers (12–14). Thus, an intriguing possibility has emerged that alteration of fatty acid metabolism in hepatocytes may be central to the pathogenesis of HCC induced by HCV core protein.
PPARs are ligand-activated nuclear receptors belonging to the steroid/thyroid hormone receptor superfamily; 3 isoforms designated as α, β/δ, and γ exist, all of which are involved in lipid homeostasis (15). PPARα regulates constitutive transcription of genes encoding fatty acid–metabolizing enzymes (16) and is associated with the maintenance of fatty acid transport and metabolism, primarily in liver, kidney, and heart. Administration of PPARα agonists, such as the widely prescribed fibrate drugs clofibrate, gemfibrozil, and fenofibrate, ameliorate hyperlipidemia in humans (17) and hepatic steatosis in mice (18).
On the other hand, long-term administration of PPARα ligands to rodents causes accelerated hepatocyte proliferation, increased ROS generation, and development of HCC (19, 20). Disruption of the PPARα gene was shown to prevent the development of HCC caused by long-term exposure to PPARα activators (21). Interestingly, accumulation of fatty acids/triglycerides in hepatocytes could lead to continuous PPARα activation because of the presence of fatty acid metabolites that serve as natural PPARα ligands. For example, mice lacking expression of the peroxisomal acyl-CoA oxidase (AOX) gene showed massive accumulation of very-long-chain fatty acids in hepatocytes, severe microvesicular steatosis, chronic PPARα activation, and development of hepatic adenoma and HCC by 15 months of age (22). These results suggest a strong contribution of activated PPARα to liver tumorigenesis.
On the basis of several lines of evidence, we hypothesized that PPARα might contribute to hepatocarcinogenesis in HCV core protein–expressing transgenic (HCVcpTg) mice. To explore this possibility, PPARα-homozygous (Ppara+/+), PPARα-heterozygous (Ppara+/–), and PPARα-null (Ppara–/–) mice bearing the HCV core protein gene, designated Ppara+/+:HCVcpTg, Ppara+/–:HCVcpTg, and Ppara–/–:HCVcpTg mice, were generated, and phenotypic changes were examined. Surprisingly, we found that severe hepatic steatosis and HCC induced by HCV core protein developed only in Ppara+/+ mice, which were related to persistent PPARα activation.
Results
Expression of HCV core protein in transgenic mice.
Ppara–/–:HCVcpTg and Ppara+/–:HCVcpTg mice appeared healthy, and body weight in both genotypes was similar to that of Ppara+/+:HCVcpTg and Ppara+/+ mice without the transgene. When hepatic expression of HCV core protein in 9-month-old transgenic mice was examined by immunoblot analysis, it was similar among Ppara+/+:HCVcpTg, Ppara+/–:HCVcpTg, and Ppara–/–:HCVcpTg mice (Figure 1A) and was also similar to expression in HCVcpTg mice reported previously (7, 9). Age and sex had only a minor influence on the hepatic expression of HCV core protein.
Figure 1. Phenotype changes in transgenic mouse liver.
(A) Immunoblot analysis of HCV core protein expression in livers of 9-month-old mice. Because no significant individual differences in the same mouse group were found in the preliminary experiments, 10 mg of liver prepared from each mouse (n = 6 /group) was mixed and homogenized. Whole-liver lysate (50 μg protein) was loaded in each well. The band of actin was used as the loading control. Results are representative of 4 independent experiments. PC, lysate prepared from COS-1 cells overexpressing HCV core protein as a positive control. (B) Histological appearance of hematoxylin- and eosin-stained liver sections from 9-month-old HCVcpTg mice. Upper and lower rows show a lower (×40) and higher (×400) magnification, respectively. Microvesicular and macrovesicular steatosis was found only in Ppara+/+:HCVcpTg mice. No inflammation or hepatocyte degeneration was evident in any of the genotypes. (C) Histological appearance of hematoxylin- and eosin-stained liver sections from 24-month-old HCVcpTg mice. Upper and lower rows show a lower (×40) and higher (×400) magnification, respectively. Hepatic steatosis was marked in Ppara+/+:HCVcpTg mice, but not in other mice. Hepatic inflammation, fibrosis, and hepatocyte degeneration were not observed. In Ppara+/–:HCVcpTg and Ppara–/–:HCVcpTg mice, dysplastic hepatocytes and precancerous lesions were not detected throughout the entire liver. (D) Content of liver triglycerides. Results are expressed as the mean ± SD (n = 6/group) and compared between genotypes at the same age. *P < 0.05 compared with Ppara+/+ nontransgenic mice; **P < 0.05 compared with Ppara+/–:HCVcpTg mice; #P < 0.05 compared with Ppara–/–:HCVcpTg mice.
Requirement of homozygous PPARα for the development of hepatic steatosis in transgenic mice.
Livers of 9-month-old male HCVcpTg mice with or without the Ppara allele were examined. Those of Ppara+/+:HCVcpTg mice were soft, slightly enlarged, and light in color and histologically showed macrovesicular and microvesicular steatosis with no apparent inflammation or hepatocyte necrosis (Figure 1B), in agreement with previous reports (7, 9). Biochemical analysis of liver extracts showed marked hepatic accumulation of triglycerides (Figure 1D). In contrast, livers of 9-month-old Ppara+/–:HCVcpTg and Ppara–/–:HCVcpTg mice showed neither histological abnormalities nor accumulation of triglycerides (Figure 1, B and D). Hepatic levels of free fatty acids in Ppara+/+:HCVcpTg mice were approximately 3 times those in Ppara+/–:HCVcpTg and Ppara–/–:HCVcpTg mice or Ppara+/+ mice not expressing the HCV core protein.
In 24-month-old Ppara+/+:HCVcpTg mice, hepatic steatosis was found (Figure 1C), and the hepatic levels of triglycerides were further increased (Figure 1D). Apparent inflammation, hepatocyte degeneration and necrosis, and fibrosis were not detected. On the other hand, Ppara+/–:HCVcpTg and Ppara–/–:HCVcpTg mice showed no steatosis (Figure 1, C and D). These results indicate that hepatic steatosis develops in Ppara+/+:HCVcpTg mice, but not in Ppara+/–:HCVcpTg and Ppara–/–:HCVcpTg mice.
Hepatic fatty acid and triglyceride metabolism in transgenic mice.
To investigate the mechanism responsible for the development of severe steatosis in Ppara+/+:HCVcpTg mice, the expression of genes associated with fatty acid and triglyceride metabolism in the livers of 9-month-old mice was analyzed using the quantitative RT-PCR method. As shown in Figure 2A, the mRNA levels of genes related to de novo lipogenesis (fatty acid synthase [FAS] and acetyl-CoA carboxylase [ACC]) and secretion of VLDL (microsomal transfer protein [MTP] and apoB) were constant in all groups. The mRNA levels of fatty acid translocase (FAT) and fatty acid transport protein (FATP), which are associated with the uptake of fatty acids into hepatocytes, were significantly increased only in Ppara+/+:HCVcpTg mice, but the mRNA levels of hepatic triglyceride lipase, another contributor to fatty acid uptake, remained unchanged (data not shown). The mRNA levels of liver fatty acid binding protein (L-FABP) were also elevated only in Ppara+/+:HCVcpTg mice. Surprisingly, the mRNA levels of AOX and medium-chain acyl-CoA dehydrogenase (MCAD), a rate-limiting enzymes in the peroxisomal and mitochondrial β-oxidation pathways, respectively, were significantly increased in Ppara+/+:HCVcpTg mice. When fatty acid uptake ability was measured in fresh liver slices, it was significantly enhanced only in Ppara+/+:HCVcpTg mice (Figure 2B). Additionally, plasma free fatty acid levels were higher in these mice than in mice in the other groups. Although there were no differences in fasting plasma glucose levels between the groups, hyperinsulinemia was observed only in Ppara+/+:HCVcpTg mice (Figure 2C), in agreement with the previous observation that significant insulin resistance developed in these mice (8). Similar results were obtained from 24-month-old mice (data not shown). These results combined show that the increased plasma fatty acid levels, which were likely due to enhanced peripheral fatty acid release caused by insulin resistance, and the increase in fatty acid uptake ability are consistent with steatogenesis in Ppara+/+:HCVcpTg mice.
Figure 2. Analyses of factors associated with hepatic fatty acid and triglyceride metabolism.
(A) Expression of genes associated with fatty acid and triglyceride metabolism in 9-month-old mouse livers. Total RNA was extracted from each mouse liver, and mRNA levels were determined by RT-PCR. mRNA levels were normalized by those of GAPDH and subsequently normalized by those in Ppara+/+ nontransgenic mice. Results are expressed as the mean ± SD (n = 6/group). *P < 0.05 compared with Ppara+/+ nontransgenic mice; **P < 0.05 compared with Ppara+/–:HCVcpTg mice; #P < 0.05 compared with Ppara–/–:HCVcpTg mice. (B) Uptake of fatty acids in 9-month-old mouse livers. Liver slices obtained from 3 mice in each group were incubated in medium containing 0.8 mM [1-14C]palmitic acid for 7 h. Fatty acid uptake ability was estimated by the sum of palmitic acid converted to CO2 and ketone bodies with that incorporated into total cellular lipids after incubation. The experiment was repeated 3 times. Results were normalized by those of Ppara+/+ nontransgenic mice and expressed as the mean ± SD. (C) Plasma concentrations of free fatty acids, glucose, and insulin. After an overnight fast, blood was obtained from each mouse and the above variables were determined. Results are expressed as the mean ± SD (n = 6/group).
Decreased mitochondrial β-oxidation in transgenic mice.
Although the transcriptional activities of major β-oxidation enzymes were markedly increased, Ppara+/+:HCVcpTg mice had severe steatosis. To explore this discrepant result, peroxisomal and mitochondrial β-oxidation activities were measured using lignoceric and palmitic acids as substrates, respectively. The lignoceric acid–degrading capacity was increased only in Ppara+/+:HCVcpTg mice, where it corresponded to an increase in AOX expression. However, the capacity for palmitic acid degradation, which occurs particularly in mitochondria, was significantly lower in Ppara+/+:HCVcpTg mice than in Ppara+/–:HCVcpTg and Ppara–/–:HCVcpTg mice (Figure 3A). Thus, decreased mitochondrial β-oxidation ability was considered to be another important mechanism for the development of steatosis induced by the core protein.
Figure 3. Analyses of mitochondrial abnormalities.
(A) Lignoceric and palmitic acid β-oxidation activities in 9-month-old mice. Results are expressed as the mean ± SD (n = 6/group). *P < 0.05 compared with Ppara+/+ nontransgenic mice; **P < 0.05 compared with Ppara+/–:HCVcpTg mice; #P < 0.05 compared with Ppara–/–:HCVcpTg mice. (B) Electron microscopic features of hepatic mitochondria of 9-month-old HCVcpTg mice. Upper and lower rows show a lower and higher magnification, respectively. In Ppara+/+:HCVcpTg mice, some mitochondria showing discontinuance of outer membranes (arrows) and amorphous inner structures were observed. In Ppara+/–:HCVcpTg and Ppara–/–:HCVcpTg mice, mitochondria appeared normal; the scale bars represent 200 nm (top) and 30 nm (bottom), respectively. (C) Immunoblot analysis of cytochrome c in 9-month-old mice. Whole-liver lysate, mitochondrial fraction, or cytosolic fraction (50 μg protein) was loaded in each well. Results are representative of 4 independent experiments. (D) Immunoblot analysis of representative mitochondrial β-oxidation enzymes using a mitochondrial fraction prepared from 9-month-old mouse livers. The mitochondrial fraction (20 μg protein) was loaded in each well. Results are representative of 4 independent experiments. The band intensity was quantified densitometrically and normalized by that in Ppara+/+ nontransgenic mouse. The mean value of the fold changes is shown under the representative band. LACS, long-chain acyl-CoA synthase; CPT, carnitine palmitoyl-CoA transferase.
We further evaluated mitochondrial abnormalities. In electron microscopic examination, discontinuous outer membranes (Figure 3B, arrows) and lack of an internal structure were observed in some mitochondria of Ppara+/+:HCVcpTg mouse livers, in agreement with the previous report (9). However, these abnormalities were not seen in Ppara+/–:HCVcpTg and Ppara–/–:HCVcpTg mice (Figure 3B). Immunoblot analysis showed that cytochrome c, which is usually localized in the mitochondrial intermembrane space, was present in the cytosolic fractions of Ppara+/+:HCVcpTg mice (Figure 3C). Moreover, immunoblot analysis using mitochondrial fractions showed that the expression levels of long-chain acyl-CoA synthase and carnitine palmitoyl-CoA transferase-I, which are enzymes indispensable to the initial step of mitochondrial β-oxidation and are localized mainly in mitochondrial outer membranes, were significantly decreased only in Ppara+/+:HCVcpTg mice (Figure 3D).
Overall, these results suggest that homozygous PPARα is essential to the pathogenesis of hepatic steatosis induced by the HCV core protein, which results from a decrease in mitochondrial fatty acid degradation capacity caused by the breakdown of mitochondrial outer membranes and a disproportionate increase in the uptake of fatty acids. Interestingly, steatosis and the related changes did not occur in Ppara+/– and Ppara–/– mice expressing the HCV core protein, which suggested that these changes were not caused by the core protein itself.
Requirement of homozygous PPARα for hepatic tumor development in transgenic mice.
At 9 months of age, hepatic nodules were not observed at all in transgenic mice, whereas, at 24 months, approximately 35% of Ppara+/+:HCVcpTg mice had macroscopically evident hepatic nodules (Table 1). Microscopically, these nodules had the appearance of well-differentiated HCC with trabecular features, which was consistent with the previous report (9). Surprisingly, Ppara+/–:HCVcpTg and Ppara–/–:HCVcpTg mice of the same ages developed no evidence of hepatic tumors, despite the expression of HCV core protein at similar levels to those found in Ppara+/+:HCVcpTg mice (Table 1). Microscopic examination showed that there were no dysplastic cells or precancerous lesions throughout the livers in Ppara+/–:HCVcpTg and Ppara–/–:HCVcpTg mice (Figure 1C). These results provide strong evidence that homozygous PPARα is essential for hepatic tumorigenesis induced by HCV core protein.
Table 1 .
Incidence of HCC in 24-month-old mice
Increased hepatocyte proliferation only in Ppara+/+:HCVcpTg mice.
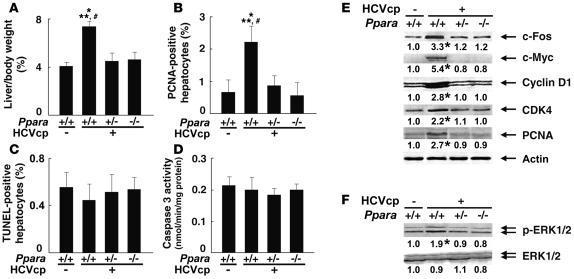
Because sustained acceleration of hepatocyte proliferation relative to apoptosis may promote the development of HCC, these opposing processes were quantified in the livers of 24-month-old mice. Both the liver-to-body weight ratio and the number of hepatocytes expressing proliferating cell nuclear antigen (PCNA) were increased only in Ppara+/+:HCVcpTg mice (Figure 4, A and B). In contrast, the number of TUNEL-positive hepatocytes and the hepatic caspase 3 activity, indicators of hepatocyte apoptosis, remained similar among the 3 mouse strains (Figure 4, C and D). Interestingly, despite the presence of HCV core protein, the amounts of these proliferative and apoptotic markers in Ppara+/–:HCVcpTg and Ppara–/–:HCVcpTg mice were similar to those in Ppara+/+ nontransgenic mice. Expression levels of several proteins, such as protooncogenes (c-Fos and c-Myc), cell-cycle regulators (cyclin D1, cyclin-dependent kinase [CDK] 4, and PCNA), and phosphorylated ERK 1 and 2, all of which are associated with hepatocyte proliferation, were elevated in Ppara+/+:HCVcpTg mice but not in other genotypes (Figure 4, E and F).
Figure 4. Increased hepatocyte proliferation in Ppara+/+:HCVcpTg mice at 24 months of age.
(A) Liver-to-body-weight ratio. Results are expressed as the mean ± SD (n = 6/group). (B) Numbers of proliferating hepatocytes. Two thousand hepatocytes were examined in each mouse, and hepatocyte nuclei positive for anti-PCNA antibody were counted. Results are expressed as the mean ± SD (n = 6/group). For A and B, comparisons are designated as follows: *P < 0.05 compared with Ppara+/+ nontransgenic mice; **P < 0.05 compared with Ppara+/–:HCVcpTg mice; #P < 0.05 compared with Ppara–/–:HCVcpTg mice. (C) Numbers of apoptotic hepatocytes. Liver sections were subjected to TUNEL staining, and TUNEL-positive hepatocyte nuclei were counted in 2,000 hepatocytes from each mouse. Results are expressed as the mean ± SD (n = 6/group). (D) Caspase 3 activity. Results are expressed as the mean ± SD (n = 6/group). (E) Immunoblot analysis of oncogene products and cell cycle regulators. The same sample used in Figure 1A (whole-liver lysate, 50 μg protein) was loaded in each well. The band of actin was used as the loading control. Results are representative of 4 independent experiments. The band intensity was quantified densitometrically, normalized by that of actin, and subsequently normalized by that in Ppara+/+ nontransgenic mice. The mean value of the fold changes is expressed under each band. (F) Immunoblot analysis of phosphorylated ERK1/2 and total ERK1/2. The same samples in Figure 4E (50 μg protein) were used.
Increased oxidative stress and DNA damage only in Ppara+/+:HCVcpTg mice.
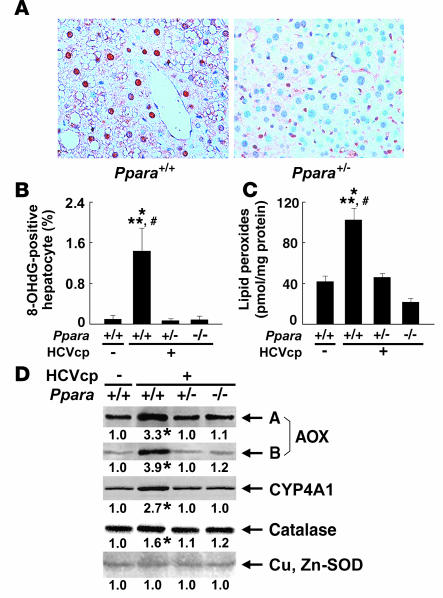
HCV core protein is associated with increased production of ROS (23). Enhanced ROS production induces nuclear DNA damage, which results in the initiation of hepatocarcinogenesis, and can also injure organelles, which can result in disorders in their function. The number of hepatocytes positive for 8-hydroxy-2′-deoxyguanosine (8-OHdG), an indicator of oxidative damage to nuclear DNA, was increased only in 24-month-old Ppara+/+:HCVcpTg mice (Figure 5, A and B). Lipid peroxides were slightly increased in the livers of 9-month-old Ppara+/+:HCVcpTg mice (data not shown) and were more abundant in the livers of 24-month-old Ppara+/+:HCVcpTg mice than in those of Ppara+/–:HCVcpTg and Ppara–/–:HCVcpTg mice or Ppara+/+ nontransgenic mice (Figure 5C). Expression of typical ROS-generating enzymes (AOX and cytochrome P450 4A1 [CYP4A1]) and ROS-eliminating enzymes (catalase and Cu, Zn-SOD) was examined. Immunoblot analysis showed marked increases in the expression of AOX and CYP4A1 and mild increases in that of catalase only in Ppara+/+:HCVcpTg mice. No changes in Cu, Zn-SOD were found in the subgroups of transgenic mice (Figure 5D). These results suggest that enhanced oxidative stress causes damage in nuclear DNA and probably in mitochondria in the Ppara+/+:HCVcpTg mice.
Figure 5. Increased oxidative stress and DNA damage in Ppara+/+:HCVcpTg mice at 24 months of age.
(A) Immunohistochemical staining using antibody against 8-OHdG. In Ppara+/+:HCVcpTg mice, some steatotic hepatocytes were positive for 8-OHdG. Original magnification, ×400. (B) Numbers of 8-OHdG–positive hepatocytes. Hepatocyte nuclei stained with anti-8-OHdG antibody were counted in 2,000 hepatocytes of each mouse. Results are expressed as the mean ± SD (n = 6/group). (C) Hepatic content of lipid peroxides. Results are expressed as the mean ± SD (n = 6 /group). *P < 0.05 compared with Ppara+/+ nontransgenic mice; **P < 0.05 compared with Ppara+/–:HCVcpTg mice; #P < 0.05 compared with Ppara–/–:HCVcpTg mice. (D) Immunoblot analysis of AOX, CYP4A1, catalase, and Cu, Zn-SOD. The whole-liver lysate used in the experiment in Figure 4E (20 μg protein for AOX and CYP4A1 and 50 μg for others) was loaded in each lane. The band of actin was used as the loading control. Results are representative of 4 independent experiments. A and B indicate full-length and truncated AOX, respectively. The band intensity was quantified densitometrically, normalized by that of actin, and subsequently normalized by that in Ppara+/+ nontransgenic mice. The mean value of the fold changes is expressed under each band. *P < 0.05 compared with Ppara+/+ nontransgenic mice.
Persistent and spontaneous PPARα activation in Ppara+/+:HCVcpTg mice.
Liver tumorigenesis induced by long-term exposure to peroxisome proliferators and the related changes, such as sustained hepatocyte proliferation and increased oxidative stress, are associated with persistent PPARα activation (19–21). To examine the activation of PPARα, we quantified the level of PPARα mRNA, which is induced by PPARα activation (24, 25). PPARα mRNA levels were higher in 9-month-old Ppara+/+:HCVcpTg mice than in Ppara+/+ nontransgenic mice (Figure 6A). These increases were more pronounced at 24 months of age. However, there were no differences in PPARα mRNA levels between Ppara+/–:HCVcpTg and Ppara+/– nontransgenic mice at either 9 or 24 months of age. The expression levels of typical PPARα target genes (16, 25, 26) — such as FAT, FATP, L-FABP, AOX, and MCAD (Figure 2); c-Myc, cyclin D1, CDK4, and PCNA (Figure 4); and CYP4A1 (Figure 5) — were simultaneously and synchronously increased in Ppara+/+:HCVcpTg mice, but not in Ppara+/–:HCVcpTg or Ppara–/–:HCVcpTg mice. These results confirm that persistent activation of PPARα occurs only in Ppara+/+:HCVcpTg mice. Various changes observed in Ppara+/+:HCVcpTg mice, i.e., increased fatty acid uptake, mitochondrial abnormalities, steatosis, ROS overproduction, accelerated hepatocyte proliferation, and hepatocarcinogenesis, were considered to be closely linked with sustained PPARα activation.
Figure 6. Persistent PPARα activation in Ppara+/+:HCVcpTg mice.
(A) PPARα mRNA levels. Total RNA was prepared from each mouse, and PPARα mRNA levels were determined by RT-PCR, normalized by those of GAPDH, and subsequently normalized by those of 9-month-old Ppara+/+ nontransgenic mice. Results are expressed as the mean ± SD (n = 6 /group). (B) Immunoblot analysis of nuclear PPARα. The nuclear fraction obtained from each mouse (100 μg protein) was loaded in each well. The band of histone H1 was used as the loading control. Results are representative of 6 independent experiments. The band intensity was quantified densitometrically, normalized by that of histone H1, and subsequently normalized by that in 9-month-old Ppara+/+ nontransgenic mice. The mean value is expressed under each band. *P < 0.05 compared with nontransgenic mice of the same age and Ppara genotype. (C) Pulse-chase experiments for 3, 6, and 12 h and pulse-label (P) experiments for nuclear PPARα using isolated mouse hepatocytes. Left: labeled PPARα bands on x-ray film. Pulse-label and pulse-chase experiments were performed as described in Methods. Results are representative of 4 independent experiments. Right: half-life of PPARα. The band intensity was measured densitometrically and subsequently normalized by that of the pulse-label experiments. The percentage of the band intensity was plotted, and the half-life of PPARα was calculated. Results obtained from 4 independent experiments are expressed as the mean ± SD. *P < 0.05 compared with nontransgenic mice in the same Ppara genotype.
Nuclear PPARα content.
The results described above suggest that persistent PPARα activation is critical to the steatogenesis and hepatocarcinogenesis induced by the HCV core protein. A question arises as to why Ppara+/–:HCVcpTg mice with an active Ppara allele do not exhibit the hallmarks of PPARα activation and do not develop HCC. To address this issue, the nuclear PPARα content was analyzed. Immunoblot analysis for PPARα showed that the amount of nuclear PPARα protein in Ppara+/+:HCVcpTg mice was approximately 2- to 3-fold that of Ppara+/+ nontransgenic mice, which was disproportionate to the higher PPARα mRNA levels (approximately 1.2- to 1.6-fold) (Figure 6, A and B). The level of nuclear PPARα in Ppara+/–:HCVcpTg mice was significantly lower than that in Ppara+/+:HCVcpTg mice and was similar to that in Ppara+/+ nontransgenic mice (Figure 6B). Thus, the lower amount of nuclear PPARα in Ppara+/–:HCVcpTg mice than in Ppara+/+:HCVcpTg mice might have heightened the threshold of expression required for long-term spontaneous PPARα activation.
The degree of an increase in nuclear PPARα levels was evidently higher than the degree of an increase in PPARα mRNA levels in HCVcpTg mice (Figure 6, A and B). To investigate this disparity, the stability of nuclear PPARα was evaluated by pulse-chase experiments using isolated hepatocytes obtained from these mice. The half-life of nuclear PPARα was significantly longer (P < 0.05) in Ppara+/+:HCVcpTg mice (11.5 ± 2.3 h) than in Ppara+/+ nontransgenic mice (5.8 ± 1.4 h) (Figure 6C). The half-life of nuclear PPARα in Ppara+/–:HCVcpTg mice tended to be prolonged compared with that in Ppara+/– nontransgenic mice (Figure 6C). These results suggest that the stability of nuclear PPARα was increased as a result of HCV core protein expression. Because it is known that the core protein interacts with retinoid X receptor α (RXRα) (27) and that PPARα influences the stability of RXRα (28), it is plausible that the core protein would affect its action in nuclei through an interaction with the PPARα-RXRα heterodimer and stabilization of PPARα.
Development of hepatic steatosis and HCC with long-term clofibrate treatment in Ppara+/–:HCVcpTg mice.
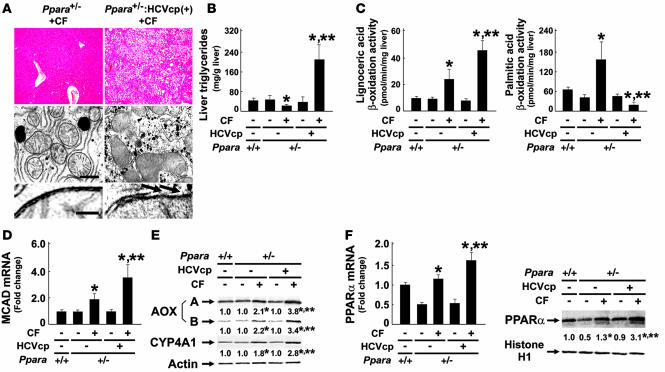
To further confirm the significance of persistent PPARα activation on core protein–induced pathological changes, Ppara+/– and Ppara+/–:HCVcpTg mice were fed a standard diet containing 0.05% clofibrate for 24 months. Interestingly, hepatic steatosis appeared in the clofibrate-treated Ppara+/–:HCVcpTg mice, but not in the Ppara+/– mice under the same treatment conditions (Figure 7, A and B). Similar to our observations in Ppara+/+:HCVcpTg mice not treated with clofibrate, aberrant mitochondria with discontinuous outer membranes and decreased palmitic acid β-oxidation activity were found only in the clofibrate-treated Ppara+/–:HCVcpTg mice (Figure 7, A and C). In addition, levels of MCAD mRNA; AOX, and CYP4A1 proteins; PPARα mRNA; and nuclear PPARα protein were higher in the clofibrate-treated Ppara+/–:HCVcpTg mice than in the clofibrate-treated Ppara+/– mice (Figure 7, D–F), which suggests that the degree of PPARα activation in the former group was greater than that in the latter group and similar to that in Ppara+/+:HCVcpTg mice not treated with clofibrate. Finally, the incidence of HCC after clofibrate treatment was higher in Ppara+/–:HCVcpTg mice (25%; 5 in 20 mice) than in Ppara+/– mice (5%; 1 in 20 mice). Therefore, these results corroborate the importance of constant PPARα activation to the pathogenesis of hepatic steatosis and HCC in the transgenic mice.
Figure 7. Development of hepatic steatosis by long-term treatment of clofibrate in Ppara+/–:HCVcpTg mice.
(A) Histological examination of Ppara+/– and Ppara+/–:HCVcpTg mice treated with diet containing 0.05% (w/w) clofibrate for 24 months (CF). Top: Histological appearance of H&E-stained liver sections. Magnification, ×40. Microvesicular and macrovesicular steatosis were detected only in clofibrate-treated Ppara+/–:HCVcpTg mice. Middle and bottom: Electron microscopic features of hepatic mitochondria. Some C-shaped mitochondria showing discontinuance of outer membranes (arrows) were found in clofibrate-treated Ppara+/–:HCVcpTg mice. Scale bars: 400 nm (middle), 30 nm (bottom). (B and C) Content of liver triglycerides and lignoceric and palmitic acid β-oxidation activities. (D) MCAD mRNA levels. mRNA levels were normalized to those of GAPDH and subsequently normalized to those in Ppara+/+ nontransgenic mice. (E) Immunoblot analysis of AOX and CYP4A1. Whole-liver lysate (20 μg protein) was loaded in each lane. Actin was used as a loading control. Results are representative of 6 independent experiments. (F) PPARα mRNA levels and nuclear PPARα contents. Left: PPARα mRNA levels. The same samples used in D were adopted. Right: Immunoblot analysis of nuclear PPARα. Nuclear fraction obtained from each mouse (100 μg protein) was loaded in each well. Histone H1 was used as a loading control. In E and F, the mean value of the fold changes is shown under each band. Results are representative of 6 independent experiments. Band intensity was quantified densitometrically, normalized to that of the loading control, and subsequently normalized to that in Ppara+/+ nontransgenic mice. *P < 0.05 compared with untreated mice of the same genotype; **P < 0.05 compared with clofibrate-treated Ppara+/– mice without core protein gene. Results are expressed as mean ± SD (n = 6/group).
Discussion
A novel and striking finding in this study is the absolute requirement of persistent PPARα activation for the development of HCV core protein–induced steatosis and HCC. Our data also show that the HCV core protein alone cannot induce steatosis and HCC in transgenic mice.
Mechanisms of development of steatosis in HCVcpTg mice were previously explained as an enhancement of de novo synthesis of fatty acids (29) and a decrease in MTP expression, the latter of which results in insufficient VLDL secretion from hepatocytes (30). In the present study, we revealed 2 novel mechanisms of steatogenesis in the transgenic mice, i.e., an impairment of mitochondrial β-oxidation due to the breakdown of mitochondrial outer membranes and an increase in fatty acid uptake into hepatocytes, associated with PPARα activation. PPARα activation, mitochondrial dysfunction, and hepatic steatosis appeared in 9-month-old Ppara+/+:HCVcpTg mice and continued until 24 months of age, clearly preceding development of HCC. These findings thereby indicate a correlation between PPARα activation, hepatic steatosis, and HCC.
We obtained the novel and rather paradoxical finding that significant PPARα activation, which generally is expected to reduce hepatic triglyceride levels, is essential for the development of severe steatosis induced by HCV core protein. According to the results of this study, the following hypothesis concerning the development of steatosis in Ppara+/+:HCVcpTg mice is proposed. First, the HCV core protein localizes partly in mitochondria (9). A recent study showed that, in isolated mitochondria, the core protein directly increased Ca2+ influx, inhibited electron transport complex I activity, and induced ROS production (31), all of which can increase the fragility of mitochondria and depress mitochondrial function. In addition, the HCV core protein also localizes in nuclei (9) and can coexist in PPARα-RXRα heterodimer through a direct interaction with the DNA-binding domain of RXRα, which enhances the transcriptional activity of PPARα target genes, such as AOX, despite the absence of PPARα ligands in cultured cells (27). The HCV core protein can also be involved in the PPARα-RXRα complex through a direct interaction with cyclic-AMP responsive element binding protein-binding protein (32), which is able to bind to PPARα (33). Thus, the core protein probably serves as a coactivator and stabilizer of PPARα in vivo, which was further confirmed in this study. Moreover, because it is also known that the core protein itself activates ERK1/2 and p38 mitogen-activated protein kinase (34), these activations might phosphorylate PPARα and thereby transactivate it (35). The core protein–induced PPARα activation enhances the basal expression of AOX and CYP4A1, which leads to increased production of ROS and dicarboxylic acids. These toxic compounds can damage mitochondrial outer membranes, which impairs the mitochondrial β-oxidation system. These damages directly induce the accumulation of long-chain fatty acids in hepatocytes. Furthermore, PPARα activation increases the expression of FAT and FATP, which promotes the influx of fatty acids from blood. Long-chain fatty acids and their CoA esters accumulated in hepatocytes are likely to act as potent detergents, which further damages the outer membranes of mitochondria. Fatty acids and their derivatives function as natural ligands of PPARα, which results in the activation of PPARα and the induction of FAT, FATP, AOX, and CYP4A1, which further accelerates mitochondrial damage, the reduction of mitochondrial β-oxidation activity, and the accumulation of fatty acids in a vicious cycle.
Persistent PPARα activation increases oxidative DNA damage because of a disproportionate increase in ROS-generating enzymes relative to the levels of degrading enzymes such as catalase and SOD, which can predispose hepatocytes to malignant transformation. In addition, persistent PPARα activation leads to increased cell division, as revealed by the expression of cell cycle regulators such as cyclin D1 and CDK4. Furthermore, there is little change in apoptosis, which, under normal circumstances, would remove damaged cells capable of undergoing transformation. Thus, under these conditions, it is plausible that some aberrant hepatocytes do not undergo apoptosis and develop into HCC.
It is well known that chronic activation of PPARα is associated with hepatocarcinogenesis in mice exposed to peroxisome proliferators or in mice lacking AOX expression. The common clinicopathological characteristics of HCC in these mice are multicentric HCC (20, 22, 36, 37), the well-differentiated appearance of HCC including trabecular features and often a “nodule-in-nodule” pattern (22, 36, 37), and no evidence of fibrosis or cirrhosis in the nonneoplastic liver parenchyma (22, 36), similar to that observed in Ppara+/+:HCVcpTg mice. However, mice chronically exposed to peroxisome proliferators are clearly distinct from Ppara+/+:HCVcpTg mice in that they have normal mitochondrial organization, increased mitochondrial β-oxidation activity, and no steatosis (16, 36). AOX-null mice are also different from Ppara+/+:HCVcpTg mice with respect to mitochondrial structure (22). These detailed comparisons between the 3 mouse models reveal the importance of mitochondrial abnormalities in the pathogenesis of HCV-related diseases.
PPARα is known to regulate the hepatic expression of many proteins associated with fatty acid and triglyceride metabolism, cell division and apoptosis, oxidative stress generation and degradation, and so forth (15, 16, 20, 21, 24–26); therefore, complete deletion of the PPARα gene from mice might cause hitherto unknown influences on the pathways involved in the development of hepatic steatosis and HCC. To consider these unknown effects, Ppara+/–:HCVcpTg mice were adopted in the current study. Surprisingly, almost all results from Ppara+/–:HCVcpTg mice were similar to those from Ppara–/–:HCVcpTg mice, which demonstrates that the presence of functional PPARα itself is not a prerequisite for the occurrence of steatosis and HCC induced by the HCV core protein. Moreover, a comparison between Ppara+/–:HCVcpTg and Ppara+/+:HCVcpTg mice uncovered an unexpected and important fact that the core protein–dependent pathological changes do not appear without significant activation of PPARα. Thus, it is not the presence of PPARα per se, but rather a high level of PPARα activation that seems to be essential for the development of HCV core protein–induced steatosis and HCC.
To reinforce the abovementioned hypothesis, Ppara+/– and Ppara+/–:HCVcpTg mice were treated with an exogenous PPAR agonist, clofibrate, for 24 months. In Ppara+/– mice, long-term clofibrate treatment caused a certain level of persistent PPARα activation and a low incidence of HCC. Interestingly, in Ppara+/–:HCVcpTg mice, clofibrate treatment induced more intensive PPARα activation and HCC at a much higher incidence, accompanied by damaged mitochondrial outer membranes, severe steatosis, and decreased mitochondrial β-oxidation activity. The results from the clofibrate-treated Ppara+/–:HCVcpTg mice were similar to those of the Ppara+/+:HCVcpTg mice not treated with clofibrate. Therefore, these findings further support the concept that a long-term and high level of PPARα activation is necessary for steatogenesis and hepatocarcinogenesis in HCVcpTg mice and emphasize the significant role of the HCV core protein as a PPARα coactivator in vivo.
A pulse-chase experiment showed that PPARα was stabilized in hepatocyte nuclei in mice expressing the HCV core protein. Many nuclear receptors, including PPARα and RXRα, are known to be degraded by the ubiquitin-proteasome system (38), which plays an important role in modulating the activity of nuclear receptors. Further studies will be needed to clarify whether the core protein influences the ubiquitin-proteasome pathway.
Recent studies have shown conflicting result, i.e., that PPARα was downregulated in the livers of chronic hepatitis C patients (39, 40). Although the association between PPARα function and chronic HCV infection remains a matter of controversy in humans, the changes observed in the transgenic mice resemble in many ways the clinicopathological features of chronically HCV-infected patients; both show a high frequency of accompanying steatosis (10, 40, 41), increased accumulation of carbon 18 monounsaturated fatty acids in the liver (42), mitochondrial dysfunction (43), increased insulin resistance (44) and oxidative stress (45, 46), male-preferential (2) and multicentric occurrence of HCC (47, 48), and the well-differentiated appearance of HCC, including trabecular features and often a “nodule-in-nodule” pattern (47, 48). Thus, it is postulated that the mechanism of steatogenesis and hepatocarcinogenesis we proposed may partially apply to patients with chronic HCV infection. If so, therapeutic interventions to alleviate persistent and excessive PPARα activation might be beneficial in the prevention of HCC. To clarify the exact relationship between PPARα activation and HCV-induced hepatocarcinogenesis in humans, further experiments using noncancerous liver tissues obtained from HCV-related HCC patients and using mice carrying human PPARα and HCV core protein genes are needed.
In conclusion, we clarified for the first time that persistent and potent PPARα activation is absolutely required for the development of severe steatosis and HCC induced by HCV core protein. In addition, we uncovered paradoxical and specific functions of PPARα in the mechanism of steatogenesis mediated by the core protein. Our results offer clues in the search for novel therapeutic and nutritional management options, especially with respect to neutral lipids, for chronically HCV-infected patients.
Methods
Mice.
The generation of HCVcpTg mice and Ppara–/– mice was described previously (7, 24, 49). Male HCVcpTg mice and female Ppara–/– mice were mated, and F1 mice bearing the HCV core protein gene were intercrossed to produce F2 mice. Ppara+/+, Ppara+/–, and Ppara–/– mice bearing the HCV core protein gene, designated as Ppara+/+:HCVcpTg, Ppara+/–:HCVcpTg, and Ppara–/–:HCVcpTg mice, in the F4 generation were subjected to serial analyses. Because HCC develops preferentially in male HCVcpTg mice (9), male mice were analyzed. Age-matched male Ppara+/+ mice without the core protein gene were used as controls. For identifying genotypes, genomic DNA was isolated from mouse tails and amplified by PCR. Primer pairs were designed as described elsewhere: 5′-GCCCACAGGACGTTAAGTTC-3′ and 5′-TAGTTCACGCCGTCCTCCAG-3′ for the HCV core gene (7) and 5′-CAGAGCAACCATCCAGATGA-3′ and 5′-AAACGCAACGTAGAGTGCTG-3′ for the PPARα gene (24). Amplified alleles for HCV core and PPARα genes were 460 and 472 base pairs, respectively. Five mice per cage were fed a routine diet and were kept free of specific pathogens according to institutional guidelines. For the clofibrate treatment experiments, 2-month-old male Ppara+/– and Ppara+/–:HCVcpTg mice were randomly divided into 2 groups (n = 20 in each group) and were fed either a routine diet or one containing 0.05% (w/w) clofibrate (Wako Pure Chemicals Industries) for 24 months. All mice were killed by cervical dislocation before their livers were excised. If a hepatic tumor was present, the tumor was removed and subjected to histological analysis, and the remaining liver tissues were used for biochemical analyses. All animal experiments were conducted in accordance with animal study protocols outlined in the Guide for the Care and Use of Laboratory Animals prepared by the National Academy of Sciences and approved by the Shinshu University School of Medicine.
Preparation of nuclear, mitochondrial, and cytosolic fractions.
Approximately 400 mg of liver tissue was minced on ice and transferred to 10% (w/v) isolation buffer (250 mM sucrose in 10 mM Tris-HCl [pH 7.4] and 0.5 mM EGTA and 0.1% bovine serum albumin [pH 7.4]). The samples were gently homogenized by 10–20 strokes with a chilled Dounce homogenizer (Wheaton) and loose-fitting pestle. The homogenate was centrifuged at 500 × g for 5 min at 4°C. The supernatant was retained, and the resulting pellet was resuspended with isolation buffer and centrifuged at 4,500 g for 10 min at 4°C. The pellet fraction was suspended again and centrifuged at 20,000 × g for 1 h at 4°C, and the resulting pellet was used as the nuclear fraction. The combined supernatant fractions were centrifuged at 7,800 × g for 10 min at 4°C to obtain a crude mitochondria pellet. This pellet was resuspended with isolation buffer, centrifuged at 7,800 × g for 10 min at 4°C, and used as the mitochondrial fraction. Finally, all supernatant fractions were collected and centrifuged at 20,000 × g for 30 min at 4°C, and the resulting supernatant was used as the cytosolic fraction.
Immunoblot analysis.
Protein concentrations were measured colorimetrically with a BCA Protein Assay kit (Pierce). For the analysis of fatty acid–metabolizing enzymes, hepatocyte mitochondrial fractions or whole-liver lysates (20 μg protein) were subjected to 10% SDS-PAGE (16). For analysis of PPARα, nuclear fractions (100 μg protein) were used. For analysis of other proteins, whole lysates or cytosolic fractions (50 μg protein) were subjected to electrophoresis. After electrophoresis, the proteins were transferred to nitrocellulose membranes, which were incubated with the primary antibody and then with alkaline phosphatase-conjugated goat anti-rabbit or anti-mouse IgG. Antibodies against HCV core protein, fatty acid–metabolizing enzymes, CYP4A1, catalase, and PPARα were described previously (9, 16, 24, 50). Antibodies against other proteins were purchased commercially: cytochrome c antibody from BD Transduction Laboratories and other antibodies from Santa Cruz Biotechnology. The band of actin or histone H1 was used as the loading control. The band intensity was measured densitometrically, normalized to that of actin or histone H1, and subsequently expressed as fold changes relative to that of Ppara+/+ nontransgenic mice.
Analysis of mRNA.
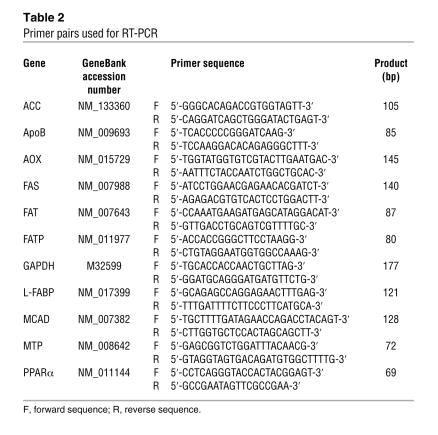
Total liver RNA was extracted using an RNeasy Mini Kit (Qiagen), and cDNA was generated by SuperScript II reverse transcriptase (Gibco BRL). Quantitative RT-PCR was performed using a SYBR green PCR kit and an ABI Prism 7700 Sequence Detection System (Applied Biosystems). The primer pairs used for RT-PCR are shown in Table 2. The mRNA level was normalized to the GAPDH mRNA level and subsequently expressed as fold changes relative to that of Ppara+/+ nontransgenic mice.
Table 2 .
Primer pairs used for RT-PCR
Light microscopy and immunohistochemical analysis.
Small blocks of liver tissue from each mouse were fixed in 10% formalin in phosphate-buffered saline and embedded in paraffin. Sections (4 μm thick) were stained with hematoxylin and eosin. For immunohistochemical localization of PCNA and 8-OHdG, other small blocks of liver tissue were fixed in 4% paraformaldehyde in phosphate-buffered saline. Sections (4 μm thick) then were affixed to glass slides and incubated overnight with mouse monoclonal antibodies against PCNA (1:100 dilution; Santa Cruz) or 8-OHdG (1:10 dilution; Japan Institute for the Control of Aging). Sections were immunostained using EnVision+ kit, with 3,3′-diaminobenzidine as a substrate (DAKO). Hepatocytes positive for PCNA or 8-OHdG were examined in 10 randomly selected ×400 microscopic fields per section. Two-thousand hepatocytes were examined for each mouse, and the number of immunostained hepatocyte nuclei was expressed as a percentage.
Assessment of hepatocyte apoptosis.
TUNEL assay was performed using a MEBSTAIN Apoptosis Kit II (Medical & Biological Laboratories). Two thousand hepatocytes were examined for each mouse, and the number of TUNEL-positive hepatocytes was expressed as a percentage.
Pulse-label and pulse-chase experiments.
Parenchymal hepatocytes were isolated by the modified in situ perfusion method (51). After perfusion with 0.05% collagenase solution (Wako), the isolated hepatocytes were washed 3 times by means of differential centrifugation and the dead cells were removed by density-gradient centrifugation at 500 g for 3 min at 4°C on Percoll (Amersham Pharmacia Biotech). The live hepatocytes were washed and suspended in William’s E medium containing 5% FBS. When the viability of the isolated hepatocytes exceeded 85% as determined by the trypan blue exclusion test, the following experiments were conducted. The isolated hepatocytes were washed twice and incubated in methionine-free medium containing 5% dialyzed FBS for 1 h at 37°C. The medium was replaced with the same medium containing 300 mCi/ml of [35S]methionine (Amersham Pharmacia Biotech). After a 3-h incubation, the labeled medium was exchanged for the standard medium, and the preparation was chased for 3, 6, or 12 h. The labeled cells were washed, homogenized, and centrifuged at 800 g for 5 min at 4°C to obtain a crude nucleus pellet. This pellet was resuspended with isolation buffer and centrifuged at 20,000 g for 1 h at 4°C to prepare the nuclear fraction. The levels of radioactivity in the homogenates of the pulse-labeled preparations were similar between the transgenic and the nontransgenic mice, which suggested that the [35S]methionine uptake capacity in the former hepatocytes was similar to that in the latter. The nuclear fraction was lysed in RIPA buffer (10 mM Tris-HCl, pH 7.4; 0.2% sodium deoxycholate, 0.2% Nonidet P-40, 0.1% SDS, 0.25 mM PMSF, and 10 mg/ml aprotinin). The lysate was incubated for 3 h at 4°C with purified anti-PPARα antibody. The immune complexes were precipitated with Staphylococcus aureus protein A bound to agarose beads. After the precipitates had been washed in RIPA buffer, the labeled proteins were resolved by 10% SDS-PAGE and visualized by autoradiography.
Analysis of fatty acid uptake ability.
Assays for fatty acid uptake were carried out according to a method reported by Graulet et al. (52) with minor modifications. Briefly, 3 mice in each group were fasted overnight. Livers were removed quickly, rinsed in ice-cold saline solution, and cut into 500-μm thick slices with an Oxford Vibratome (Oxford Laboratories). Approximately 150 mg of fresh liver (6–8 liver slices) was placed on stainless steel grids positioned in a 25-ml flask equipped with suspended plastic center wells (Kontes) and incubated in RPMI-1640 medium (Sigma-Aldrich) devoid of fatty acids for 2 h at 37°C. The medium was then replaced with fresh RPMI-1640 medium supplemented with an antibiotic-antimycotic cocktail and 0.8 mM [1-14C]palmitic acid (4 mCi/mmol) (American Radiolabeled Chemicals) complexed to BSA (palmitic acid:albumin molar ratio of 4:1). After a 7-h incubation, the medium was collected and slices were washed with 2 ml of saline solution and homogenized in Tris buffer (25 mM Tris-HCl, pH 8.0; 50 mM NaCl). Fatty acid uptake ability was calculated as the sum of palmitic acid converted to CO2 and ketone bodies with that incorporated into total cellular lipids after incubation. For measurement of CO2 production by the liver slices, the center wells were placed into scintillation vials containing 4 ml of scintillation cocktail, and radioactivity was counted. For measurement of ketone body generation, aliquots of medium (500 μl) and liver homogenates (250 μl) were treated with ice-cold perchloric acid to make final concentrations of 200 mM and were centrifuged at 3,000 g for 20 min at 4°C. Aliquots of the supernatant containing the ketone bodies were introduced into the scintillation vials, and radioactivity was counted. Total cellular lipids were extracted from the liver homogenates according to a modified method developed by Folch et al. (53), collected into scintillation vials, and evaporated to dryness under an air stream; radioactivity was then counted. The experiment was repeated 3 times, and palmitic acid uptake ability was expressed as fold changes relative to that of Ppara+/+ nontransgenic mice.
Other methods.
To determine the hepatic content of lipids and lipid peroxides, lipids were extracted according to a method by Folch et al. (53). Triglycerides and free fatty acids were measured with a Triglyceride E-test kit and a NEFA C-test kit (Wako), respectively. Lipid peroxides (malondialdeyde and 4-hydroxyalkenals) were measured using an LPO-586 kit (OXIS International). Hepatic β-oxidation activity was determined as described previously (16). Hepatic caspase 3 activity was measured as described elsewhere (54). Plasma glucose and insulin levels were determined using a Glucose CII-test kit (Wako) and a mouse insulin ELISA kit (U-type, AKRIN-031; Shibayagi), respectively.
Statistics.
Statistical analysis was performed with a 2-tailed Student’s t test for quantitative variables or with a chi-square test for qualitative variables. Quantitative data are expressed as the mean ± SD. P < 0.05 was considered to be statistically significant.
Acknowledgments
We thank Trevor Ralph for editorial assistance and Chikako Tanaka for helpful suggestions.
Footnotes
Nonstandard abbreviations used: ACC, acetyl-CoA carboxylase; AOX, acyl-CoA oxidase; CDK, cyclin-dependent kinase; CYP4A1, cytochrome P450 4A1; FAS, fatty acid synthase; FAT, fatty acid translocase; FATP, fatty acid transport protein; HCC, hepatocellular carcinoma; HCVcpTg, HCV core protein–expressing transgenic; L-FABP, liver fatty acid-binding protein; MCAD, medium-chain acyl-CoA dehydrogenase; MTP, microsomal transfer protein; 8-OHdG, 8-hydroxy-2′-deoxyguanosine; PCNA, proliferating cell nuclear antigen; RXRα, retinoid X receptor α.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 118:683–694 (2008). doi:10.1172/JCI33594
References
- 1.Kiyosawa K., et al. Interrelationship of blood transfusion, non-A, non-B hepatitis and hepatocellular carcinoma: analysis by detection of antibody to hepatitis C virus. Hepatology. 1990;12:671–675. doi: 10.1002/hep.1840120409. [DOI] [PubMed] [Google Scholar]
- 2.Kiyosawa K., et al. Hepatocellular carcinoma: recent trends in Japan. Gastroenterology. 2004;127(5 Suppl. 1):S17–S26. doi: 10.1053/j.gastro.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka Y., et al. Inaugural article: a comparison of the molecular clock of hepatitis C virus in the United States and Japan predicts that hepatocellular carcinoma incidence in the United States will increase over the next two decades. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15584–15589. doi: 10.1073/pnas.242608099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okuda K., Fujimoto I., Hanai A., Urano Y. Changing incidence of hepatocellular carcinoma in Japan. Cancer Res. 1987;47:4967–4972. [PubMed] [Google Scholar]
- 5.El-Serag H.B., Mason A.C. Rising incidence of hepatocellular carcinoma in the United States. N. Engl. J. Med. 1999;340:745–750. doi: 10.1056/NEJM199903113401001. [DOI] [PubMed] [Google Scholar]
- 6.Shimotohno K. Hepatitis C virus and its pathogenesis. Semin. Cancer Biol. 2000;10:233–240. doi: 10.1006/scbi.2000.0322. [DOI] [PubMed] [Google Scholar]
- 7.Moriya K., et al. Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J. Gen. Virol. 1997;78:1527–1531. doi: 10.1099/0022-1317-78-7-1527. [DOI] [PubMed] [Google Scholar]
- 8.Shintani Y., et al. Hepatitis C virus infection and diabetes: direct involvement of the virus in the development of insulin resistance. Gastroenterology. 2004;126:840–848. doi: 10.1053/j.gastro.2003.11.056. [DOI] [PubMed] [Google Scholar]
- 9.Moriya K., et al. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat. Med. 1998;4:1065–1068. doi: 10.1038/2053. [DOI] [PubMed] [Google Scholar]
- 10.Powell E.E., Jonsson J.R., Clouston A.D. Steatosis: co-factor in other liver diseases. Hepatology. 2005;42:5–13. doi: 10.1002/hep.20750. [DOI] [PubMed] [Google Scholar]
- 11.Ohata K., et al. Hepatic steatosis is a risk factor for hepatocellular carcinoma in patients with chronic hepatitis C virus infection. Cancer. 2003;97:3036–3043. doi: 10.1002/cncr.11427. [DOI] [PubMed] [Google Scholar]
- 12.Browning J.D., Horton J.D. Molecular mediators of hepatic steatosis and liver injury. J. Clin. Invest. 2004;114:147–152. doi: 10.1172/JCI22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le T.H., et al. The zonal distribution of megamitochondria with crystalline inclusions in nonalcoholic steatohepatitis. Hepatology. 2004;39:1423–1429. doi: 10.1002/hep.20202. [DOI] [PubMed] [Google Scholar]
- 14.Yang S., Lin H.Z., Hwang J., Chacko V.P., Diehl A.M. Hepatic hyperplasia in noncirrhotic fatty livers: is obesity-related hepatic steatosis a premalignant condition? Cancer Res. 2001;61:5016–5023. [PubMed] [Google Scholar]
- 15.Desvergne B., Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr. Rev. 1999;20:649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 16.Aoyama T., et al. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor α (PPARα). J. Biol. Chem. 1998;273:5678–5684. doi: 10.1074/jbc.273.10.5678. [DOI] [PubMed] [Google Scholar]
- 17.Staels B., et al. Mechanism of action of fibrates on lipid and lipoprotein metabolism. Circulation. 1998;98:2088–2093. doi: 10.1161/01.cir.98.19.2088. [DOI] [PubMed] [Google Scholar]
- 18.Harano Y., et al. Fenofibrate, a peroxisome proliferator-activated receptor α agonist, reduces hepatic steatosis and lipid peroxidation in fatty liver Shionogi mice with hereditary fatty liver. Liver Int. 2006;26:613–620. doi: 10.1111/j.1478-3231.2006.01265.x. [DOI] [PubMed] [Google Scholar]
- 19.Yeldandi A.V., Rao M.S., Reddy J.K. Hydrogen peroxide generation in peroxisome proliferator-induced oncogenesis. Mutat. Res. 2000;448:159–177. doi: 10.1016/s0027-5107(99)00234-1. [DOI] [PubMed] [Google Scholar]
- 20.Yu S., Rao M.S., Reddy J.K. Peroxisome proliferator-activated receptors, fatty acid oxidation, steatohepatitis and hepatocarcinogenesis. Curr. Mol. Med. 2003;3:561–572. doi: 10.2174/1566524033479537. [DOI] [PubMed] [Google Scholar]
- 21.Peters J.M., Cattley R.C., Gonzalez F.J. Role of PPARα in the mechanism of action of the nongenotoxic carcinogen and peroxisome proliferator Wy-14,643. Carcinogenesis. 1997;18:2029–2033. doi: 10.1093/carcin/18.11.2029. [DOI] [PubMed] [Google Scholar]
- 22.Fan C.Y., et al. Steatohepatitis, spontaneous peroxisome proliferation and liver tumors in mice lacking peroxisomal fatty acyl-CoA oxidase. Implications for peroxisome proliferator-activated receptor α natural ligand metabolism. J. Biol. Chem. 1998;273:15639–15645. doi: 10.1074/jbc.273.25.15639. [DOI] [PubMed] [Google Scholar]
- 23.Moriya K., et al. Oxidative stress in the absence of inflammation in a mouse model for hepatitis C virus-associated hepatocarcinogenesis. Cancer Res. 2001;61:4365–4370. [PubMed] [Google Scholar]
- 24.Lee S.S., et al. Targeted disruption of the α isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol. Cell. Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandard S., Muller M., Kersten S. Peroxisome proliferator-activated receptor α target genes. Cell. Mol. Life Sci. 2004;61:393–416. doi: 10.1007/s00018-003-3216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters J.M., et al. Role of peroxisome proliferator-activated receptor α in altered cell cycle regulation in mouse liver. Carcinogenesis. 1998;19:1989–1994. doi: 10.1093/carcin/19.11.1989. [DOI] [PubMed] [Google Scholar]
- 27.Tsutsumi T., et al. Interaction of hepatitis C virus core protein with retinoid X receptor α modulates its transcriptional activity. Hepatology. . 2002;35:937–946. doi: 10.1053/jhep.2002.32470. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka N., et al. In vivo stabilization of nuclear retinoid X receptor α in the presence of peroxisome proliferator-activated receptor α. FEBS Lett. 2003;543:120–124. doi: 10.1016/s0014-5793(03)00423-x. [DOI] [PubMed] [Google Scholar]
- 29.Moriishi K., et al. Critical role of PA28γ in hepatitis C virus-associated steatogenesis and hepatocarcinogenesis. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1661–1666. doi: 10.1073/pnas.0607312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perlemuter G., et al. Hepatitis C virus core protein inhibits microsomal triglyceride transfer protein activity and very low density lipoprotein secretion: a model of viral-related steatosis. FASEB J. 2002;16:185–194. doi: 10.1096/fj.01-0396com. [DOI] [PubMed] [Google Scholar]
- 31.Korenaga M., et al. Hepatitis C virus core protein inhibits mitochondrial electron transport and increases reactive oxygen species (ROS) production. J. Biol. Chem. 2005;280:37481–37488. doi: 10.1074/jbc.M506412200. [DOI] [PubMed] [Google Scholar]
- 32.Gomez-Gonzalo M., et al. Hepatitis C virus core protein regulates p300/CBP co-activation function. Possible role in the regulation of NF-AT1 transcriptional activity. Virology. 2004;328:120–130. doi: 10.1016/j.virol.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 33.Yu S., Reddy J.K. Transcription coactivators for peroxisome proliferator-activated receptors. Biochim. Biophys. Acta. 2007;1771:936–951. doi: 10.1016/j.bbalip.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 34.Spaziani A., Alisi A., Sanna D., Balsano C. Role of p38 MAPK and RNA-dependent protein kinase (PKR) in hepatitis C virus core-dependent nuclear delocalization of cyclin B1. J. Biol. Chem. 2006;281:10983–10989. doi: 10.1074/jbc.M512536200. [DOI] [PubMed] [Google Scholar]
- 35.Diradourian C., Girard J., Pegorier J.P. Phosphorylation of PPARs: from molecular characterization to physiological relevance. Biochimie. 2005;87:33–38. doi: 10.1016/j.biochi.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 36.Reddy J.K., Rao M.S., Azarnoff D.L., Sell S. Mitogenic and carcinogenic effects of a hypolipidemic peroxisome proliferator, [4-chloro-6-(2,3-xylidino)-2- pyrimidinylthio]acetic acid (Wy-14,643), in rat and mouse liver. Cancer Res. 1979;39:152–161. [PubMed] [Google Scholar]
- 37.Rao M.S., Reddy J.K. Hepatocarcinogenesis of peroxisome proliferators. Ann. N. Y. Acad. Sci. 1996;804:573–587. doi: 10.1111/j.1749-6632.1996.tb18646.x. [DOI] [PubMed] [Google Scholar]
- 38.Genini D., Catapano C.V. Control of peroxisome proliferator-activated receptor fate by the ubiquitin-proteasome system. J. Recept. Signal. Transduct. Res. . 2006;26:679–692. doi: 10.1080/10799890600928202. [DOI] [PubMed] [Google Scholar]
- 39.Dharancy S., et al. Impaired expression of the peroxisome proliferator-activated receptor alpha during hepatitis C virus infection. Gastroenterology. 2005;128:334–342. doi: 10.1053/j.gastro.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 40.de Gottardi A., et al. Peroxisome proliferator-activated receptor-alpha and -gamma mRNA levels are reduced in chronic hepatitis C with steatosis and genotype 3 infection. Aliment. Pharmacol. Ther. 2006;23:107–114. doi: 10.1111/j.1365-2036.2006.02729.x. [DOI] [PubMed] [Google Scholar]
- 41.Lefkowitch J.H., et al. Pathological diagnosis of chronic hepatitis C: a multicenter comparative study with chronic hepatitis B. Gastroenterology. 1993;104:595–603. doi: 10.1016/0016-5085(93)90432-c. [DOI] [PubMed] [Google Scholar]
- 42.Moriya K., et al. Increase in the concentration of carbon 18 monounsaturated fatty acids in the liver with hepatitis C: analysis in transgenic mice and humans. Biochem. Biophys. Res. Commun. 2001;281:1207–1212. doi: 10.1006/bbrc.2001.4523. [DOI] [PubMed] [Google Scholar]
- 43.Barbaro G., et al. Hepatocellular mitochondrial alterations in patients with chronic hepatitis C: ultrastructural and biochemical findings. Am. J. Gastroenterol. . 1999;94:2198–2205. doi: 10.1111/j.1572-0241.1999.01294.x. [DOI] [PubMed] [Google Scholar]
- 44.Hui J.M., et al. Insulin resistance is associated with chronic hepatitis C virus infection and fibrosis progression [corrected]. Gastroenterology. 2003;125:1695–1704. doi: 10.1053/j.gastro.2003.08.032. [DOI] [PubMed] [Google Scholar]
- 45.Kato J., et al. Normalization of elevated hepatic 8-hydroxy-2’-deoxyguanosine levels in chronic hepatitis C patients by phlebotomy and low iron diet. Cancer Res. 2001;61:8697–8702. [PubMed] [Google Scholar]
- 46.Horiike S., et al. Accumulation of 8-nitroguanine in the liver of patients with chronic hepatitis C. J. Hepatol. 2005;43:403–410. doi: 10.1016/j.jhep.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 47.Takenaka K., et al. Possible multicentric occurrence of hepatocellular carcinoma: a clinicopathological study. Hepatology. 1994;19:889–894. [PubMed] [Google Scholar]
- 48.Oikawa T., et al. Multistep and multicentric development of hepatocellular carcinoma: histological analysis of 980 resected nodules. J. Hepatol. 2005;42:225–229. doi: 10.1016/j.jhep.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 49.Akiyama T.E., et al. Peroxisome proliferator-activated receptor-α regulates lipid homeostasis, but is not associated with obesity. J. Biol. Chem. 2001;276:39088–39093. doi: 10.1074/jbc.M107073200. [DOI] [PubMed] [Google Scholar]
- 50.Nakajima T., et al. Peroxisome proliferator-activated receptor α protects against alcohol-induced liver damage. Hepatology. 2004;40:972–980. doi: 10.1002/hep.20399. [DOI] [PubMed] [Google Scholar]
- 51.Ni R., et al. Fas-mediated apoptosis in primary cultured mouse hepatocytes. Exp. Cell Res. 1994;215:332–337. doi: 10.1006/excr.1994.1349. [DOI] [PubMed] [Google Scholar]
- 52.Graulet B., Gruffat D., Durand D., Bauchart D. Fatty acid metabolism and very low density lipoprotein secretion in liver slices from rats and preruminant calves. J. Biochem. 1998;124:1212–1219. doi: 10.1093/oxfordjournals.jbchem.a022240. [DOI] [PubMed] [Google Scholar]
- 53.Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 54.Gurtu V., Kain S.R., Zhang G. Fluorometric and colorimetric detection of caspase activity associated with apoptosis. Anal. Biochem. 1997;251:98–102. doi: 10.1006/abio.1997.2220. [DOI] [PubMed] [Google Scholar]