Abstract
The current Irish biota has controversial origins. Ireland was largely covered by ice at the Last Glacial Maximum (LGM) and may not have had land connections to continental Europe and Britain thereafter. Given the potential difficulty for terrestrial species to colonize Ireland except by human introduction, we investigated the stoat (Mustela erminea) as a possible cold-tolerant model species for natural colonization of Ireland at the LGM itself. The stoat currently lives in Ireland and Britain and across much of the Holarctic region including the high Arctic. We studied mitochondrial DNA variation (1771 bp) over the whole geographical range of the stoat (186 individuals and 142 localities), but with particular emphasis on the British Isles and continental Europe. Irish stoats showed considerably greater nucleotide and haplotype diversity than those in Britain. Bayesian dating is consistent with an LGM colonization of Ireland and suggests that Britain was colonized later. This later colonization probably reflects a replacement event, which can explain why Irish and British stoats belong to different mitochondrial lineages as well as different morphologically defined subspecies. The molecular data strongly indicate that stoats colonized Ireland naturally and that their genetic variability reflects accumulation of mutations during a population expansion on the island.
Keywords: cytochrome b, D-loop, last glacial maximum, mitochondrial DNA, phylogeography
1. Introduction
Although Britain and Ireland are neighbouring islands, the difference in their current biota is astonishing. Britain has a fauna and flora that is somewhat restricted but broadly similar to the nearby areas of continental Europe. By contrast, Ireland is very species poor, but has some unexpected forms among those present. Most noteworthy are those that otherwise occur in southwest Europe, collectively known as the ‘Lusitanian element’, including species such as the Kerry slug (Geomalacus maculosus) and Mackay's Heath (Erica mackaiana; Corbet 1961). Moore (1987) articulated these unusual features of the biogeography of Ireland as ‘the Irish question’, recognizing that there is a need to understand how the Irish fauna and flora could have developed in this way.
The unusual nature of the Irish biota, and particularly its distinctiveness from the British, is particularly seen clearly in mammals (Yalden 1999). For instance, in mainland Britain, non-commensal small mammals are represented by three species of vole, three species of shrew, three species of mouse and two small carnivores, all of which apparently colonized naturally at around the end of the last glaciation. Only four of these eleven species are found in Ireland, three with a long-standing presence (the pygmy shrew (Sorex minutus), the wood mouse (Apodemus sylvaticus) and the stoat (Mustela erminea)) and the fourth (the bank vole; Clethrionomys glareolus) is a twentieth century introduction. Furthermore, phylogeographic studies have shown that the pygmy shrews in Ireland have greater molecular similarity with populations in northern Iberia than those in Britain (Mascheretti et al. 2003). A comparable result has been obtained with the pine marten (Martes martes; Davison et al. 2001), providing further examples of the Lusitanian element, as applied to genetic forms within species of mammal. Mountain hares (Lepus timidus) in Ireland also have more molecular similarity to populations in continental Europe than to those in Britain (Hamill et al. 2006).
Few of the species currently present in Britain and Ireland would have been present at the Last Glacial Maximum (LGM; 19–23 kyr ago), when both landmasses were predominantly covered by ice (Andersen & Borns 1997; Mix et al. 2001), so colonization has been largely since that time. This colonization to Britain was over a land bridge to continental Europe which persisted until 7500 years ago at the very latest (Yalden 1999). The restricted fauna of Ireland clearly suggests that there was not such a long-lasting land connection between Ireland and either Britain and/or continental Europe. Indeed, while there would have been a land connection between Ireland, Britain and continental Europe at the LGM (owing to lowered sea levels; Andersen & Borns 1997), there are doubts whether Ireland was connected to either landmass at all thereafter (Lambeck & Purcell 2001). If there was no land bridge, that would explain the restricted fauna and flora in Ireland (Stuart & Van Wijngaarden-Bakker 1985). It would also suggest that the organisms which do live there have, to a large extent, been brought by humans who first appeared in Ireland in the Mesolithic (Woodman et al. 1997). The Lusitanian element would therefore suggest a particular importance of cultural links in early human history between Ireland and southwest Europe in introducing species to Ireland (Corbet 1961).
The Irish question can be fully answered only by the consideration of the colonization history of a wide range of species of animals and plants. However, given the uncertainty surrounding natural overland colonization, it is worthwhile to study the best candidates for this and establish that it occurred at all. From the palaeontological record, two such candidates for natural colonization are the mountain hare and the stoat. Single fossil specimens of the mountain hare and the stoat have been dated to the Late Glacial period, i.e. after the LGM but before the known occurrence of Mesolithic people in Ireland (Woodman et al. 1997; McCormick 1999). Clearly, if these two species occurred in Ireland before the records of people, it suggests natural colonization. Both these cases underline the uncertainty of the presence of a land bridge to Ireland after the LGM. Given their cold tolerance, the stoat and mountain hare are species that may have been able to survive LGM conditions in Ireland (Stuart & Van Wijngaarden-Bakker 1985). If these species did manage to colonize Ireland during the LGM, then the speculation about whether or not there was a land bridge subsequent to the LGM is irrelevant to their colonization, i.e. they would already have been there. Indeed, when we suggest ‘colonization’ at the LGM, it is also possible that the species were already present in Ireland at this time and the colonization was an even earlier event.
While single radiocarbon-dated records of the two species are encouraging, further evidence of natural colonization is desirable. Here, we adopt a complementary phylogeographic approach, i.e. we examine whether the molecular variation in modern specimens is consistent with natural colonization, as opposed to human introduction. Our study is based on the stoat because we believe it to be a particularly good model, and unlike the mountain hare (Hamill et al. 2006), it has not been subjected to any previous phylogeographic analysis relating to Ireland (Kurose et al. 2005).
Stoats are found over a very wide range of temperature conditions from warm temperate to arctic (King 1991). They currently occur in the high Arctic of Greenland and Canada, feeding on lemmings (Gilg et al. 2003), and at the LGM, it is known from fossils that lemmings survived in Ireland (Woodman et al. 1997), providing a potential food supply. Although fossils of stoats have yet to be found from the LGM in Ireland, the fossil record in continental Europe indicates their probable presence because the species is one of the glacial faunal elements, occurring in assemblages together with mammoth, woolly rhinoceros, spotted hyena and reindeer (Sommer & Benecke 2004). A stoat vertebra has also been found in a deposit believed to be ca 15 kyr old on the island of Andøya off northern Norway (Fjellberg 1978), at a time when Fennoscandia still had substantial ice cover (Andersen & Borns 1997).
One of the great advantages of the stoat for phylogeographic analysis is its wide and continuous contemporary distribution in Ireland, Britain and continental Europe. Not only it is thermally adaptable but the species is also generalized and common; in Britain, for instance, the stoat is the most abundant wild carnivore (Harris et al. 1995). Although there have been some recent introductions of the stoat to Shetland and New Zealand for biological control (King 1991), there is no reason to believe there have been accidental or deliberate introductions of the stoat to Britain or Ireland, or movements around continental Europe. Therefore, there is every expectation that the stoats present during the Late Glacial (based on the radiocarbon-dated specimens) survived in Ireland through to the present-day without any extinctions or introductions. Here, we examine whether the molecular data are consistent with these expectations of natural colonization based on a comparison of Irish and British stoats within the context of the European and Holarctic ranges of the species.
2. Material and methods
(a) Sample collection, processing and sequencing
A total of 197 tissue and skin samples collected from stoats from 153 localities in Eurasia and Greenland successfully yielded sequences. Details of these samples, which largely came from museum skin collections, are given in the electronic supplementary material. DNA isolation, PCR amplification and sequencing were described in detail previously (Martínková & Searle 2006). The full set of primers used for amplification and sequencing of mitochondrial genes for cytochrome b (cyt b), Thr-tRNA, Pro-tRNA and partial D-loop are listed in the table 1. We took particular care to ensure authenticity of sequences from the museum samples using methods derived from laboratory protocols for handling ancient DNA (Martínková & Searle 2006).
Table 1.
Primers and PCR conditions. (The new primers designed for this study include the indication of the position of their 3′ end within the dog complete mitochondrial sequence (Kim et al. 1998). Ta, annealing temperature; Cb, cytochrome b; D, partial D-loop (and flanking tRNA sequences); F, forward; R, reverse.)
| primer name | sequence (5′–3′) | Ta | reference |
|---|---|---|---|
| Cb-M1-F | CTC ACA TGG AAT CTA ACC ATG AC | 56 | Kurose et al. (2000) |
| Cb-L14474-F | CTT TAT CTG CCT ATT CCT ACA CG | 56 | this study |
| Cb-H14549-R | CTA TGA ATG CAG TTG CTA TGA C | 55 | this study |
| Cb-Mustela07-F | TTC ATC ATT TCA GCA CTA GCA GCA GTC | 55 | Fleming & Cook (2002) |
| Cb-Mustela06-R | GTG GAA TGG GAT TTT GTC AGA GTC GGA | 55 | Fleming & Cook (2002) |
| Cb-L14897-F | GCC CTA TTC CTT ATT CTA ACA C | 55 | this study |
| Cb-H14939-R | GGC TGG GAT ATA GTT GTC TGG | 56 | this study |
| D-Cbz-F | ATG AAT TGG AGG ACA ACC AGT | 55 | Kurose et al. (1999) |
| Cb-MR1-R | TCT TCC TTG AGT CTT AGG GAG | 56 | Kurose et al. (2000) |
| D-L15454-F | CTG ACA TTC TAA CTA AAC TAT TCC | 55 | this study |
| D-H15777-R | TGA AGT AAG AAC CAG ATG CCA G | 55 | this study |
| D-MER-R | CGC GGG TGG TGT ATA AAT AT | 55 | Kurose et al. (1999) |
(b) Data analysis
Chromatogram contigs were assembled in Sequencher v. 4.2 (Gene Codes) and sequences were aligned manually with BioEdit v. 7.0.5.2 (Hall 1999). Haplotypes of mitochondrial (mt) DNA sequences of stoats were identified by Dambe v. 4.2.13 (Xia & Xie 2001), and haplotype and nucleotide diversity and total and net divergence were calculated with DnaSP v. 4.00.5 (Rozas et al. 2003). For network construction, the 49 cyt b haplotypes that were obtained from our 197 specimens were combined with published haplotypes from Russia, Japan and North America (14 GenBank sequences, 12 new haplotypes and 13 new localities). From the 197 stoats, we were able to obtain concatenated mtDNA sequences from 186 individuals, yielding 76 haplotypes. Median-joining networks were constructed in Network v. 4.1.1.0 (Bandelt et al. 1999) using either the complete cyt b sequences or the concatenated sequence of the cyt b, Thr-tRNA, Pro-tRNA and the partial D-loop excluding an ambiguous part of the TnCn stretch. The sequences reported in this paper have been deposited in the GenBank database (Accession numbers: EF088939–EF089135).
(c) Analysis of population size changes
Mismatch distributions were constructed from the stoat sequence data with Arlequin v. 3.00 (Excoffier et al. 2005) and compared with those expected under the sudden population expansion model (Rogers & Harpending 1992) using goodness-of-fit statistics based on the sum of square deviations from 10 000 bootstrap replicates (Schneider & Excoffier 1999). The population expansion hypothesis was further tested with Arlequin by Fu's (1997) Fs index of selective neutrality that is sensitive also to demographic population expansion.
(d) Estimation of mutation rate
The mutation rate of the whole mtDNA concatenated region of the stoat was estimated under a molecular clock assumption from the Irish haplotype dataset by Bayesian coalescent analysis in Beast v. 1.3 (Drummond & Rambaut 2006). The lower constraint of the age of the Irish population was set to 13 kyr ago, which is the age of the Late Glacial fossil found in Ireland (Woodman et al. 1997). This relates to a stoat from Killavullen Cave, Co. Cork that Woodman et al. (1997) dated as 10 680±110 14C years old. In terms of calendar years, this is ca 12 750–12 900 years old based on the IntCal04 radiocarbon calibration curve (Reimer et al. 2004). The upper age constraint was selected as the time when Ireland was fully covered by an ice-sheet 40 kyr ago (Bowen et al. 2002), an event also supported by a gap in the Irish fossil record (Woodman et al. 1997). The mutation rate was then calculated from a range of plausible alternative trees in the Markov chain Monte Carlo (MCMC) algorithm search in which root height fits the constraint range. The MCMC was run for 50 million steps and the first 10% were discarded as burn-in.
(e) Bayesian molecular dating
As the mismatch distribution and Fu's (1997) Fs indicated that the Irish, British and continental European populations of stoat exhibited independent demographic histories, their ages were estimated using an exponential growth model. They were calculated by Bayesian coalescent analysis using MCMC in Beast with the previously estimated mutation rate and the age constraints on the Irish population. Beast records the age of a particular node in various trees after the MCMC convergence in which the node appears. Hence, the age estimate is not constrained to a precisely defined evolutionary history (tree topology and branch lengths), but rather explores the available tree space making the method particularly suitable for dating recent divergence events. Five independent MCMC runs were made with 10 million steps each, sampled every 1000th step. The results of all runs were then combined to obtain the ages of the most recent common ancestors of Irish, British and European stoat populations, respectively, with the first 10% discarded as burn-in.
3. Results
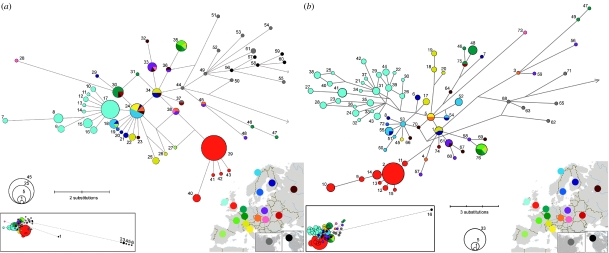
We analysed complete cyt b sequences from a total of 211 stoats (166 localities), which yielded 61 haplotypes (figure 1a and electronic supplementary material). All British cyt b haplotypes (which came from stoats collected throughout the mainland and in Islay and Shetland) formed a monophyletic lineage (figure 1a), 85% of which shared the same haplotype (no. 39). Similarly, Irish stoats (also collected throughout the country and in the Isle of Man) formed a monophyletic but distinct lineage with a single dominant haplotype (no. 17). There is little indication of genetic structure in continental Eurasia. Some of the haplotypes identified in Alaska (nos 56–60) were similar to those from Siberia and Japan (no. 44, nos 49–55, no. 61). Fleming & Cook (2002) identified this ‘Beringian’ clade, and we found that it forms part of an extensive continental Eurasian lineage. Other North American haplotypes remain distinct and consistent with Fleming and Cook's ‘Continental (American)’ and ‘British Columbia islands’ clades (figure 1a).
Figure 1.
The relatedness and diversity of genetic lineages of the stoat, depicted by median-joining networks of sequences obtained from individuals throughout the species range. (a) cyt b sequences (1140 bp), the arrow indicates the connection to Beringian and North American cyt b haplotypes (inset), haplotype no. 44 is separated from haplotype no. 1 by 14 substitutions and from haplotype no. 2 by 37 substitutions, respectively. (b) Concatenated sequences of cyt b, Thr-tRNA, Pro-tRNA and partial D-loop, excluding the ambiguous part of the TnCn region (1771 bp), the arrow indicates the connection to the Greenland haplotype no. 16 (inset) separated from haplotype no. 68 by 50 substitutions. Colour coding is used to distinguish individual European countries, Asia and North America. Further details of the numbered haplotypes are available in the electronic supplementary material.
We identified 76 haplotypes in 186 stoats (142 localities) from a concatenated sequence of mtDNA, comprising 1771 bp of cyt b, Thr-tRNA, Pro-tRNA and the partial D-loop, excluding an ambiguous part of the TnCn stretch (figure 1b and electronic supplementary material). British and Irish populations retained their distinctiveness and each remained monophyletic (figure 1b).
Considering the concatenated sequence, we found markedly lower nucleotide and haplotype diversity in the British population than in Irish or continental European populations. Nucleotide (π±s.d.) and haplotype (h±s.d.) diversity were as follows: Irish population: π=0.0026±0.0002, h=0.96±0.01, n=52; British population: π=0.0007±0.0001, h=0.58±0.08, n=53; and continental European population: π=0.0031±0.0002, h=0.97±0.01, n=74. The total (Dxy) and net (Da) divergences between Irish and continental European populations of 0.43 and 0.15%, respectively, were similar in magnitude to the divergences between Britain and continental Europe (Dxy=0.45% and Da=0.26%). However, the British and Irish populations were more divergent from each other (Dxy=0.61% and Da=0.45%) than either was from the continental European population. There is no reason to believe that the greater genetic diversity of Irish stoats is the result of multiple colonization events; mismatch distributions of all populations significantly fitted the sudden population expansion model, which describes growth of a single founder population (Ireland: sum of square deviations SSD=0.0021, p=0.46; Britain: SSD=0.0131, p=0.56; continental Europe: SSD=0.00067, p=0.87). Also consistent with population expansion, Fu's (1997) Fs values were negative and significantly different from zero for the Irish (Fs=−10.188, p=0) and continental European lineages (Fs=−28.548, p=0) but non-significant for the British lineage (Fs=−2.207, p=0.13).
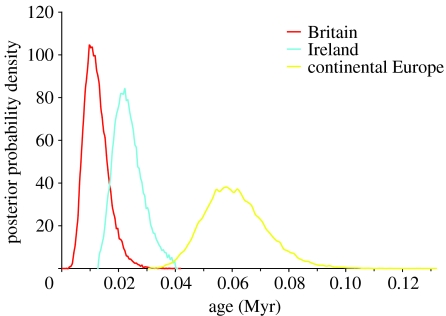
The mutation rate of the concatenated mtDNA region consistent with the age constraints of the Irish population sampled over a range of plausible alternative trees was 7.244×10−8 substitutions site−1 yr−1 (14.5% per Myr). This high rate is expected when measuring within-species divergences younger than 1 Myr (Ho et al. 2005). Analysis with this mutation rate showed that the age of the most recent common ancestor of the Irish lineage is greater than that for the British lineage, though non-significantly so (figure 2). With 95% confidence, the most recent common ancestor of the Irish lineage is 14 270–34 050 years old (mean estimate, 23 510), whereas that of the British lineage is 4858–20 710 years old (mean estimate, 12 240).
Figure 2.
Estimates of the age of the Irish, British and continental European stoat populations. These are shown as posterior probability density functions estimated by the Bayesian coalescent analysis using a mutation rate of 0.07244 bp−1 Myr−1 and age constraints of the Irish population: 13–40 kyr. Five independent runs of MCMC were performed for 10 million steps each, with the first 1 million steps discarded as burn-in.
4. Discussion
The molecular diversity data that we obtained are extraordinary. Even though the landmass of Britain is 2.5 times larger than Ireland and is located closer to continental Europe, we found stoats to be far less genetically variable in Britain than Ireland. Indeed, for the concatenated cyt b and D-loop dataset, the haplotype and nucleotide diversities for Ireland are similar to those for continental Europe, while those in Britain are considerably lower.
These data argue strongly against the likelihood of introduction of stoats to Ireland. Generally, populations arising from introductions show lower genetic diversity than those that have been colonized naturally. The numbers of the individuals colonizing tend to be smaller and introductions tend to be more recent than natural colonizations. We have, for instance, found low levels of genetic diversity among stoats introduced to New Zealand (N. Martínková et al. 2007, unpublished data) and bank voles introduced to Ireland (P. Stuart et al. 2007, unpublished data). On the basis of the molecular data, it would seem positively perverse to suggest that stoats in Ireland are an introduction while those in Britain are a natural colonization (and, to our knowledge, there has never been any suggestion that stoats were introduced to Britain).
Therefore, the molecular data are supportive of the fossil evidence for a natural colonization of Ireland by stoats. We applied a Bayesian method to date the origins of the Irish and British populations of stoats from the molecular data, using the palaeontological record for Ireland to set age constraints. The Irish and British populations fit into separate monophyletic lineages. The constraints determined the 95% CI that we obtained for the age of the Irish lineage: 14 270–34 050 years, which is, of course, compatible with an LGM origin for the lineage. These dates are also compatible with colonization over a Late Glacial land bridge from continental Europe to southwest England and Ireland, which is the most probable time for such a land bridge if it existed (Lambeck & Purcell 2001).
The estimate of the age of the British lineage is 4858–20 710 years old, suggesting a more recent colonization than the Irish one. Again, given that the British colonization is natural, it is difficult to argue that an earlier colonization of Irish stoats should be an introduction. Clearly, the dating of the British lineage is consistent with the natural colonization of Britain by stoats before the flooding of the English Channel.
Some of our most interesting data relate to stoats from the Isle of Man, an island in the Irish Sea, approximately halfway between Ireland and Britain. The Isle of Man has a similar range of non-commensal small mammals as Ireland, with the same three species (wood mouse, pygmy shrew and stoat) out of the eleven found in Britain (Yalden 1999). However, the Isle of Man was connected longer to Britain than to Ireland (Innes et al. 2004). Hence, the Manx stoats might be expected to be more similar to the British than the Irish. In fact, in terms of mtDNA, the Manx stoats fall within the Irish lineage. Morphologically, the Irish and Manx stoats are also more similar. They are classified together as a distinct subspecies, Mustela erminea hibernica, on the basis of pelage characteristics (Miller 1912).
A scenario can be developed which best explains our molecular data for stoats in the British Isles. Following Stuart & Van Wijngaarden-Bakker (1985), we believe that stoats are sufficiently cold tolerant to have survived close to the British–Irish ice sheet at the LGM, and therefore were present on the exposed landmass of Ireland, having colonized at that time or earlier. Thus, we propose that the progenitors of the Irish mtDNA lineage and morphotype were cold tolerant. Consistent with that, mtDNA haplotypes from high latitudes (Scandinavia) and altitudes (Swiss and Italian Alps) are among those most closely related to modern Irish haplotypes (figure 1). We believe that these cold-tolerant stoats followed the retreating ice at the end of the LGM and spread throughout Britain, Ireland and the Isle of Man. The stoats in Ireland would have become isolated with rising sea level after the LGM and would have increased in population size from a small number of founders (hence the signal for population expansion). At a later time, the Isle of Man population of stoats would also have become separated from the British. However, Ireland and the Isle of Man would have become islands long before Britain was itself separated from continental Europe. They perhaps became islands early enough that cold-sensitive species were totally unable to colonize them overland, hence the paucity of species in both Ireland and the Isle of Man.
There is a need to explain why British stoats are currently a different mtDNA lineage and morphotype from those found in Ireland and the Isle of Man. We believe that the continued land bridge with continental Europe is the key. This land bridge would have been available to stoats during the warm periods of the Late Glacial and Postglacial. For instance, during the Postglacial period between the end of the Younger Dryas (11 400 years ago; Walker et al. 2003) and the flooding of the English Channel (7500 years ago at the very latest; Yalden 1999), the climate in Britain would have been similar to today (Andersen & Borns 1997). Therefore, we suggest that an mtDNA lineage and morphotype of stoats adapted to such warm conditions would have been able to enter Britain and replace the cold-tolerant lineage that was originally present and continues to survive in the isolated populations of Ireland and the Isle of Man. In this way, we can not only explain the morphological and mitochondrial differences between British and Irish stoats, but also clarify why the British mtDNA lineage is younger and less variable than the mtDNA lineage in Ireland.
There is evidence from the literature for replacement processes of the type we suggest. A phylogeographic study by Piertney et al. (2005) indicated partial replacement of one mtDNA lineage of water voles (Arvicola terrestris) by another in Britain, and ancient DNA studies have demonstrated sequential replacement of mtDNA lineages in brown bears (Ursus arctos) occupying Beringia (Barnes et al. 2002). There is also evidence that different mtDNA haplotypes may adapt individuals to different temperature conditions (Fontanillas et al. 2005). Different external morphologies may also of course be related to different temperature adaptations. Within the crow species (Corvus corone), the all-black carrion crow (Corvus corone corone) is expanding in Britain at the expense of the grey-and-black hooded crow (C. c. cornix) apparently in response to climate change (Cook 1975). The hooded crow is now found only in the north of Scotland. However, in a clear parallel with what we suggest for the stoat, it is the hooded crow (i.e. the form that is being displaced) that is found in Ireland, even though the climate in Ireland is similar to southern Britain.
In terms of the Irish question, we do not claim to have ‘answered’ it. There is a need to study a wide variety of species before there will be a comprehensive understanding of the peculiarities of Ireland's biota. Human introductions have undoubtedly been very important in generating the Irish fauna and flora, and it is actually very difficult to demonstrate convincingly that any particular terrestrial species did manage to colonize naturally. We believe that the weight of evidence strongly favours such a natural colonization by the stoat and that this colonization was very early, probably around the LGM. Our data suggest that there is no need to propose a Late Glacial land bridge to explain the natural occurrence of stoats in Ireland.
Our studies of the stoat provide another very interesting example of a terrestrial mammal with strong genetic differences between Irish and British populations to add to pine martens (Davison et al. 2001), pygmy shrews (Mascheretti et al. 2003) and mountain hares (Hamill et al. 2006). However, in the case of the stoat, we suggest a new explanation for this discrepancy between Britain and Ireland: that there was originally the same genetic type in Britain and Ireland, and that the much long-lasting connection with continental Europe allowed a replacement event in Britain. Replacement may be important in other examples of within-species genetic differences between Ireland and Britain. It could also explain some of the differences in the species lists for the two countries.
Acknowledgments
We are grateful to all people who provided samples for this study as listed in the electronic supplementary material. We thank I. Barnes, D. Förster, İ. Gündüz, J. Provan, A. Rambaut and B. Shapiro for their technical advice, S. Martínek for help with the figures and S. Bearhop, A. Douglas, J. Herman, C. Maggs and J. Provan for their comments on earlier versions of the manuscript. This work was supported by the Natural Environment Research Council. R.M. is supported by the Quercus partnership between Queen's University Belfast and the Environment & Heritage Service, Northern Ireland.
Supplementary Material
List of Mustela erminea samples, localities and haplotype identification numbers
References
- Andersen B.G, Borns H.W., Jr . Scandinavian University Press; Oslo, Norway: 1997. The Ice Age world. [Google Scholar]
- Bandelt H.-J, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Barnes I, Matheus P, Shapiro B, Jensen D, Cooper A. Dynamics of Pleistocene population extinctions in Beringian brown bears. Science. 2002;295:2267–2270. doi: 10.1126/science.1067814. doi:10.1126/science.1067814 [DOI] [PubMed] [Google Scholar]
- Bowen D.Q, Philips F.M, McCabe A.M, Knutz P.C, Sykes G.A. New data for the Last Glacial Maximum in Great Britain and Ireland. Quaternary Sci. Rev. 2002;21:89–101. doi:10.1016/S0277-3791(01)00102-0 [Google Scholar]
- Cook A. Changes in carrion-hooded crow hybrid zone and possible importance of climate. Bird Study. 1975;22:165–168. [Google Scholar]
- Corbet G.B. Origin of the British insular races of small mammals and of the ‘Lusitanian’ fauna. Nature. 1961;191:1037–1040. doi:10.1038/1911037a0 [Google Scholar]
- Davison A, Birks J.D.S, Brookes R.C, Messenger J.E, Griffiths H.I. Mitochondrial phylogeography and population history of pine martens Martes martes compared with polecats Mustela putorius. Mol. Ecol. 2001;10:2479–2488. doi: 10.1046/j.1365-294x.2001.01381.x. doi:10.1046/j.1365-294X.2001.01381.x [DOI] [PubMed] [Google Scholar]
- Drummond, A. J. & Rambaut, A. 2006 Beast v1.0, http://evolve.zoo.ox.ac.uk/beast/
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fjellberg A. Fragments of Middle Weichselian fauna on Andøya, north Norway. Boreas. 1978;7:39. [Google Scholar]
- Fleming M.A, Cook J.A. Phylogeography of endemic ermine (Mustela erminea) in southeast Alaska. Mol. Ecol. 2002;11:795–807. doi: 10.1046/j.1365-294x.2002.01472.x. doi:10.1046/j.1365-294X.2002.01472.x [DOI] [PubMed] [Google Scholar]
- Fontanillas P, Dépraz A, Giorgi M.S, Perrin N. Nonshivering thermogenesis capacity associated to mitochondrial DNA haplotypes and gender in the greater white-toothed shrew, Crocidura russula. Mol. Ecol. 2005;14:661–670. doi: 10.1111/j.1365-294X.2004.02414.x. doi:10.1111/j.1365-294X.2004.02414.x [DOI] [PubMed] [Google Scholar]
- Fu Y.-X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilg O, Hanski I, Sittler B. Cyclic dynamics in a simple vertebrate predator–prey community. Science. 2003;302:866–868. doi: 10.1126/science.1087509. doi:10.1126/science.1087509 [DOI] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hamill R.M, Doyle D, Duke E.J. Spatial patterns of genetic diversity across European subspecies of the mountain hare, Lepus timidus L. Heredity. 2006;97:355–365. doi: 10.1038/sj.hdy.6800880. doi:10.1038/sj.hdy.6800880 [DOI] [PubMed] [Google Scholar]
- Harris S, Morris P, Wray S, Yalden D. Joint Nature Conservation Committee; Peterborough, UK: 1995. A review of British mammals: population estimates and conservation status of British mammals other than cetaceans. [Google Scholar]
- Ho S.Y.W, Phillips M.J, Cooper A, Drummond A.J. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 2005;22:1561–1568. doi: 10.1093/molbev/msi145. doi:10.1093/molbev/msi145 [DOI] [PubMed] [Google Scholar]
- Innes J.B, Chiverrell R.C, Blackford J.J, Davey P.J, Gonzalez S, Rutherford M.M, Tomlinson P.R. Earliest Holocene vegetation history and island biogeography of the Isle of Man, British Isles. J. Biogeogr. 2004;31:761–772. doi:10.1111/j.1365-2699.2003.01048.x [Google Scholar]
- Kim K.S, Lee S.E, Jeong H.W, Ha J.H. The complete nucleotide sequence of the domestic dog (Canis familiaris) mitochondrial genome. Mol. Phylogenet. Evol. 1998;10:210–220. doi: 10.1006/mpev.1998.0513. doi:10.1006/mpev.1998.0513 [DOI] [PubMed] [Google Scholar]
- King, C. M. 1991 Stoat. In The handbook of British mammals (eds G. B. Corbet & S. Harris), pp. 377–387, 3rd edn. Oxford, UK: Blackwell.
- Kurose N, Masuda R, Yoshida M.C. Phylogeographic variation in two mustelines, the least weasel Mustela nivalis and the ermine M. erminea of Japan, based on mitochondrial DNA control region sequences. Zool. Sci. 1999;16:971–977. doi:10.2108/zsj.16.971 [Google Scholar]
- Kurose N, Abramov A.V, Masuda R. Intrageneric diversity of the cytochrome b gene and phylogeny of Eurasian species of the genus Mustela (Mustelidae, Carnivora) Zool. Sci. 2000;17:673–679. doi: 10.2108/zsj.17.673. doi:10.2108/zsj.17.673 [DOI] [PubMed] [Google Scholar]
- Kurose N, Abramov A.V, Masuda R. Comparative phylogeography between the ermine Mustela erminea and the least weasel M. nivalis of Palaearctic and Nearctic regions, based on analysis of mitochondrial DNA control region sequences. Zool. Sci. 2005;22:1069–1078. doi: 10.2108/zsj.22.1069. doi:10.2108/zsj.22.1069 [DOI] [PubMed] [Google Scholar]
- Lambeck K, Purcell A.P. Sea-level change in the Irish Sea since the Last Glacial Maximum: constraints from isostatic modelling. J. Quaternary Sci. 2001;16:497–506. doi:10.1002/jqs.638 [Google Scholar]
- Martínková N, Searle J.B. Amplification success rate of DNA from museum skin collections: a case study of stoats from 18 museums. Mol. Ecol. Notes. 2006;6:1014–1017. doi:10.1111/j.1471-8286.2006.01482.x [Google Scholar]
- Mascheretti S, Rogatcheva M.B, Gündüz I, Fredga K, Searle J.B. Phylogeography of the Eurasian pygmy shrew Sorex minutus: clues on its mode of colonisation of Ireland. Proc. R. Soc. B. 2003;270:1593–1599. doi: 10.1098/rspb.2003.2406. doi:10.1098/rspb.2003.2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick F. Early evidence for wild animals in Ireland. In: Benecke N, editor. The Holocene history of the European vertebrate fauna. Modern aspects of research. Verlag Marie Leidorf GmbH; Rahden, Germany: 1999. pp. 355–371. [Google Scholar]
- Miller G.S. British Museum; London, UK: 1912. Catalogue of the mammals of western Europe. [Google Scholar]
- Mix A.C, Bard E, Schneider R. Environmental processes of the ice age: land, oceans, glaciers (EPILOG) Quaternary Sci. Rev. 2001;20:627–657. doi:10.1016/S0277-3791(00)00145-1 [Google Scholar]
- Moore P.D. Snails and the Irish question. Nature. 1987;328:381–382. doi:10.1038/328381a0 [Google Scholar]
- Piertney S.B, Stewart W.A, Lambin X, Telfer S, Aars J, Dallas J.F. Phylogeographic structure and postglacial evolutionary history of water voles (Arvicola terrestris) in the United Kingdom. Mol. Ecol. 2005;14:1435–1444. doi: 10.1111/j.1365-294X.2005.02496.x. doi:10.1111/j.1365-294X.2005.02496.x [DOI] [PubMed] [Google Scholar]
- Reimer P.J, et al. IntCal04 terrestrial radiocarbon age calibration, 0–26 cal ky BP. Radiocarbon. 2004;46:1029–1058. [Google Scholar]
- Rogers A.R, Harpending H. Population-growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sánchez-DelBarrio J.C, Messeguer X, Rozas R. DnaSP, DNA polymorphism analysis by coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. doi:10.1093/bioinformatics/btg359 [DOI] [PubMed] [Google Scholar]
- Schneider S, Excoffier L. Estimation of demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics. 1999;152:1079–1089. doi: 10.1093/genetics/152.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer R, Benecke N. Late- and post-glacial history of the Mustelidae in Europe. Mammal Rev. 2004;34:249–284. doi:10.1111/j.1365-2907.2004.00043.x [Google Scholar]
- Stuart A.J, Van Wijngaarden-Bakker L. Quaternary vertebrates. In: Edwards K.J, Warren W.P, editors. The Quaternary history of Ireland. Academic Press; London, UK: 1985. pp. 221–250. [Google Scholar]
- Walker M.J.C, Coope G.R, Sheldrick C, Turney C.S.M, Lowe J.J, Blockley S.P.E, Harkness D.D. Devensian lateglacial environmental changes in Britain: a multi-proxy environmental record from Llanilid, South Wales, UK. Quaternary Sci. Rev. 2003;22:475–520. doi:10.1016/S0277-3791(02)00247-0 [Google Scholar]
- Woodman P, McCarthy M, Monaghan N. The Irish quaternary fauna project. Quaternary Sci. Rev. 1997;16:129–159. doi:10.1016/S0277-3791(96)00037-6 [Google Scholar]
- Xia X, Xie Z. DAMBE: data analysis in molecular biology and evolution. J. Hered. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. doi:10.1093/jhered/92.4.371 [DOI] [PubMed] [Google Scholar]
- Yalden D. Poyser; London, UK: 1999. The history of British mammals. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of Mustela erminea samples, localities and haplotype identification numbers