Abstract
Several significant steps have been completed toward a general method for the site-specific incorporation of unnatural amino acids into proteins in vivo. An “orthogonal” suppressor tRNA was derived from Saccharomyces cerevisiae tRNA2Gln. This yeast orthogonal tRNA is not a substrate in vitro or in vivo for any Escherichia coli aminoacyl-tRNA synthetase, including E. coli glutaminyl-tRNA synthetase (GlnRS), yet functions with the E. coli translational machinery. Importantly, S. cerevisiae GlnRS aminoacylates the yeast orthogonal tRNA in vitro and in E. coli, but does not charge E. coli tRNAGln. This yeast-derived suppressor tRNA together with yeast GlnRS thus represents a completely orthogonal tRNA/synthetase pair in E. coli suitable for the delivery of unnatural amino acids into proteins in vivo. A general method was developed to select for mutant aminoacyl-tRNA synthetases capable of charging any ribosomally accepted molecule onto an orthogonal suppressor tRNA. Finally, a rapid nonradioactive screen for unnatural amino acid uptake was developed and applied to a collection of 138 amino acids. The majority of glutamine and glutamic acid analogs under examination were found to be uptaken by E. coli. Implications of these results are discussed.
Our recent efforts to expand the scope of protein mutagenesis have focused on engineering an organism capable of site-specifically incorporating unnatural amino acids directly from the growth media into proteins (1–3). Our strategy involves (i) generation of an “orthogonal” suppressor tRNA that is not a substrate for any endogenous Escherichia coli aminoacyl-tRNA synthetase (aaRS) yet functions with the E. coli ribosomal machinery; (ii) evolution of a mutant aaRS that aminoacylates this orthogonal tRNA with any natural amino acid; and (iii) evolution of a mutant aaRS that charges the orthogonal tRNA with an unnatural amino acid. We previously have reported the design of an orthogonal suppressor tRNA derived from E. coli tRNA2Gln (1) as well as the evolution of mutant E. coli glutaminyl-tRNA synthetase (GlnRS) enzymes capable of efficiently charging this tRNA with glutamine (2). Despite these efforts, no mutant E. coli GlnRS has yet been isolated that charges the E. coli tRNA2Gln-derived orthogonal tRNA more efficiently than wild-type E. coli tRNA2Gln under physiological conditions. Aminoacylation of any endogenous E. coli tRNA renders a synthetase unsuitable for charging an unnatural amino acid because the resulting misacylation of endogenous tRNA is known to be lethal to the host organism (4). An orthogonal tRNA/synthetase pair has been developed based on the Saccharomyces cerevisiae tRNA2Gln and yeast GlnRS and is described herein.
A strategy also has been developed to evolve mutant synthetases capable of charging unnatural amino acids onto the orthogonal tRNA. Such a scheme poses unique challenges because unnatural amino acids are not required for the growth of a cell. We describe a general in vivo selection for mutant aaRS enzymes capable of charging any nontoxic, ribosomally accepted small molecule (including α-hydroxy acids, β-amino acids, etc.) onto an orthogonal suppressor tRNA. Because this selection does not rely on the unique chemical reactivity of a given amino acid, a large variety of unnatural substrates may be evaluated with libraries of mutant aaRSs for their ability to be incorporated into proteins.
Finally, we have examined the uptake of unnatural amino acids directly from the growth media into the host organism. Traditional methods for assaying amino acid uptake involve the synthesis of an amino acid in a radiolabeled form, growth of cells in the presence of labeled amino acid, and scintillation of the resulting cultures after filtration and washing. Because very few radiolabeled unnatural amino acids are commercially available, this method proves extremely labor intensive for large collections of dozens or hundreds of amino acids. To address this shortcoming, a rapid and nonradioactive screen for unnatural amino acid uptake is described below that uses a strategy borrowed from genetics using amino acids as lethal “alleles.”
MATERIALS AND METHODS
Strains and Plasmids.
E. coli strains BT235 (5), X3R2 (6), and DH10B were obtained from Hachiro Inokuchi (Kyoto University, Japan), Dieter Söll (Yale University, New Haven, CT), and GIBCO/BRL, respectively. Plasmid pMT416(7) was provided by Robert Hartley (National Institutes of Health, Bethesda, MD). Plasmids for runoff transcription of suppressor tRNAs were derived from pYPhe2 (8) as described below. Suppressor tRNA overexpression plasmids were derived from pAC123(2).
Construction and in Vitro Assays of Suppressor tRNAs.
DNA encoding tRNAs for runoff transcription were constructed from two overlapping synthetic oligonucleotides (Genosys, The Woodlands, TX) and inserted between the KpnI and HindIII sites of pYPhe2 as described (8, 9). Genes encoding tRNAs for in vivo expression were similarly constructed from overlapping oligonucleotides and inserted between the EcoRI and PstI sites of pAC123 (2) to generate pACYsupA38. Sequences of the oligonucleotides used to construct the yeast suppressor tRNA2Gln(A36) gene for runoff transcription are as follows with the tRNA sequence underlined: 5′-GCGGGGTACCTAATACGACTCACTATAGGTCCTATAGTGTAGTGGTTATCACTTTCGGTTCTAATCCGAAC-3′; 5′-GGCGGCAAGCTTGGATGGATCACCTGGAGGTCCCACCCGGATTCGAACTGGGGTTGTTCGGATTAGAACCGAAAG-3′. Runoff transcription of the tRNAs was carried out by using T7 RNA polymerase as described (9). In vitro transcription and translation reactions were performed by using 3 μg of plasmid containing the E. coli chorismate mutase gene bearing an amber mutation at site Gln-88 and 10 μg of suppressor tRNA per 30-μl reaction at a final magnesium concentration of 7 mM (9). Valine-acylated tRNA was generated as reported (10). Amounts of truncated and full-length proteins from in vitro suppression were quantitated by using a Molecular Dynamics 445SI PhosphorImager.
Cloning and Purification of Yeast GlnRS.
The gene encoding yeast GlnRS was cloned by PCR from S. cerevisiae genomic DNA (Promega) using the following synthetic oligonucleotide primers: 5′-GGAATACCATATGTCTTCTGTAGAAGAAT-3′; 5′-AAACTGCAGCACATTAAATCATTCACT-3′. DNA encoding the E. coli GlnRS promoter and terminator were cloned by PCR from E. coli genomic DNA prepared from strain X3R2 by using the A.S.A.P. Genomic DNA Isolation Kit (Roche Molecular Biochemicals, Mannheim, Germany). The 87-bp and 200-bp PCR products representing the promoter and terminator were ligated together with the 2.5-kb yeast GlnRS gene and cloned into pBR322 to afford pBRYQRS.
The yeast GlnRS-encoding gene was subcloned by PCR between the XhoI and SmaI sites of plasmid pCAL-n (Stratagene). Purification of cell lysates using calmodulin-bound resin following the manufacturer’s instructions yielded nearly pure yeast GlnRS free from contaminating E. coli GlnRS. The enzyme was stored in 50% glycerol, 20 mM Hepes, 50 mM KCl, 2 mM DTT, pH 7.2 at −20°C.
In Vitro Assays of GlnRS Enzymes.
Concentrations of GlnRS enzymes were determined by SDS/PAGE and staining with Coomassie blue R250, followed by comparison with known concentrations of BSA (Sigma). Specific activities were determined at physiological levels of 3 mM ATP (11), 150 μM glutamine (12), and 2 μM tRNA (13). Assays were carried out at 30°C as described (2, 14). Wild-type E. coli tRNA was used as a mix of whole tRNA from E. coli (Boehringer Mannheim); tRNAGln constitutes 1.8% of whole E. coli tRNA (15). Wild-type yeast tRNAGln was generated by runoff transcription. E. coli GlnRS enzyme was purified as described (2).
Positive Selection.
Plasmid pBLAM was constructed from pBR322 by using the QuikChange Site-Directed Mutagenesis kit (Stratagene). Plasmid pBLAM-YQRS was constructed by subcloning the yeast GlnRS expression cassette (including the E. coli GlnRS promoter and terminator) between the AvaI and AflIII sites of pBLAM. Plasmid pBLAM-YQRSΔ500 was generated by excision of the DNA between the AvrII and PstI sites of pBLAM-YQRS, deleting GlnRS amino acids 224–809.
Competent DH10B cells harboring pACYsupA38 were transformed by electroporation (18 kV/cm, τ = 4.5–5.3 msec) with mixtures of pBLAM-YQRS and pBLAM-YQRSΔ500 (or with pBLAM-YQRS libraries encoding mutant yeast GlnRS enzymes). Cells were centrifuged, washed once with minimal media containing 1% glycerol and 0.3 mM leucine (GMML; found to permit the efficient uptake of natural amino acids; unpublished results), and resuspended in GMML supplemented optionally with 1 mM each of one or more unnatural amino acids of interest. Unnatural amino acids suspected of entering E. coli via the same pathway were separated into different cultures to avoid cross-inhibition of amino acid uptake (see below). After incubation for 2–3 h at 30°C, ampicillin was added. Cultures were incubated at 30°C until saturation (usually 18–36 h). Plasmid DNA was harvested from saturated cultures by using standard methods and analyzed by restriction digestion.
Negative Selection.
DNA encoding barnase containing an amber nonsense mutation at residue Gln-2 was generated by PCR from pMT416 (7) using the following oligonucleotides: 5′-GGAATTCCATATGGCATAGGTTATCAACACGTTTG-3′ and 5′-CCGGCGGCATGCTTATCTGATTTTTGTAAAG-3′. Efforts to insert the resulting PCR product between the NdeI and SphI sites of pAC123LamB (2) (placing expression of the barnase gene under arabinose induction) were unsuccessful, presumably because read-through of a single amber codon at this position is lethal. The PCR product therefore was inserted between the NdeI and SphI sites of pBR322 (resulting in a promoterless barnase gene), mutagenized to install amber codons at position Asp-44 or at positions Asp-44 and Gly-65, and subcloned into pAC123LamB to afford pYsupA38B2 and pYsupA38B3, respectively.
Competent DH10B cells harboring pYsupA38B2 or pYsupA38B3 were transformed by electroporation with mixtures of pBLAM-YQRS and pBLAM-YQRSΔ500 (or with pBLAM-YQRS libraries encoding mutant yeast GlnRS enzymes). Cells were harvested by centrifugation, washed once with GMML, and resuspended in GMML lacking unnatural amino acids. After incubation for 2–3 h at 30°C with agitation, tetracycline (25 μg/ml), chloramphenicol (40 μg/ml), and arabinose (10 mM) were added to induce barnase expression. Cultures were incubated at 30°C until saturation (usually 24–48 h).
Amino Acid Toxicity and Uptake Screen.
DH10B cells harboring pBLAM-YQRS and pACYsupA38 were inoculated at a cell density of approximately 2 × 106 cells/ml into GMML containing 100 μg/ml of ampicillin. For toxicity assays, unnatural amino acids were added at concentrations ranging from 8 μM to 1 mM. For uptake assays, an unnatural amino acid representing a “lethal allele” (see text) was added at its IC50 concentration together with 2 mM of the amino acid of interest. Cultures were grown at 30°C until control cultures lacking any added unnatural amino acid reached an OD600 between 0.5 and 1.0 (typically 18–24 h). Survival rates relative to the control culture were determined either by OD600 or by plating dilutions of the cultures on nonselective media.
RESULTS AND DISCUSSION
Yeast tRNA2Gln(CUA)A38 Is an “Orthogonal tRNA” in E. coli.
Recent reports that E. coli GlnRS does not charge S. cerevisiae tRNAGln (16) prompted us to determine whether the yeast suppressor tRNA2Gln(CUA) was also not a substrate for E. coli GlnRS. In addition, the mutation of base 38 from U to A was investigated as a possible means of increasing suppression efficiency in accord with previous observations (1, 17). Two yeast amber (TAG) suppressor tRNAs, tRNA2Gln(A36) and tRNA2Gln(A36 A38), were generated by in vitro runoff transcription of the corresponding DNA templates. These tRNAs, along with wild-type E. coli tRNA2Gln, were assayed in vitro for their ability to be aminoacylated with 3H-glutamine by purified E. coli GlnRS. Concentrations of tRNA, amino acid, and ATP in each assay approximated physiological levels in E. coli. The E. coli GlnRS aminoacylation rates of both yeast tRNA2Gln(A36) (6.4 pmol product per mg enzyme per min) and yeast tRNA2Gln(A36 A38) (5.6 pmol/mg per min) were approximately 100,000-fold lower than that of wild-type E. coli tRNA2Gln (6.8 × 105 pmol/mg per min), demonstrating that these yeast suppressors are very poor substrates for E. coli GlnRS.
The inability of E. coli GlnRS, or any E. coli aaRS, to aminoacylate these yeast suppressor tRNAs was verified in vivo by complementation of two nonsense mutations. E. coli strain BT235 (5) harbors an amber mutation at an essential glutamine position in the genomic β-galactosidase gene (lacZam1000). Suppression of this stop codon with a glutamine-inserting suppressor tRNA is required for growth on lactose minimal media. BT235 transformed with plasmid pACsupE [expressing the E. coli tRNA2Gln(A36), or supE, suppressor tRNA, which is an efficient substrate for E. coli GlnRS] grew readily on lactose, whereas BT235 transformed with pACYsupA38 [expressing the yeast tRNA2Gln(A36 A38) suppressor] failed to grow on lactose, even when E. coli GlnRS is overexpressed from a plasmid. Similarly, an amber nonsense mutation was introduced by site-directed mutagenesis into position Ala-184 in the TEM-1 β-lactamase gene from pBR322 to afford plasmid pBLAM. Residue 184 is not conserved among β-lactamases and enzymes containing Ala, Arg, Asn, Cys, Gln, Gly, Ile, Leu, Lys, Ser, or Tyr at this position all possess wild-type activity (18). The suppression of any amino acid at this position arising from aminoacylation of a suppressor tRNA by any endogenous synthetase likely would lead to functional β-lactamase and consequently the ability to grow in the presence of ampicillin. E. coli doubly transformed with pBLAM and pACsupE grew equally well in the presence or absence of 100 μg/ml ampicillin, whereas E. coli harboring pBLAM and pACYsupA38 did not survive in 100 μg/ml of ampicillin even with concomitant plasmid-based E. coli GlnRS overexpression. Together, these results demonstrate that yeast tRNA2Gln(A36 A38) is not an efficient substrate in vivo for any E. coli aaRS.
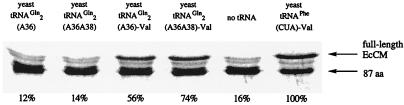
To assess the ability of yeast tRNA2Gln(A36) and tRNA2Gln(A36 A38) to function with the E. coli protein biosynthetic machinery, these tRNAs were generated by runoff transcription in truncated forms lacking their 3′ terminal CA bases. Enzymatic ligation of these truncated tRNAs with valyl-acylated pdCpA afforded full-length, chemically aminoacylated suppressor tRNAs (10). Addition of the preacylated yeast tRNA2Gln(A36) or tRNA2Gln(A36 A38) to an in vitro translation reaction containing E. coli chorismate mutase DNA bearing an amber stop codon at position Gln-88 provided significant quantities of full-length protein (57% and 74%, respectively, relative to a yeast Phe-derived suppressor tRNA acylated with valine, Fig. 1). These suppression efficiencies are comparable to those observed with the E. coli tRNA2Gln-derived orthogonal suppressor (1) and indicate that yeast tRNA2Gln(A36) and tRNA2Gln(A36 A38) are able to function with the E. coli ribosomal machinery. In contrast, reactions containing unacylated yeast tRNA2Gln(A36) or (A36 A38) failed to provide full-length protein relative to the negative control lacking any suppressor tRNA, consistent with the above findings that these yeast suppressor tRNAs are not efficient substrates for any endogenous E. coli aaRS.
Figure 1.
In vitro suppression with yeast glutamine-derived suppressor tRNAs. The amount of full-length protein produced relative to the reaction containing valine-acylated yeast tRNAPhe(CUA) was quantitated by using a PhosphorImager and is listed below each lane.
These results collectively demonstrate that yeast tRNA2Gln(A36) and tRNA2Gln(A36 A38) satisfy the first criteria of an orthogonal tRNA suitable for the delivery of unnatural amino acids into proteins in vivo. Neither suppressor tRNA is significantly acylated by any endogenous E. coli aaRS, yet both suppressors are compatible with the E. coli ribosome.
Yeast GlnRS Aminoacylates Yeast tRNA2Gln(CUA) in Vitro and in E. coli.
The ability of yeast tRNA2Gln(A36 A38) to serve as an orthogonal suppressor tRNA raises the possibility of using yeast GlnRS in conjunction with this suppressor as an functional aaRS/tRNA pair in E. coli. Active site identity between yeast and E. coli GlnRS genes (19) coupled with the high-resolution crystal structure of E. coli GlnRS (20) provides a subsequent rationale for altering amino acid specificity. Purified yeast GlnRS was assayed in vitro for its ability to aminoacylate both wild-type yeast tRNA2Gln as well as the yeast tRNA2Gln(A36 A38) suppressor (both generated by run-off transcription). Under physiological concentrations of substrates, yeast GlnRS was found to aminoacylate the yeast tRNA2Gln(A36 A38) suppressor 20 times slower than wild-type yeast tRNA2Gln (Table 1). The degree to which yeast GlnRS tolerates the CUA amber suppressor anticodon is markedly higher than the tolerance demonstrated by E. coli GlnRS for the analogous E. coli tRNA2Gln(A36 A38) suppressor [charged 1,000-fold slower than wild-type E. coli tRNA2Gln (1), Table 1]. This disparity is not surprising given the relatively modest level of sequence conservation between the E. coli and yeast GlnRS proteins in the region contacting the tRNA anticodon. The activity under physiological conditions of yeast GlnRS toward the yeast tRNA2Gln(A36 A38) suppressor (180 pmol/mg per min) is comparable with the activity of E. coli GlnRS toward the efficient E. coli supE suppressor (63 pmol/mg per min) and the activity of E. coli GlnRS toward the very efficient E. coli tRNA2Gln(A36 A38) suppressor (590 pmol/mg per min) (1). These findings suggest that yeast GlnRS can acylate the yeast tRNA2Gln(A36 A38) suppressor at a rate sufficient for nonsense suppression in vivo.
Table 1.
Activity of E. coli and yeast GlnRS toward various wild-type and suppressor tRNAs
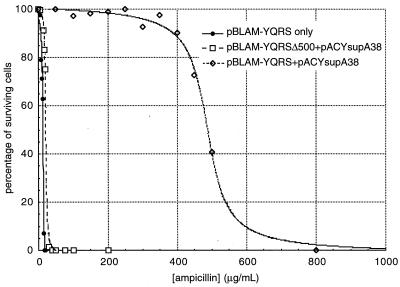
The in vitro results described above were confirmed by in vivo suppression of amber nonsense codons. Although strain BT235 transformed with pACYsupA38 [expressing the yeast tRNA2Gln(A36 A38) suppressor] cannot survive on lactose minimal media because of the inability of E. coli GlnRS to charge the yeast suppressor, the introduction of a second plasmid expressing the yeast GlnRS gene (pBRYQRS) rescued the ability of the cells to grow on lactose. Yeast GlnRS expressed in E. coli therefore is able to charge the yeast suppressor tRNA with glutamine leading to suppression of the lacZ nonsense mutation. The degree to which yeast GlnRS is able to aminoacylate the yeast suppressor in E. coli was evaluated by using the pBLAM plasmid expressing the β-lactamase gene containing an amber nonsense mutation at Ala-184. Survival of E. coli strain DH10B transformed with various plasmids was titrated against a wide range of ampicillin concentrations added to the growth media (Fig. 2). DH10B cells lacking any plasmids exhibited an IC50 of <2.5 μg/ml ampicillin, whereas DH10B cells transformed with pBR322 (expressing wild-type β-lactamase that represents 100% suppression efficiency) manifested an IC50 of approximately 2,000 μg/ml of ampicillin (data not shown). Introduction of the pBLAM plasmid resulted in an IC50 of 12.7 μg/ml, reflecting a small amount of read-through of the amber mutation in the β-lactamase gene. Cells doubly transformed with pACYsupA38 and pBLAM-YQRSΔ500 (expressing a nonfunctional yeast GlnRS gene lacking the C-terminal 500 amino acids, including nearly all active site residues) showed an IC50 of 21.2 μg/ml, suggesting that the overexpression of the yeast suppressor tRNA results in slightly increased levels of amber codon read-through. Cells harboring both pACYsupA38 and pBLAM-YQRS (expressing wild-type yeast GlnRS) demonstrated an IC50 of 488 μg/ml of ampicillin, indicating that coexpression of the yeast orthogonal suppressor and yeast GlnRS in E. coli results in yeast suppressor aminoacylation and efficient amber suppression (Fig. 2). The yeast tRNA2Gln(A36 A38) suppressor and yeast GlnRS therefore constitute a functional tRNA-synthetase pair in vitro and in E. coli.
Figure 2.
Suppression in vivo by yeast GlnRS and yeast tRNA2Gln(A36 A38) (see text).
Yeast GlnRS Does Not Aminoacylate E. coli tRNA.
The final criterion of a completely orthogonal tRNA-synthetase pair is the inability of the synthetase to charge any endogenous tRNA. Failure to satisfy this requirement while attempting to use such a pair for unnatural amino acid delivery in vivo would lead to misacylation of endogenous tRNA, massive mutagenesis throughout the proteome, and cell death (4). Purified yeast GlnRS was assayed in vitro under physiological conditions for its ability to acylate whole E. coli tRNA with 3H-glutamine. No significant quantities of glutamine acylated tRNA could be detected above a control reaction lacking enzyme (<9 pmol/mg per min, Table 1), indicating that yeast GlnRS charges the yeast tRNA2Gln(A36 A38) suppressor at least 20 times faster than all endogenous E. coli tRNAs. The ability of yeast GlnRS to acylate E. coli tRNAGln in vivo also was assessed. E. coli strain X3R2 contains a genomic deletion of GlnRS (ΔglnS) and harbors a heat-inducible λ prophage that supplies GlnRS to the cell (6). Because GlnRS is an essential protein, curing of the resident prophage by heat induction is lethal to X3R2 unless an enzyme capable of aminoacylating E. coli tRNAGln is expressed from a plasmid. X3R2 cells transformed with pBREQRS (expressing E. coli GlnRS) are viable after heat induction, whereas X3R2 transformed with pBRYQRS (expressing yeast GlnRS) are not viable upon induction. These complementation results further demonstrate that yeast GlnRS cannot efficiently aminoacylate E. coli tRNAGln. Taken together, the in vitro and in vivo findings presented above demonstrate that yeast tRNA2Gln(A36 A38) in conjunction with yeast GlnRS constitute a completely orthogonal tRNA-synthetase pair suitable for the delivery of unnatural amino acids in response to amber nonsense codons.
A General Selection for Mutant GlnRS Enzymes that Charge Orthogonal Suppressor tRNAs with Unnatural Amino Acids.
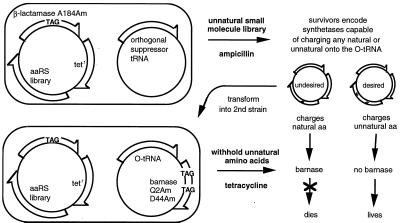
The next step in our efforts requires the selection of mutant yeast GlnRS enzymes capable of charging unnatural amino acids onto the yeast orthogonal suppressor. One approach toward such a selection is to exploit the unique chemical reactivity of unnatural amino acid side chains. However, many amino acids of interest do not have unique reactivity, and thus a more general selection scheme is desirable. Toward this end, we developed a selection for mutant aminoacyl-tRNA synthetases capable of charging any ribosomally accepted molecule, including unnatural amino acids, α-hydroxy acids, and β-amino acids, onto an orthogonal suppressor tRNA. The paradox of selecting for acceptance of an unnatural substrate that is not required for any cellular function was resolved by using both positive and negative selections. In the positive selection, an amber nonsense codon is placed in an essential gene at a nonessential position (i.e., one that tolerates any amino acid but lies before one or more critical residues). Cells expressing mutant synthetase enzymes are selected in the presence of a library of unnatural amino acids added to the growth media for their ability to insert any natural or unnatural amino acid in response to this amber codon. The vast majority of survivors express synthetase enzymes that maintain their ability to charge cognate natural amino acids onto the suppressor tRNA. Rarely, a mutant synthetase enzyme may emerge that acylates one or more unnatural amino acids onto the orthogonal suppressor (Fig. 3). Synthetase genes surviving the positive selection then are subjected to a negative selection in which an amber nonsense codon is placed in a toxic gene at a nonessential position. Cells expressing mutant synthetases now are selected for their ability to survive in the absence of unnatural amino acids. Those cells expressing synthetases capable of charging natural amino acids onto the orthogonal suppressor will produce full-length toxic protein, inhibiting cell growth (Fig. 3). Any cell that expresses a mutant synthetase capable of exclusively acylating an unnatural amino acid onto the orthogonal tRNA will survive both selections. Cells harboring synthetases with mixed substrate specificities will experience growth advantages proportional to their ability to charge unnatural amino acids and inversely proportional to their ability to charge natural amino acids.
Figure 3.
A general selection for mutant aaRS enzymes that charge any ribosomally accepted small molecule onto an orthogonal tRNA.
The positive selection was implemented by using the pBLAM β-lactamase suppressor system described above. Because position 184 in β-lactamase tolerates a wide range of natural amino acids (18), the suppression of nearly any natural or unnatural amino acid in response to the amber codon at this position likely yields active β-lactamase. The presence of several amino acids critical to β-lactamase activity following residue 184 (18), ensures that the truncated enzyme does not confer resistance to β-lactam antibiotics. A series of enrichment experiments was used to demonstrate the ability of this positive selection to select for functional yeast GlnRS enzymes. Plasmids pBLAM-YQRS (expressing wild-type yeast GlnRS) and pBLAM-YQRSΔ500 (expressing the nonfunctional truncated yeast GlnRS gene) were mixed in ratios of 1:103, 1:105, or 1:107, transformed into DH10B cells harboring pACYsupA38 [expressing yeast tRNA2Gln(A36 A38)], and grown in liquid media containing varying concentrations of ampicillin. Upon saturation of the cultures, plasmid DNA was purified and the ratios of pBLAM-YQRS to pBLAM-YQRSΔ500 were determined by restriction digestion and gel electrophoresis (Table 2). Maximum enrichment resulted from media supplemented with 400–500 μg/ml of ampicillin. Under these conditions, pBLAM-YQRS was enriched by factors ranging from 130 to 2 × 105, with the largest enrichment factors arising from cultures with the smallest initial ratio of functional to nonfunctional GlnRS plasmids (Table 3). Successive application of this selection by retransforming harvested plasmid DNA rapidly leads to dominance of the plasmids encoding functional yeast GlnRS enzymes (data not shown). This positive selection is thus an effective method for the selection of functional synthetases, regardless of their substrate specificities, from mixed populations of cells expressing functional and nonfunctional enzymes. Importantly, the stringency of this positive selection may be tuned by varying the concentration of ampicillin added to the growth media as demonstrated in Fig. 2.
Table 2.
Enrichment of functional GlnRS genes
| Ampicillin, (μg/ml) | Starting ratio of YQRS to YQRSΔ500 | Ending ratio of YQRS to YQRSΔ500 | Enrichment factor |
|---|---|---|---|
| 200 | 1:103 | 1:25 | 40 |
| 200 | 1:105 | 1:40 | 2,500 |
| 200 | 1:107 | <1:50 | <200,000 |
| 400 | 1:103 | 1:8 | 130 |
| 400 | 1:105 | 1:20 | 5,000 |
| 400 | 1:107 | 1:50 | 200,000 |
| 500 | 1:103 | 1:12 | 83 |
| 500 | 1:105 | 1:25 | 4,000 |
| 500 | 1:107 | 1:50 | 200,000 |
| 1,000 | 1:103 | 1:30 | 33 |
| 1,000 | 1:105 | 1:50 | 2,000 |
| 1,000 | 1:107 | <1:50 | <200,000 |
Table 3.
Enrichment of nonfunctional GlnRS genes
| Barnase amber codons | Starting ratio of YQRS to YQRSΔ500 | Ending ratio of YQRS to YQRSΔ500 | Enrichment factor |
|---|---|---|---|
| Gln-2 Asp-44 | 103:1 | 1:4 | 4,000 |
| Gln-2 Asp-44 | 105:1 | 1:3 | 300,000 |
| Gln-2 Asp-44 | 107:1 | 1:3 | 3 × 107 |
| Gln-2 Asp-44 Gly-65 | 103:1 | 1:4 | 4,000 |
| Gln-2 Asp-44 Gly-65 | 105:1 | 1:1.5 | 150,000 |
| Gln-2 Asp-44 Gly-65 | 107:1 | 3:1 | 3 × 106 |
The negative selection uses the extreme toxicity of barnase, the extracellular RNase of Bacillus amyloliquefaciens (21, 22). Amber nonsense mutations were introduced into the barnase gene at two or three positions (Gln-2/Asp-44 or Gln-2/Asp-44/Gly-65) and the resulting constructs subcloned into the plasmid expressing the yeast orthogonal suppressor tRNA (pACYsupA38) to afford plasmids pYsupA38B2 and pYsupA38B3, respectively. The positions of these amber mutations were hypothesized to be nonessential to enzyme activity based on an analysis of the three-dimensional structure of barnase (23). Charging of the yeast orthogonal tRNA by a functional synthetase results in the suppression of these amber codons and subsequent production of barnase leading to cell death. The stringency of this negative selection can be modulated by varying the number of amber codons in the barnase gene; cells harboring pYsupA38B2 with two nonsense codons will die from a lower degree of orthogonal tRNA charging than cells harboring pYsupA38B3 and therefore are under more stringent selection conditions.
Cells harboring pBLAM-YQRS and pYsupA38B2 (or pYsupA38B3) were not viable, whereas cells harboring pBLAM-YQRSΔ500 and pYsupA38B2 (or pYsupA38B3) were completely viable. This finding confirms that the negative selection eliminates cells expressing synthetases capable of charging the yeast orthogonal tRNA with a natural amino acid. To assess the ability of the negative selection to enrich nonfunctional synthetases from mixed populations of functional and nonfunctional enzymes, pBLAM-YQRS and pBLAM-YQRSΔ500 were mixed in ratios of 103:1, 105:1, or 107:1 and transformed into DH10B cells harboring pYsupA38B2 or pYsupA38B3. After induction of barnase gene expression, cells were grown to saturation in liquid media. Restriction analysis of harvested plasmid DNA revealed strong enrichment of the plasmid containing the nonfunctional GlnRS gene (pBLAM-YQRSΔ500) (Table 3). Enrichment factors varied between 4,000 and 3 × 107, with the more stringent selection affording 2- to 10-fold higher enrichment than the selection using the triply mutated barnase gene. As observed with the positive selection, lower starting ratios of desired to undesired plasmids yielded higher enrichment factors. These results demonstrate that the barnase-based negative selection strongly selects for synthetases that are nonfunctional under a given set of growth conditions. In the absence of exogenously added unnatural amino acids, synthetases that use unnatural amino acids as substrates would experience a growth advantage over synthetases that exclusively charge natural amino acids (which are constantly provided by the cell). Moreover, the addition of undesired substrates to this negative selection would provide strong selective pressure against acceptance of these substrates.
The substantially higher enrichment resulting from the negative selection compared with the positive selection may arise from several factors. The barnase-based selection is a dominant negative system in which cells harboring mixtures of desired and undesired plasmids (arising from transformation of multiple plasmids into one competent cell) will suffer growth inhibition. In contrast, undesired plasmids are not directly harmful to the cell in the β-lactamase-based selection provided that sufficient quantities of plasmids encoding functional synthetases are present. Spontaneous single base mutation of the amber codons introduced into the barnase gene is lethal and therefore does not contribute to a false positive background, whereas single base mutation of the β-lactamase amber codon results in a strong growth advantage and therefore contributes to the background of the selection. Finally, β-lactamase is secreted gradually into the growth media and therefore can act in trans to rescue cells that may not harbor desired plasmids. Rescue in trans is not possible in the negative selection.
A Rapid Screen for Unnatural Amino Acid Uptake.
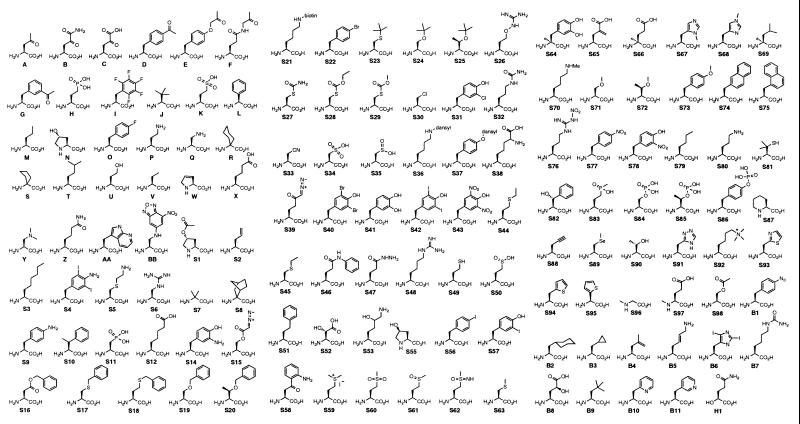
Unnatural amino acid uptake is a critical issue that we have not previously addressed. The high charge density of α-amino acids suggests that these compounds are unlikely to be cell permeable. Natural amino acids are uptaken into bacteria via a collection of protein-based transport systems displaying varying degrees of amino acid specificity. We therefore set out to develop a rapid screen for assessing which unnatural amino acids, if any, were being uptaken by cells. A total of 138 unnatural amino acids and α-hydroxy acids of interest were obtained commercially or by short syntheses from available starting materials (I. Shin, A. Varvak, and P.G.S., unpublished results) and are shown in Fig. 4. Each amino acid was screened at 1 mM in glycerol minimal media for toxicity to DH10B harboring pBLAM-YQRS and pACYsupA38. Toxicities were sorted into five groups: no toxicity (no significant change in doubling times), low toxicity (doubling times increased by less than 10%: S63, S69, S74, S75, S81, S95), moderate toxicity (doubling times increased by 10–50%: B, M, P, S12, S14, S22, S41, S49, S52, S62, S64, S65, S71, S91, S93, B10), high toxicity (doubling times increased by 50–100%: C, Q, V, BB, S2, S5, S50, S60, S78, S83, S89, S90), and extreme toxicity (doubling times increased by more than 100%: W, S15, S26, S27, S30, S31, S39, S47, S88, S94). The toxicity of amino acids scoring as highly or extremely toxic was measured as a function of their concentration to obtain IC50 values (Table 4). In general, amino acids that are very close analogs of natural amino acids (e.g., Q, W, S5, S26, S27, S50, S90, S94) or that display reactive functionality (e.g., S15, S39, S47) demonstrated the highest toxicities. The former trend suggests that mechanisms of toxicity for these unnatural amino acids may be incorporation into proteins or inhibition of essential enzymes that process natural amino acids.
Figure 4.
Library of unnatural amino acids and α-hydroxy acids used in this work. S1-S98 were purchased from Sigma; B1-B11 were purchased from Bachem.
Table 4.
Toxic unnatural amino acids as uptake pathway-specific “lethal alleles”
| Amino acid | IC50, μM | Rescued by | Amino acid | IC50, μM | Rescued by |
| C | 60 | Glu | S31 | 15 | Tyr |
| Q | 20 | S39 | <8 | ||
| V | 50 | Ala | S47 | 20 | Gln |
| W | <8 | Pro | S50 | 60 | Glu |
| BB | 400 | S60 | 200 | Gln, Glu | |
| S2 | 500 | Leu | S78 | 100 | Tyr |
| S5 | 50 | Lys | S83 | 20 | Glu |
| S15 | <8 | S88 | <8 | ||
| S26 | 40 | Arg | S89 | <8 | Met |
| S27 | 200 | Gln | S90 | 60 | Thr |
| S30 | <8 | Ala | S94 | <8 | |
To identify possible uptake pathways for each toxic amino acid, toxicity assays were repeated at IC50 levels (typically 3–500 μM) in media supplemented with an excess (2 mM) of one structurally similar natural amino acid. For 16 toxic amino acids, the presence of excess natural amino acid rescued the ability of the cells to grow in the presence of the toxin, presumably because the natural amino acid effectively outcompeted the toxin for either cellular uptake or for binding to essential enzymes (Table 4). In these cases, the toxic amino acid could be assigned a possible uptake pathway and represented a “lethal allele” whose complementation is required for cell survival. The 16 lethal alleles identified in this manner span 10 possible amino acid uptake groups: Ala, Glu, Lys, Leu, Met, Pro, Gln, Arg, Thr, and Tyr.
These lethal alleles are extremely useful for assaying the ability of cells to uptake nontoxic unnatural amino acids. Each nontoxic unnatural amino acid was added at 2 mM to media containing IC50 levels of each lethal allele. Complementation of the toxic allele, evidenced by the restoration of cell growth, suggests that the nontoxic amino acid is uptaken by the cell (although intracellular levels are not revealed), possibly by the same uptake pathway as that assigned to the lethal allele. A lack of complementation is inconclusive. By using this method, the ability of 22 glutamine and glutamic acid analogs to be uptaken by DH10B was evaluated. Amino acids S27 and S47 were used as toxic glutamine alleles at 100 μM and 30 μM, respectively, whereas S50 was used as a toxic glutamic acid allele at 150 μM. Results from S27 and S47 complementation were in complete agreement and identified amino acids B, Z, S6, S60, S61, and S62 (in addition to S27 and S47) as being uptaken by cells possibly via the glutamine uptake pathway. Similarly, complementation of S50 identified B, C, K, X, S60, S65, and S84 as being uptaken into DH10B, possibly via the glutamic acid transport system. These findings indicate that the E. coli glutamine and glutamic acid transport pathways may tolerate significant perturbations in amino acid structure, including side-chain elongation (X and Z), ketone or methylene placement at the γ-position (B, C, S65), carboxamide replacement with a sulfoxide (S61, a known substrate for a bacterial glutamine transporter; ref. 24) or hydrazide (S47, also a known glutamine transporter substrate; ref. 25), as well as a variety of hybridization changes at the side-chain terminus (S60, S62, K, S84). The lack of glutamine allele complementation by S29, S60, S98, or H1 and similarly the lack of glutamic acid complementation by H, S66, S85, S97, or H1 is inconclusive because downstream targets of these toxic alleles are unknown. The ease of obtaining these results demonstrates that complementation of lethal unnatural amino acid alleles is an efficient method for qualitatively assessing amino acid uptake, requiring far less effort than radiolabeling large numbers of compounds. This general strategy may be used to rapidly evaluate the cellular uptake of a wide range of nonnatural molecules such as nucleic acid base analogs, carbohydrate analogs, or peptide analogs.
Implications for the Evolution of an Organism with an Expanded Genetic Code.
We have shown that yeast GlnRS together with yeast tRNA2Gln(A36 A38) represent a completely orthogonal and functional tRNA-synthetase pair in vitro and in E. coli. This system therefore satisfies the major requirements to begin the evolution of an organism that inserts unnatural amino acids in response to amber codons in vivo. The general selection described above for synthetases capable of acylating nearly any unnatural small molecule onto an orthogonal suppressor tRNA not only provides a promising route toward this goal, but also expands enormously the range of substrates suitable for this methodology. The concept of coupling positive and negative selections may prove to be a very powerful general strategy for selecting mutagenized proteins and nucleic acids with the ability to interact with unnatural substrates. The identification of 16 unnatural amino acid “lethal alleles,” which may be specific to 10 amino acid transport pathways, addresses a key uncertainty in our efforts. By using these toxic alleles in an efficient screen for cellular uptake, we have shown that a wide variety of glutamine and glutamic acid analogs are capable of being transported into E. coli. Efforts to evolve an organism with an expanded genetic code by subjecting large libraries of mutant yeast GlnRS enzymes to the positive and negative selections described above are well underway, and preliminary phenotypes emerging from these selections are promising.
Acknowledgments
Christina Marchetti assisted with the purification of yeast GlnRS. We thank Thomas Magliery and Miro Pastrnak for helpful comments. D.R.L. is a Howard Hughes Predoctoral Fellow. This research was supported by U.S. Department of Defense Grant DAAG55-98-1-0467 and the U.S. Department of Energy under Contract DE-AC03-76SF00098.
ABBREVIATIONS
- GlnRS
glutaminyl-tRNA aminoacyl synthetase
- aaRS
aminoacyl-tRNA synthetase
- GMML
minimal media containing 1% glycerol and 0.3 mM leucine
References
- 1.Liu D R, Magliery T J, Schultz P G. Chem Biol. 1997;4:685–691. doi: 10.1016/s1074-5521(97)90224-6. [DOI] [PubMed] [Google Scholar]
- 2.Liu D R, Magliery T J, Pastrnak M, Schultz P G. Proc Natl Acad Sci USA. 1997;94:10092–10097. doi: 10.1073/pnas.94.19.10092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornish V W, Mendel D, Schultz P G. Angew Chem Int Ed Engl. 1995;34:621–633. [Google Scholar]
- 4.Yarus M. Prog Nucleic Acids Res Mol Biol. 1979;23:195–225. doi: 10.1016/s0079-6603(08)60134-8. [DOI] [PubMed] [Google Scholar]
- 5.Yamao F, Inokuchi H, Ozeki H. Jpn J Genet. 1988;63:237–249. doi: 10.1266/jjg.63.237. [DOI] [PubMed] [Google Scholar]
- 6.Uemura H, Rogers M J, Swanson R, Watson L, Söll D. Protein Eng. 1988;2:293–296. doi: 10.1093/protein/2.4.293. [DOI] [PubMed] [Google Scholar]
- 7.Hartley R W. J Mol Biol. 1988;202:913–915. doi: 10.1016/0022-2836(88)90568-2. [DOI] [PubMed] [Google Scholar]
- 8.Noren C, Anthony-Cahill S J, Suich D J, Noren K A, Griffith M C, Schultz P G. Nucleic Acids Res. 1990;18:83–88. doi: 10.1093/nar/18.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cload S T, Liu D R, Froland W A, Schultz P G. Chem Biol. 1996;3:1033–1038. doi: 10.1016/s1074-5521(96)90169-6. [DOI] [PubMed] [Google Scholar]
- 10.Thorson J S, Cornish V W, Barrett J E, Cload S T, Yano T, Schultz P G. In: Methods in Molecular Biology. Martin R, editor. Clifton, NJ: Humana; 1995. pp. 43–73. [DOI] [PubMed] [Google Scholar]
- 11.Neidhardt F, Umbarger H E. In: Escherichia coli and Salmonella. Neidherdt F, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 13–16. [Google Scholar]
- 12.Raunio R, Rosenqvist H. Acta Chim Scand. 1970;24:2737–2744. doi: 10.3891/acta.chem.scand.24-2737. [DOI] [PubMed] [Google Scholar]
- 13.Ikemura T. Mol Biol Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 14.Hoben P, Söll D. Methods Enzymol. 1985;113:55–59. doi: 10.1016/s0076-6879(85)13011-9. [DOI] [PubMed] [Google Scholar]
- 15.Lloyd A J, Thomann H-U, Ibba M, Söll D. Nucleic Acids Res. 1995;23:2886–2892. doi: 10.1093/nar/23.15.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whelihan E F, Schimmel P. EMBO J. 1997;16:2966–2974. doi: 10.1093/emboj/16.10.2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bradley D, Park J V, Soll L. J Bacteriol. 1981;145:704–712. doi: 10.1128/jb.145.2.704-712.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang W, Petrosino J, Hirsch M, Shenkin P S, Palzkill T. J Mol Biol. 1996;258:688–703. doi: 10.1006/jmbi.1996.0279. [DOI] [PubMed] [Google Scholar]
- 19.Lamour V, Quelvillon S, Diriong S, N′guyen V C, Lipinski M, Mirande M. Proc Natl Acad Sci USA. 1994;91:8670–8674. doi: 10.1073/pnas.91.18.8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rould M A, Perona J J, Steitz T A. Nature (London) 1991;352:213–218. doi: 10.1038/352213a0. [DOI] [PubMed] [Google Scholar]
- 21.Axe D D, Foster N W, Fersht A R. Proc Natl Acad Sci USA. 1996;93:5590–5594. doi: 10.1073/pnas.93.11.5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jucovic M, Hartley R W. Proc Natl Acad Sci USA. 1996;93:2343–2347. doi: 10.1073/pnas.93.6.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buckle A M, Schreiber G, Fersht A R. Biochemistry. 1994;33:8878–8889. doi: 10.1021/bi00196a004. [DOI] [PubMed] [Google Scholar]
- 24.Ayling P D. J Bacteriol. 1981;148:514–520. doi: 10.1128/jb.148.2.514-520.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiner J H, Heppel L A. J Biol Chem. 1971;246:6933–6941. [Google Scholar]
- 26.Ludmerer S W, Wright D J, Schimmel P. J Biol Chem. 1993;268:5519–5523. [PubMed] [Google Scholar]
- 27.Sherman J M, Thomann H, Söll D. J Mol Biol. 1996;256:818–828. doi: 10.1006/jmbi.1996.0128. [DOI] [PubMed] [Google Scholar]
- 28.Ibba M, Hong K-W, Sherman J M, Sever S, Söll D. Proc Natl Acad Sci USA. 1996;93:6953–6958. doi: 10.1073/pnas.93.14.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]