Abstract
The TATA box-binding protein (TBP) is an essential component of the RNA polymerase II transcription apparatus in eukaryotic cells. Until recently, it was thought that the general transcriptional machinery was largely invariant and relied on a single TBP, whereas a large and diverse collection of activators and repressors were primarily responsible for imparting specificity to transcription initiation. However, it now appears that the “basal” transcriptional machinery also contributes to specificity via tissue-specific versions of TBP-associated factors as well as a tissue-specific TBP-related factor (TRF1) responsible for gene selectivity in Drosophila. Here we report the cloning of a TBP-related factor (TRF2) that is found in humans, Drosophila, Caenorhabditis elegans, and other metazoans. Like TRF1 and TBP, TRF2 binds transcription factor IIA (TFIIA) and TFIIB and appears to be part of a larger protein complex. TRF2’s primary amino acid structure suggests divergence in the putative DNA binding domain, and not surprisingly, it fails to bind to DNA containing canonical TATA boxes. Most importantly, TRF2 is associated with loci on Drosophila chromosomes distinct from either TBP or TRF1, so it may have different promoter specificity and regulate a select subset of genes. These findings suggest that metazoans have evolved multiple TBPs to accommodate the vast increase in genes and expression patterns during development and cellular differentiation.
Gene expression is largely controlled at the level of transcription (1). Recent evidence suggests that there are three classes of proteins involved in regulation of transcription: sequence-specific DNA binding proteins (activators and repressors), coregulators (i.e., coactivators and corepressors), and components of the basal machinery (2). Previously it was thought that the combinatorial action of activators and repressors was responsible for regulating transcription initiation, whereas the basal machinery remained invariant and simply responded to the enhancer binding factors.
Now it has become apparent that the basal factors themselves are combinatorial, and that they may play a bigger role in determining tissue- and cell type-specific patterns of gene transcription than previously appreciated. For example, the composition of the transcription factor IID (TFIID) complex, which is composed of TBP (TATA box-binding protein) and multiple TBP-associated factors (TAFs) is not fixed. Indeed, TAFII105, a TAFII130 homolog, appears to be restricted to select TFIID complexes found in certain cell types and may contribute to tissue-specific regulation (3). Similarly, human TAFII30 is associated with a subset of TFIID complexes (4). TAFs can apparently also function apart from TFIID, as they were recently found in the SAGA complex, although its role in transcription initiation remains to be elucidated (5). Remarkably, TBP itself may not be part of all transcriptionally active TAF complexes (6), consistent with the notion of specificity within the preinitiation complex. In addition to variations in the basal machinery, it has become evident that there are multiple coregulators with many subunits in common (7–9), leading to the hypothesis of regulation by mixing and matching subunits into multiple coregulating complexes.
As further evidence of gene selectivity by the general transcription apparatus, it was found that Drosophila TBP-related factor (TRF, now called TRF1), which was originally thought to be an enhancer binding factor (10), most likely functions as a cell type-specific or gene-selective TBP (11). Like TBP, TRF1 binds the basal transcription factors TFIIA and TFIIB and can direct transcription from TATA-containing promoters. TRF1 is also part of a protein complex, but the complex appears to be distinct from TFIID. This TBP-related factor is expressed primarily in neuronal and germ cells and was originally isolated in a genetic screen for mutants with a “shaker” phenotype, which is associated with defects in potassium channel gene expression (10). Additionally, immunostaining of Drosophila polytene chromosomes revealed that TRF1 appears to regulate a unique subset of genes, many of which are associated with neuronal function, fertility, or tRNA genes (11). It is thus possible that TRF1 is a tissue-specific component of the initiation complex involved in regulating select classes of genes.
The discovery of one cell type-specific TBP homolog led us to ask whether there were other TBP homologs. By analyzing the available expressed sequence tags (ESTs) we found that there are indeed other TBP related factors. This additional TBP-related factor, called TRF2, was found in human, mouse, Drosophila, Caenorhabditis elegans, rat, Xenopus, and the nematode Brugia malayi, but neither yeast nor archaea. The primary structure of this protein varies substantially across species, but each contains a conserved bipartite repeat core domain homologous to that of TBP. In the case of Drosophila TRF2, it is 39% identical to the core domain of TBP. TBP residues that contact TFIIA and TFIIB are highly conserved with TRF2, and indeed by using pull-down assays we found that hTRF2 and dTRF2 bind to TFIIA and TFIIB. Furthermore, we find that TRF2, like TBP and TRF1, appears to be part of a larger protein complex. Finally, immunostaining of Drosophila polytene chromosomes suggests that TRF2 may regulate a subset of genes distinct from those of TBP and TRF1.
MATERIALS AND METHODS
Database Searches.
By conducting a BLAST search (12) of the available EST database with human TBP, we found human TRF2. Subsequent searches using the TRF2 sequence yielded Drosophila, mouse, C. elegans, rat, Xenopus, and B. malayi versions of TRF2. To date, 33 human TRF2 ESTs have been found, including GenBank accession nos. H94774, AI126163, and AI075193. Our sequencing efforts revealed that there is an in-frame stop codon (TAG) 192 bp upstream from the start codon, which is not apparent in the EST database because of sequencing ambiguity.
The partial Drosophila TRF2 EST has the accession no. AA392067. The full-length C. elegans sequence is contained in the cosmid F39H11. A partial list of accession numbers for TRF2 EST’s from other organisms follows: B. malayi, AA109303; mouse, AA840611; Xenopus, AI031140; and rat, AI102800.
Cloning and Sequencing of Drosophila TRF2 cDNA.
A 50-base probe from the 5′ end of the Drosophila EST was chemically synthesized and radiolabeled with [γ-32P]ATP. We used this probe to screen a Drosophila cDNA library and isolated five independent clones. Both strands of Drosophila TRF2 were sequenced, and the sequence has been deposited in GenBank (accession no. AF136569).
Northern Blot Analysis.
Northern blots were purchased (CLONTECH) and probed with a human TRF2 cDNA probe made by random-priming the 1,100-bp NotI/XhoI fragment of the EST H94774, which contains the entire coding region. The blots were washed with 0.1× SSC at 50°C. The hTRF2 cDNA signal (1.5 kb) was quantitated, and the blot was stripped and reprobed using actin cDNA. The actin signal was quantitated and used to normalize the TRF2 signal. In the case of heart and muscle, the standard actin transcript (2 kb), not the overexpressed, smaller transcript (1.5 kb) was used. In the case of testis, the 1.5-kb TRF2 transcript was quantitated, not the 2.3-kb transcript.
TFIIA and TFIIB Interaction Assay.
Human TFIIA (large and small subunits) and TFIIB were expressed in Escherichia coli and purified to homogeneity (13). Drosophila TFIIA (large and small subunits) and TFIIB were expressed in E. coli and purified to homogeneity (11). Glutathione Sepharose beads (Amersham Pharmacia) were loaded with either a fusion of glutathione S-transferase (GST) and hTRF2, GST and dTRF2, or GST alone. TFIIA and TFIIB were incubated with the protein beads for 2 hours at 4°C in HEMG (25 mM Hepes, pH 7.9/0.1 mM EDTA/12.5 mM MgCl2/10% glycerol) with 100 mM NaCl, 0.1% CHAPS, and 0.1% Nonidet P-40, and then washed extensively. Blots were probed with antibodies recognizing human or Drosophila TFIIA (large) or TFIIB, as appropriate.
Antibodies.
The peptide from residues 149–192 of dTRF2 was chemically synthesized and coupled to rabbit serum albumin using dimethyl adipimidate. Rabbits were immunized with 100 μg of conjugated peptide in Ribi adjuvant (Ribi Immunochem) and boosted twice. Antibodies were affinity-purified against peptide coupled to vinylsulfone-activated agarose beads (Sigma).
Recombinant Protein Production.
pGST-hTRF2 was constructed by inserting human TRF2 into pVL1392GST (14) using NdeI and XbaI sites generated by PCR. pGST-dTRF2 was constructed by inserting residues 149–375 of dTRF2 into pVL1392GST by using NdeI and XbaI sites generated by PCR.
pGST-hTRF2 and pGST-dTRF2 were used for production of recombinant baculovirus (PharMingen) which was used to infect Sf9 cells and produce GST-hTRF2 and GST-dTRF2. Infected Sf9 cells were lysed 2 days posttransfection, and the soluble extract was incubated with glutathione Sepharose beads (Amersham Pharmacia) for 2 hours. The beads were subsequently washed with HEMG buffer containing 1M NaCl, 1% Nonidet P-40, and 1% CHAPS.
Size Exclusion Chromatography.
Drosophila embryo nuclear extract was prepared essentially as described (11) and fractionated by POROS heparin chromatography (PerSeptive Biosystems, Framingham, MA) by using a gradient of 0.1–1.0 M NaCl in HEMG with 0.5% CHAPS. The peak of TRF2 eluted at 0.6 M NaCl. The 0.6 M NaCl fraction was loaded on a Superose 6 (Amersham Pharmacia) gel filtration column in HEMG with 0.6 M NaCl and 0.5% CHAPS. Fractions were collected and tested for the presence of Drosophila TRF2 by Western blot analysis using anti-Drosophila TRF2 antibody.
Immunostaining.
Drosophila polytene chromosomes from the Bg61–4.1 fly line were stained by using indirect immunofluorescence as described (15). dTAF250 was detected by using anti-dTAF250 (mouse monoclonal), and dTRF2 was detected by using the polyclonal rabbit antibody described above, and then with an affinity-purified secondary antibody (donkey anti-rabbit or anti-mouse IgG conjugated with FITC and Texas red, respectively.) Samples were examined by using a Zeiss Universal fluorescence microscope with a ×63 Neofluor objective. An Image Point cooled charge-coupled device camera (Photometrics, Tuscon, AZ) was used to collect digital images.
RESULTS
TRF2, a Third TBP Homolog.
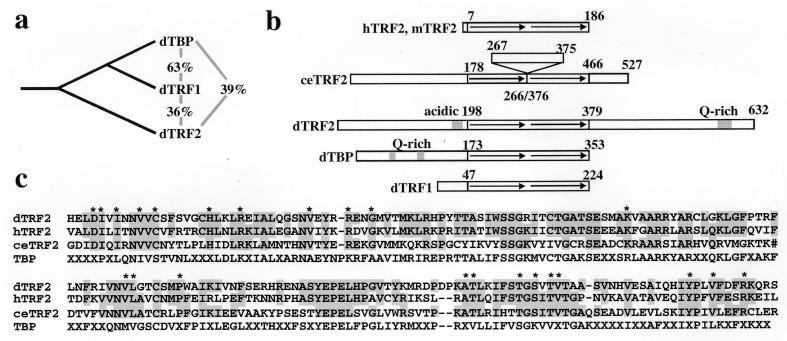
A blast search (12) of the available genomic and EST databases revealed human, mouse, Drosophila, C. elegans, rat, Xenopus, and B. malayi TBP homologs distinct from cell type-specific Drosophila TRF1 (see Materials and Methods). Because only a partial Drosophila TRF2 sequence was available in the database, we cloned full-length Drosophila TRF2. A Drosophila cDNA library was screened by using a probe derived from the Drosophila TRF2 EST, and five independent clones were found. A comparison of the three Drosophila TBP family members indicates that TBP and TRF1 are most closely related, and TRF2 is more related to TBP than TRF1 (Fig. 1 a and c, and Fig. 2). This result suggests that TRF2 diverged from TBP and later, TRF1 evolved from TBP. This hypothesis is supported by the observation that yeast contains TBP but no TRF family members, whereas C. elegans has only TBP and TRF2 and Drosophila contains TBP, TRF1, and TRF2.
Figure 1.
Comparison of TBP family members. (a) Percent identity in the bipartite repeat domain between the Drosophila TBP family members. (b) Organization of domains in TBP family members. The arrows represent the bipartite repeats of the core domain. (c) Alignment of the core domains of dTRF2, hTRF2, ceTRF2, and the TBP consensus sequence, with “X” denoting nonconserved residues. ∗ denotes residues conserved between TRF2 family members that are different from the TBP consensus. The TBP consensus was obtained by comparing Drosophila, human, and C. elegans TBP. Shown are residues 198–379 of dTRF2, 7–186 of hTRF2, and 178–466 of C. elegans TRF2. Full-length Drosophila TRF2 and full-lentgh human TRF2 have been deposited in the GenBank database (accession nos. AF136569 and AF136570). # in the ceTRF2 sequence denotes the 109-residue insertion.
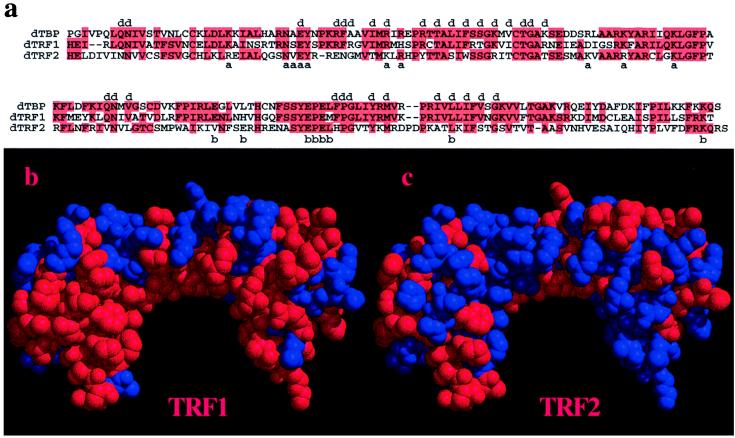
Figure 2.
Analysis of dTBP, dTRF1, and dTRF2 conserved residues. (a) Alignment of dTBP, dTRF1, and dTRF2. Red boxes highlight identical residues; a, b, and d indicate TFIIA-, TFIIB-, and DNA-binding residues, respectively (18, 23, 25, 26). (b and c) Location of residues conserved in dTRF1 (a) and dTRF2 (b) visualized on the TBP model. Red residues are conserved with TBP, and blue residues are nonconserved. The TBP model is as described in ref. 26, with the N-terminal repeat on the right.
Although human and Drosophila TRF2 both have a conserved bipartite repeat core (Fig. 1 b and c and Fig. 2), they vary in size and domain structure. Human TRF2 contains essentially only the core domain (Fig. 1b), whereas Drosophila TRF2 has an N-terminal domain and, unlike either TBP or TRF1, a novel C-terminal domain. The bipartite repeats of human and Drosophila TRF2 are 55% identical (Fig. 1c). Several residues that are conserved among the TRF2 proteins are different from the TBP consensus sequence (Fig. 1c). Most notable are the sequences DIXIXNVVC and TGSXTVT, which are near the N and C termini, respectively, of the bipartite repeat (Fig. 1a). These regions have both been implicated in transcriptional activation in yeast (16, 17).
C. elegans TRF2 is 39% identical to dTRF2, 33% identical to TRF1, and 31% identical to dTBP in the core domain. A BLAST search of available ESTs and GenBank sequences revealed that the human protein most similar is TRF2. C. elegans TRF2 has a unique domain structure; there is a 109-aa insertion between the bipartite repeats of the core domain. When compared with TBP, the insertion is located in the interrepeat strand between helix 2 and sheet 1′, which is on the top of the “saddle” (18). Mutations in this region of TBP affect activation, but not DNA, TFIIA, or TFIIB binding (19). TAFs are thought to bind to this region of TBP (20), so it is intriguing to speculate that this insertion is a domain with TAF-like function, or is an interface for interaction with novel TAF-like subunits.
TRF2 Binds Both TFIIA and TFIIB.
As part of preinitiation complex assembly, TBP binds both TFIIA and TFIIB (2). In vitro, both TBP and TRF1 have been shown to form stable complexes with TFIIA and TFIIB (11, 21, 22). Not surprisingly, Drosophila TBP residues that contact TFIIA and TFIIB (23–25) are well conserved in Drosophila TRF1 (70% and 75%, respectively). TRF2 residues corresponding to residues in TBP that contact TFIIA and TFIIB are also well conserved. Although Drosophila TRF2 is only 39% identical to Drosophila TBP in the core domain, the residues corresponding to the TFIIA and TFIIB binding residues are 60% and 75% identical, respectively. Furthermore, the TFIIA-binding residues are 90% similar, as three TFIIA-binding residues in TBP are changed from lysine to arginine or from arginine to lysine in TRF2.
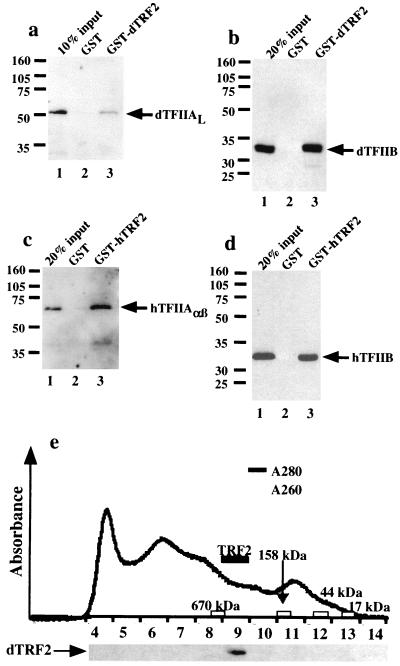
Because of the high degree of conservation with the putative TFIIA- and TFIIB-binding residues in TRF2, we expected that TRF2 would interact with TFIIA and TFIIB, much like TBP and TRF1 bind these components of the preinitiation complex. Therefore, we generated GST fusion proteins containing either human or Drosophila TRF2 and immobilized them on glutathione Sepharose beads. Next, we incubated human TFIIA and TFIIB with immobilized human TRF2 and Drosophila TFIIA and TFIIB with immobilized Drosophila TRF2. After washing extensively with buffer containing 0.1% CHAPS and 0.1% Nonidet P-40, TFIIA and TFIIB were retained on both human TRF2 and Drosophila TRF2 beads but not control beads (Fig. 3 a–d), although Drosophila TRF2 shows lower affinity for TFIIA than does human TRF2. Thus, as predicted by sequence homology, TRF2 can bind directly to both TFIIA and TFIIB.
Figure 3.
Biochemical analysis of TRF2. (a–d) Western blot analysis of TFIIA and TFIIB binding to TRF2. A GST fusion of dTRF2 (a and b) or hTRF2 (c and d) and GST (a–d) was bound to glutathione beads and then incubated with dTFIIA (a), dTFIIB (b), hTFIIA (c), or hTFIIB (d). Unbound protein was then washed away, and the bound protein was detected by Western blot analysis. (e) Elution of endogenous dTRF2 from a Superose 6 gel filtration column (Amersham Pharmacia). Endogenous Drosophila TRF2 eluted slightly after the 670-kDa molecular mass standard. The elution profile of the molecular mass standards is indicated along the axis.
TRF2’s Putative DNA-Binding Domain Is Distinct from That of TRF1 and TBP, and It Fails to Bind Canonical TATA-Box Elements.
Visualization of the conserved and nonconserved residues of TRF1 and TRF2 on the TBP structure leads to the striking observation that the underside of the “saddle,” which directly contacts DNA, is much more conserved in TRF1 than TRF2 (see Fig. 2b). In fact, 84% of TBP’s DNA-binding residues (18) are conserved in TRF1, as compared with 63% overall conservation. By contrast, only 32% of TBP’s DNA-binding residues are conserved in TRF2, as compared with 39% overall conservation. Thus, TRF2 may not bind to TATA-containing promoters. Indeed, thus far, we have not observed either human or Drosophila TRF2 binding to TATA-containing promoters in vitro, either in the presence or absence of TFIIA and TFIIB (data not shown). We have found that TRF2 binds DNA but have not yet identified a specific DNA sequence recognized by TRF2.
Endogenous TRF2 Is Part of a Large Protein Complex.
In addition to TBP, TAFs also are required to reconstitute activated transcription and promoter recognition in vitro (27). Biochemical and in vivo evidence suggest that some activators recruit TFIID to the promoter via the TAFs (28–30). TBP and at least 9 TAFs form the Drosophila TFIID complex, whereas human TFIID contains as many as 13 TAFs, together forming a complex of >800 kDa. Thus, TRF2 may also be part of a large multisubunit complex. To address this question, we analyzed dTRF2 from Drosophila embryo nuclear extracts by gel filtration chromatography to determine whether it migrates as a monomeric 68-kDa protein or whether it elutes as a large complex. When compared with molecular weight standards, native TRF2 eluted from a Superose 6 column slightly after the 670-kDa marker but well before the 158-kDa marker; we therefore estimate that TRF2 is part of a native complex of approximately 500 kDa (Fig. 3e). To date, we have not been able to immunoprecipitate endogenous TRF2.
TRF2 Is Differentially Expressed.
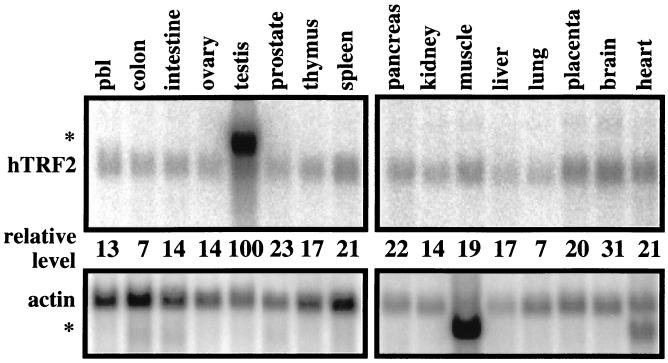
Cell type specificity and developmental specificity is caused by differential expression of genes and is largely controlled by activators and repressors (1). However, variations in the basal machinery may also play an important role in imparting specificity to the initiation of transcription, so we determined whether TRF2 itself might be differentially expressed in certain tissues, as was the case for TRF1. We used Northern blot analysis to determine the relative expression levels of TRF2 in multiple human tissues, normalized to actin expression. The highest levels of TRF2 expression was observed in testis (Fig. 4), as is the case for TBP (31). This result is in agreement with the source of human TRF2 ESTs; 10 of 33 ESTs are from testis. Furthermore, an abundant alternative transcript is also observed in testis, which is expressed 4-fold more than the normal transcript. Aside from testis, there is some variation in the level of expression in other organs tested but no obvious tissue restriction. Brain has the highest levels of expression, and colon the lowest, with a 4.3-fold difference between the two (Fig. 4). Larger differences may exist between specific cell types that would not be detected at this level of analysis.
Figure 4.
Northern blot analysis of hTRF2 expression in multiple human tissues. (Upper) Blots were probed with TRF2 and actin. An alternative TRF2 transcript was observed in testis (∗), and an alternative actin transcript was observed in both muscle and heart (∗). (Lower) Comparison of relative hTRF2 levels. The hTRF2 signal was normalized against actin, and the testis level was arbitrarily set to 100. pbl, peripheral blood leukocyte.
TRF2 Targets a Set of Genes Distinct from Those Associated with TRF1 and TBP.
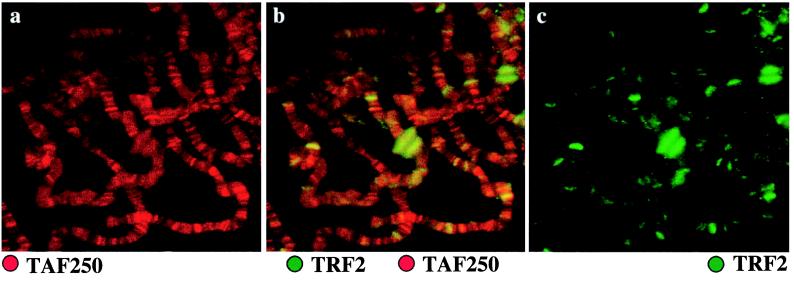
If transcriptional selectivity can be achieved at the level of basal factors, then different TBP family members might be expected to be associated with different loci of the genome. Immunostainings of Drosophila salivary gland polytene chromosomes show that two components of TFIID, TAFII250 (Fig. 5a) and TBP (11), are found to be located at a large number of loci. In contrast, TRF2 is found at fewer sites (Fig. 5b). Some of the loci containing TRF2 appear to have little or no TFIID present (Fig. 5b). Likewise, antibody staining revealed that TRF1 is specifically associated with a unique subset of loci, many of which represent loci that contain neuronal or fertility genes (11). Most importantly, TRF2 is associated with an entirely different set of loci than TRF1, indicating that these two factors are likely involved with expression of distinct sets of genes.
Figure 5.
TRF2 and TAFII250 localization on Drosophila polytene chromosomes. Anti-TAFII250 (raised in a mouse) antibodies visualized with Texas red-conjugated secondary antibody is shown in red, and anti-dTRF2 (raised in a rabbit) antibodies visualized with FITC-conjugated secondary antibodies is shown in green. (a) Anti-TAFII250 only. (b) Both anti-TAFII250 and anti-dTRF2. (c) Anti-dTRF2 only.
The major sites of staining during puff stage 6–7 include the large early ecdysone puffs at 74EF and 75B of chromosome 3L, as well as puff sites at 2B, 45F-46A, 55E, 71DE, 72D, 78D, 85EF, 88D, and 93D. Additionally, over 40 other sites show labeling over background. Staining of developmentally important genes raises the possibility that TRF2 may be involved in temporal regulation of transcription. This notion is consistent with the observation that Drosophila embryos contain a high maternal dose of TRF2 (data not shown).
DISCUSSION
A third TBP-related factor (TRF2) was identified by searching databases for TBP homologs. The identification of human, mouse, Drosophila, C. elegans, and B. Malayi sequences has allowed a cross-species comparison of the protein. There is a significant degree of variation between species, both within the conserved bipartite repeat and within the N- and C-terminal domains. While the identity between dTRF2 and dTBP within the bipartite repeat core is 39%, TFIIA-binding residues are 90% similar, and TFIIB-binding residues are 75% identical. Consistent with conservation of these residues, TFIIA and TFIIB bound to TRF2 in vitro, suggesting that TRF2, like TBP and TRF1, may participate in transcription in conjunction with TFIIA and TFIIB. Furthermore, just as TBP is part of the TFIID complex, endogenous TRF2 appears to be part of a large, multisubunit complex (500 kDa). Most importantly, endogenous TRF2 is associated with a select set of loci on the Drosophila chromosome in salivary glands distinct from TBP and TRF1, including developmental puffs, which suggests a role for TRF2 in development.
Have Metazoans Evolved Multiple Core Promoter Recognition Complexes to Accommodate Gene Selectivity?
There is a growing body of evidence suggesting that instead of a single TFIID complex that directs all RNA polymerase II transcription, there are multiple versions of TFIID, and furthermore, other complexes can substitute for TFIID. For example, TAFII105 (3) and TAFII 30 (4) are not present in all TFIID complexes, suggesting that different variants of TFIID may be involved with regulating tissue- and developmental-stage-specific transcription. In addition to TAFII105, other TAFs have diverged and evolved tissue- or developmental-stage-specific homologs (M. Fuller, personal communication). Surprisingly, a transcriptionally active TFIID-like complex lacking TBP has been reported (6). In addition to alternate versions of TFIID, TRF1 in vitro (11) can substitute for TBP. Finally, the transcription factor YY1, which is unrelated to TBP, can direct transcription from the initiator element of the adeno-associated virus P5 promoter, among others (32). It is possible that other general factors such as TFIIA, TFIIB, TFIIF, TFIIH, and TFIIS may also have variants that contribute to new specificities.
A sequence comparison of TBP, TRF1, and TRF2 indicates that the TBP DNA-binding residues are 84% conserved in TRF1 but only 32% conserved in TRF2. Not surprisingly, TRF1 can bind both the AdML and Adh promoters, whereas we have been unable to demonstrate TRF2 binding to TATA-containing promoters in vitro. This could be because of experimental factors rather than lack of intrinsic binding activity. For example, although our current human and Drosophila TRF2 preparations bind TFIIA and TFIIB, they may not bind TATA-containing promoters because they are partially misfolded. Alternatively, some TRF2-associated factors may be necessary for specific DNA binding. It may ultimately be necessary to identify target genes regulated by TRF2 before a cognate recognition sequence can be identified and tested for binding. Thus, TRF2 may bind to a novel set of core promoters that remain to be identified. This could lead to control of gene expression with specificity partially determined by the core promoter.
The variety of TFIID-like complexes raises the possibility of transcriptional regulation at the level of the core promoter recognition and basal factor interactions. Different versions of TFIID, the TRF1 complex, and the TRF2 complex may be different core promoter-recognizing factors analogous to multiple σ factors in bacteria. Therefore, in eukaryotes there appears to be a requirement for transcriptional activators, repressors, coactivators, and core promoter-recognizing factors to be assembled to direct gene-specific, temporal, and spatial patterns of gene expression. This multilevel combinatorial mechanism of transcription regulation might have evolved to direct the elaborate and exquisitely controlled networks of gene expression needed for both housekeeping functions and developmental programs in higher eukaryotes.
It appears that only a single TBP family member is necessary for transcription regulation in yeast. By contrast, seven metazoan organisms have been found to contain at least two distinct TBP homologs, TBP and TRF2. Interestingly, Drosophila apparently uses at least three TBP family members, including TRF1 and TRF2. Thus far, mammalian TRF1 has not been identified, either by available EST database searching or by traditional experimental homology cloning procedures (B. Lemon & R.T., unpublished results). However, given that mammalian genomes generally contain larger gene families than Drosophila, we anticipate that mammals will use multiple TBP family members and that future experiments will reveal additional TRFs. It will be intriguing to ascertain how TBP family members collaborate to coordinately regulate gene expression.
Acknowledgments
We thank Janis Werner for preparation of Drosophila polytene chromosome spreads, David King for peptide synthesis, and Mallory Haggart for DNA sequencing. For critical reading of the manuscript we thank Bryan Lemon, Jörk Zwicker, Andreas Ladurner, and Anders Näär. We also thank Mike Holmes, Soojin Ryu, Carla Inouye, Andreas Hochheimer, Jörk Zwicker, Tom O’Brien, Shinako Takada, Ray Jacobson, and Shane Albright for various reagents, as well as the rest of the Tjian laboratory for helpful suggestions. M.D.R. is a Leukemia Society Fellow. This work was supported in part by grants from the National Institute of Health to J.T.L. (GM25232) and R.T. (CA25417).
ABBREVIATIONS
- TBP
TATA box-binding protein
- TF
transcription factor
- TRF
TBP-related factor
- TAF
TBP-associated factor
- EST
expressed sequence tag
- GST
glutathione S-transferase
Note Added in Proof
Ohbayashi et al. recently reported the sequences of mouse (33) and human (34) TRF2.
Footnotes
References
- 1.Tjian R, Maniatis T. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 2.Goodrich J A, Cutler G, Tjian R. Cell. 1996;84:825–830. doi: 10.1016/s0092-8674(00)81061-2. [DOI] [PubMed] [Google Scholar]
- 3.Dikstein R, Zhou S, Tjian R. Cell. 1996;87:137–146. doi: 10.1016/s0092-8674(00)81330-6. [DOI] [PubMed] [Google Scholar]
- 4.Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Cell. 1994;79:107–117. doi: 10.1016/0092-8674(94)90404-9. [DOI] [PubMed] [Google Scholar]
- 5.Grant P A, Schieltz D, Pray-Grant M G, Steger D J, Reese J C, Yates J R, III, Workman J L. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 6.Wieczorek E, Brand M, Jacq X, Tora L. Nature (London) 1998;393:187–191. doi: 10.1038/30283. [DOI] [PubMed] [Google Scholar]
- 7.Jiang Y W, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway J W, Conaway R C, Kornberg R D. Proc Natl Acad Sci USA. 1998;95:8538–8543. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun X, Zhang Y, Cho H, Rickert P, Lees E, Lane W, Reinberg D. Mol Cell. 1998;2:213–222. doi: 10.1016/s1097-2765(00)80131-8. [DOI] [PubMed] [Google Scholar]
- 9.Ryu S, Zhou S, Ladurner A G, Tjian R. Nature (London) 1999;397:446–450. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- 10.Crowley T E, Hoey T, Liu J K, Jan Y N, Jan L Y, Tjian R. Nature (London) 1993;361:557–561. doi: 10.1038/361557a0. [DOI] [PubMed] [Google Scholar]
- 11.Hansen S K, Takada S, Jacobson R H, Lis J T, Tjian R. Cell. 1997;91:71–83. doi: 10.1016/s0092-8674(01)80010-6. [DOI] [PubMed] [Google Scholar]
- 12.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Näär A M, Beaurang P A, Robinson K M, Oliner J D, Avizonis D, Scheek S, Zwicker J, Kadonaga J T, Tjian R. Genes Dev. 1998;12:3020–3031. doi: 10.1101/gad.12.19.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruppert S, Tjian R. Genes Dev. 1995;9:2747–2755. doi: 10.1101/gad.9.22.2747. [DOI] [PubMed] [Google Scholar]
- 15.Shopland L S, Lis J T. Chromosoma. 1996;105:158–171. doi: 10.1007/BF02509497. [DOI] [PubMed] [Google Scholar]
- 16.Kim T K, Hashimoto S, Kelleher R J, III, Flanagan P M, Kornberg R D, Horikoshi M, Roeder R G. Nature (London) 1994;369:252–255. doi: 10.1038/369252a0. [DOI] [PubMed] [Google Scholar]
- 17.Arndt K M, Ricupero-Hovasse S, Winston F. EMBO J. 1995;14:1490–1497. doi: 10.1002/j.1460-2075.1995.tb07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J L, Nikolov D B, Burley S K. Nature (London) 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 19.Stargell L A, Struhl K. Mol Cell Biol. 1996;16:4456–4464. doi: 10.1128/mcb.16.8.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bryant G O, Martel L S, Burley S K, Berk A J. Genes Dev. 1996;10:2491–2504. doi: 10.1101/gad.10.19.2491. [DOI] [PubMed] [Google Scholar]
- 21.Yokomori K, Admon A, Goodrich J A, Chen J L, Tjian R. Genes Dev. 1993;7:2235–2245. doi: 10.1101/gad.7.11.2235. [DOI] [PubMed] [Google Scholar]
- 22.Malik S, Hisatake K, Sumimoto H, Horikoshi M, Roeder R G. Proc Natl Acad Sci USA. 1991;88:9553–9557. doi: 10.1073/pnas.88.21.9553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan S, Hunziker Y, Sargent D F, Richmond T J. Nature (London) 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 24.Buratowski S, Zhou H. Science. 1992;255:1130–1132. doi: 10.1126/science.1546314. [DOI] [PubMed] [Google Scholar]
- 25.Geiger J H, Hahn S, Lee S, Sigler P B. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 26.Nikolov D B, Chen H, Halay E D, Usheva A A, Hisatake K, Lee D K, Roeder R G, Burley S K. Nature (London) 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 27.Pugh B F, Tjian R. Cell. 1990;61:1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- 28.Chen J L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 29.Thut C J, Chen J L, Klemm R, Tjian R. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 30.Zhou J M, Zwicker J, Szymanski P, Levine M, Tjian R. Proc Natl Acad Sci USA. 1998;95:13483–13488. doi: 10.1073/pnas.95.23.13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt E E, Schibler U. Development. 1995;121:2373–2383. doi: 10.1242/dev.121.8.2373. [DOI] [PubMed] [Google Scholar]
- 32.Usheva A, Shenk T. Cell. 1994;76:1115–1121. doi: 10.1016/0092-8674(94)90387-5. [DOI] [PubMed] [Google Scholar]
- 33.Ohbayashi T, Makino Y, Tamura T. Nucleic Acids Res. 1999;27:750–755. doi: 10.1093/nar/27.3.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohbayashi T, Kishimoto T, Yasutaka M, Shimada M, Nakadai T, Aoki T, Kawata T, Niwa S, Tamura T. Biochem Biophys Res Commun. 1999;255:137–142. doi: 10.1006/bbrc.1999.0159. [DOI] [PubMed] [Google Scholar]