Abstract
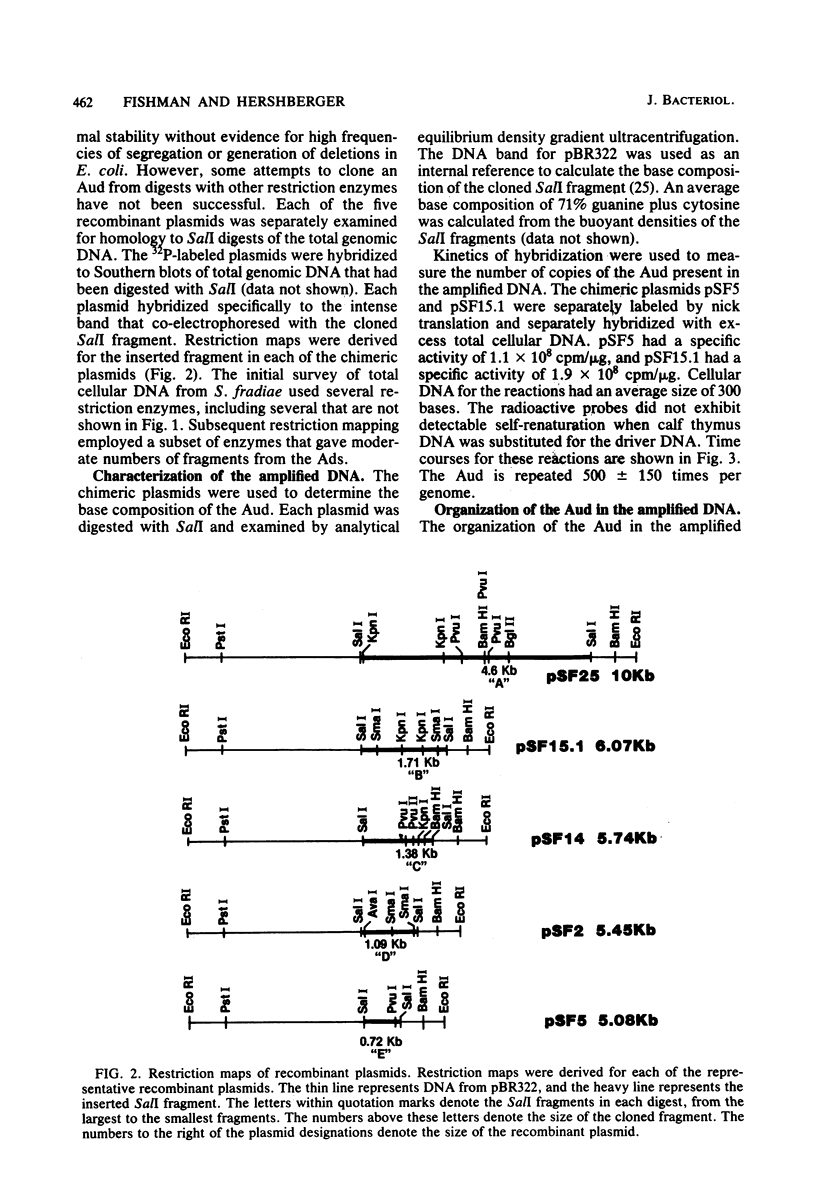
A spontaneous mutant of Streptomyces fradiae contained an amplifiable unit of DNA with a sequence length of approximately 10.5 kilobases that was amplified to approximately 500 copies per chromosome. The amplified DNA appears to be cryptic. SalI fragments of the amplified DNA were cloned into Escherichia coli to construct a restriction map and characterize the amplified DNA. The amplified DNA contained tandem repeats of the amplifiable unit of DNA. The unit had an average base composition of 71% guanine plus cytosine, similar to the chromosomal DNA of Streptomyces species. At least a portion of the amplifiable unit of DNA was present at a low copy number in the wild-type strain. The phenotype of amplified DNA was designated Ads1SF for amplified DNA sequence 1 in S. fradiae.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson R. P., Roth J. R. Tandem genetic duplications in phage and bacteria. Annu Rev Microbiol. 1977;31:473–505. doi: 10.1146/annurev.mi.31.100177.002353. [DOI] [PubMed] [Google Scholar]
- Baltz R. H. Genetic recombination in Streptomyces fradiae by protoplast fusion and cell regeneration. J Gen Microbiol. 1978 Jul;107(1):93–102. doi: 10.1099/00221287-107-1-93. [DOI] [PubMed] [Google Scholar]
- Baltz R. H., Seno E. T. Properties of Streptomyces fradiae mutants blocked in biosynthesis of the macrolide antibiotic tylosin. Antimicrob Agents Chemother. 1981 Aug;20(2):214–225. doi: 10.1128/aac.20.2.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benigni R., Petrov P. A., Carere A. Estimate of the genome size by renaturation studies in Streptomyces. Appl Microbiol. 1975 Aug;30(2):324–326. doi: 10.1128/am.30.2.324-326.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Chater K. F., Hopwood D. A., Kieser T., Thompson C. J. Gene cloning in Streptomyces. Curr Top Microbiol Immunol. 1982;96:69–95. doi: 10.1007/978-3-642-68315-2_5. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Properties of a supercoiled deoxyribonucleic acid-protein relaxation complex and strand specificity of the relaxation event. Biochemistry. 1970 Oct 27;9(22):4428–4440. doi: 10.1021/bi00824a026. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Hashimoto H., Rownd R. H. Transition of the R factor NR1 and Proteus mirabilis: level of drug resistance of nontransitioned and transitioned cells. J Bacteriol. 1975 Jul;123(1):56–68. doi: 10.1128/jb.123.1.56-68.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G. H. On the efficacy of commonly used ribonuclease inhibitors. Biochem Biophys Res Commun. 1976 Mar 22;69(2):469–474. doi: 10.1016/0006-291x(76)90545-3. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Meselson M., Yuan R. DNA restriction enzyme from E. coli. Nature. 1968 Mar 23;217(5134):1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- Molin S., Diaz R., Uhlin B. E., Nordström K. Runaway replication of plasmid R1 is not caused by loss of replication inhibitor activity of gene cop. J Bacteriol. 1980 Aug;143(2):1046–1048. doi: 10.1128/jb.143.2.1046-1048.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono H., Hintermann G., Crameri R., Wallis G., Hütter R. Reiterated DNA sequences in a mutant strain of Streptomyces glaucescens and cloning of the sequence in Escherichia coli. Mol Gen Genet. 1982;186(1):106–110. doi: 10.1007/BF00422920. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Robinson M., Lewis E., Napier E. Occurrence of reiterated DNA sequences in strains of Streptomyces produced by an interspecific protoplast fusion. Mol Gen Genet. 1981;182(2):336–340. doi: 10.1007/BF00269680. [DOI] [PubMed] [Google Scholar]
- Rownd R., Mickel S. Dissociation and reassociation of RTF and r-determinants of the R-factor NR1 in Proteus mirabilis. Nat New Biol. 1971 Nov 10;234(45):40–43. doi: 10.1038/newbio234040a0. [DOI] [PubMed] [Google Scholar]
- SCHILDKRAUT C. L., MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its buoyant density in CsCl. J Mol Biol. 1962 Jun;4:430–443. doi: 10.1016/s0022-2836(62)80100-4. [DOI] [PubMed] [Google Scholar]
- Schrempf H. Plasmid loss and changes within the chromosomal DNA of Streptomyces reticuli. J Bacteriol. 1982 Aug;151(2):701–707. doi: 10.1128/jb.151.2.701-707.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seno E. T., Baltz R. H. S-Adenosyl-L-methionine: macrocin O-methyltransferase activities in a series of Streptomyces fradiae mutants that produce different levels of the macrolide antibiotic tylosin. Antimicrob Agents Chemother. 1982 May;21(5):758–763. doi: 10.1128/aac.21.5.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seno E. T., Pieper R. L., Huber F. M. Terminal stages in the biosynthesis of tylosin. Antimicrob Agents Chemother. 1977 Mar;11(3):455–461. doi: 10.1128/aac.11.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavik N. S., Hershberger C. L. Internal structural organization of chloroplast DNA from Euglena gracilis Z. J Mol Biol. 1976 May 25;103(3):563–581. doi: 10.1016/0022-2836(76)90217-5. [DOI] [PubMed] [Google Scholar]
- Slavik N. S., Hershberger C. L. The kinetic complexity of Euglena gracilis chloroplast DNA. FEBS Lett. 1975 Apr 1;52(2):171–174. doi: 10.1016/0014-5793(75)80799-x. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Veltkamp E., Stuitje A. R. Replication and structure of the bacteriocinogenic plasmids Clo DF13 and CoI E1. Plasmid. 1981 Jan;5(1):76–99. doi: 10.1016/0147-619x(81)90078-0. [DOI] [PubMed] [Google Scholar]
- Wetmur J. G. Hybridization and renaturation kinetics of nucleic acids. Annu Rev Biophys Bioeng. 1976;5:337–361. doi: 10.1146/annurev.bb.05.060176.002005. [DOI] [PubMed] [Google Scholar]





