Abstract
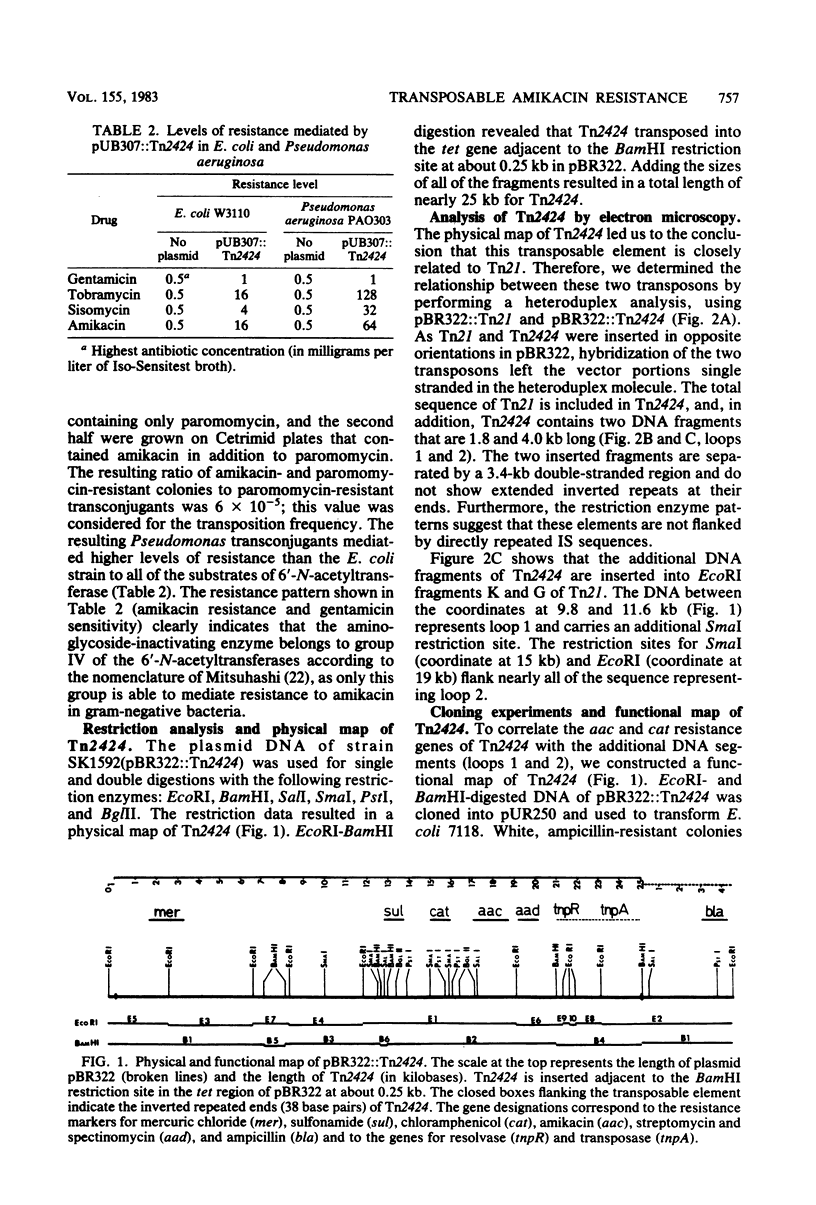
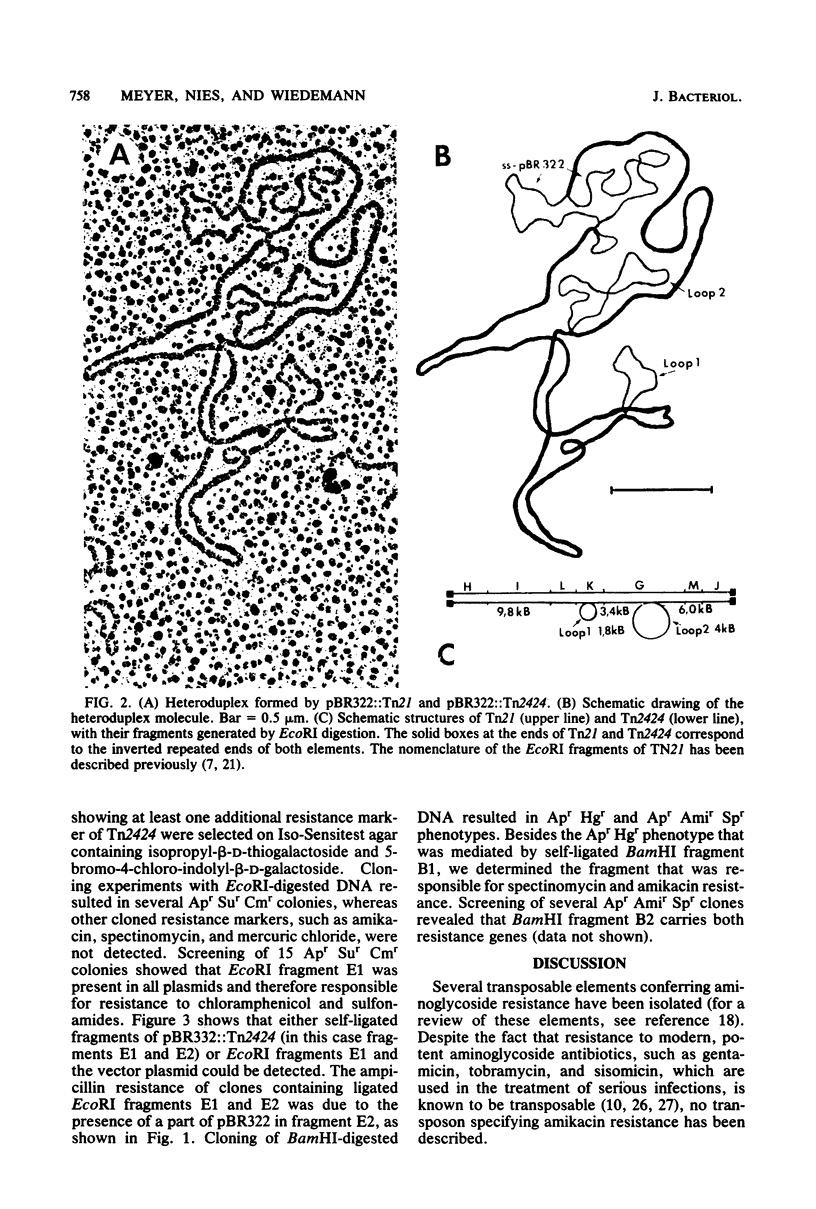
Tn2424, a multiresistance transposon 25 kilobases long, was isolated from IncFII plasmid NR79. Tn2424 transposed resistance to sulfonamides, streptomycin and spectinomycin, mercuric chloride, chloramphenicol, and amikacin with a frequency of 6 X 10(-5). Resistance to amikacin was mediated by a 6'-N-acetyltransferase, which conferred higher levels of resistance in Pseudomonas aeruginosa than in Escherichia coli. A restriction analysis and cloning experiments resulted in a physical and functional map of Tn2424. Comparison by a heteroduplex technique revealed that Tn2424 includes the total sequence of Tn21 and two additional DNA fragments that are 1.8 and 4 kilobases long.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altenbuchner J., Choi C. L., Grinsted J., Schmitt R., Richmond M. H. The transposons Tn501(Hg) and Tn1721(Tc) are related. Genet Res. 1981 Jun;37(3):285–289. doi: 10.1017/s0016672300020280. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett P. M., Grinsted J., Choi C. L., Richmond M. H. Characterisation of Tn501, a transposon determining resistance to mercuric ions. Mol Gen Genet. 1978 Feb 7;159(1):101–106. doi: 10.1007/BF00401753. [DOI] [PubMed] [Google Scholar]
- Bennett P. M., Grinsted J., Richmond M. H. Transposition of TnA does not generate deletions. Mol Gen Genet. 1977 Jul 20;154(2):205–211. doi: 10.1007/BF00330839. [DOI] [PubMed] [Google Scholar]
- Benveniste R., Davies J. Enzymatic acetylation of aminoglycoside antibiotics by Escherichia coli carrying an R factor. Biochemistry. 1971 May 11;10(10):1787–1796. doi: 10.1021/bi00786a009. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- COETZEE J. N. TRANSDUCTION OF SWARMING IN PROTEUS MIRABILIS. J Gen Microbiol. 1963 Oct;33:1–7. doi: 10.1099/00221287-33-1-1. [DOI] [PubMed] [Google Scholar]
- Chandler M., Silver L., Lane D., Caro L. Properties of an autonomous r-determinant from R100.1. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1223–1231. doi: 10.1101/sqb.1979.043.01.139. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Nugent M., Richards H. Transposons encoding trimethoprim or gentamicin resistance in medically important bacteria. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):45–51. doi: 10.1101/sqb.1981.045.01.009. [DOI] [PubMed] [Google Scholar]
- Davies J., Smith D. I. Plasmid-determined resistance to antimicrobial agents. Annu Rev Microbiol. 1978;32:469–518. doi: 10.1146/annurev.mi.32.100178.002345. [DOI] [PubMed] [Google Scholar]
- Grinsted J., Saunders J. R., Ingram L. C., Sykes R. B., Richmond M. H. Properties of a R factor which originated in Pseudomonas aeruginosa 1822. J Bacteriol. 1972 May;110(2):529–537. doi: 10.1128/jb.110.2.529-537.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas M. J., Davies J. Characterization of the plasmids comprising the "R factor" R5 and their relationships to other R plasmids. Plasmid. 1980 May;3(3):260–277. doi: 10.1016/0147-619x(80)90040-2. [DOI] [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981 Mar;145(3):1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsu K., Inoue M., Mitsuhashi S. Transposition of the carbenicillin-hydrolyzing beta-lactamase gene. J Bacteriol. 1982 May;150(2):483–489. doi: 10.1128/jb.150.2.483-489.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Messing J., Gronenborn B., Müller-Hill B., Hans Hopschneider P. Filamentous coliphage M13 as a cloning vehicle: insertion of a HindII fragment of the lac regulatory region in M13 replicative form in vitro. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3642–3646. doi: 10.1073/pnas.74.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Easton A. M., Rownd R. H. Mapping of the resistance genes of the R plasmid NR1. Mol Gen Genet. 1978 Jan 17;158(3):217–224. doi: 10.1007/BF00267192. [DOI] [PubMed] [Google Scholar]
- Pemberton J. M., Holloway B. W. Chromosome mapping in Pseudomonas aeruginosa. Genet Res. 1972 Jun;19(3):251–260. doi: 10.1017/s0016672300014518. [DOI] [PubMed] [Google Scholar]
- Rubens C. E., McNeill W. F., Farrar W. E., Jr Evolution of multiple-antibiotic-resistance plasmids mediated by transposable plasmid deoxyribonucleic acid sequences. J Bacteriol. 1979 Nov;140(2):713–719. doi: 10.1128/jb.140.2.713-719.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubens C. E., McNeill W. F., Farrar W. E., Jr Transposable plasmid deoxyribonucleic acid sequence in Pseudomonas aeruginosa which mediates resistance to gentamicin and four other antimicrobial agents. J Bacteriol. 1979 Sep;139(3):877–882. doi: 10.1128/jb.139.3.877-882.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüther U. Construction and properties of a new cloning vehicle, allowing direct screening for recombinant plasmids. Mol Gen Genet. 1980;178(2):475–477. doi: 10.1007/BF00270503. [DOI] [PubMed] [Google Scholar]
- Schmitt R., Altenbuchner J., Wiebauer K., Arnold W., Pühler A., Schöffl F. Basis of transposition and gene amplification by Tn1721 and related tetracycline-resistance transposons. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):59–65. doi: 10.1101/sqb.1981.045.01.011. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Harafuji H., Yamamoto T. A gene and its product required for transposition of resistance transposon Tn2603. J Bacteriol. 1982 Aug;151(2):723–728. doi: 10.1128/jb.151.2.723-728.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Tanaka M., Baba R., Yamagishi S. Physical and functional mapping of Tn2603, a transposon encoding ampicillin, streptomycin, sulfonamide, and mercury resistance. Mol Gen Genet. 1981;181(4):464–469. doi: 10.1007/BF00428737. [DOI] [PubMed] [Google Scholar]
- Younghusband H. B., Inman R. B. The electronmicroscopy of DNA. Annu Rev Biochem. 1974;43(0):605–619. doi: 10.1146/annurev.bi.43.070174.003133. [DOI] [PubMed] [Google Scholar]
- Zheng Z. X., Chandler M., Hipskind R., Clerget M., Caro L. Dissection of the r-determinant of the plasmid R100.1: the sequence at the extremities of Tn21. Nucleic Acids Res. 1981 Dec 11;9(23):6265–6278. doi: 10.1093/nar/9.23.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz F., Grinsted J. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J Bacteriol. 1982 Jul;151(1):222–228. doi: 10.1128/jb.151.1.222-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]