Abstract
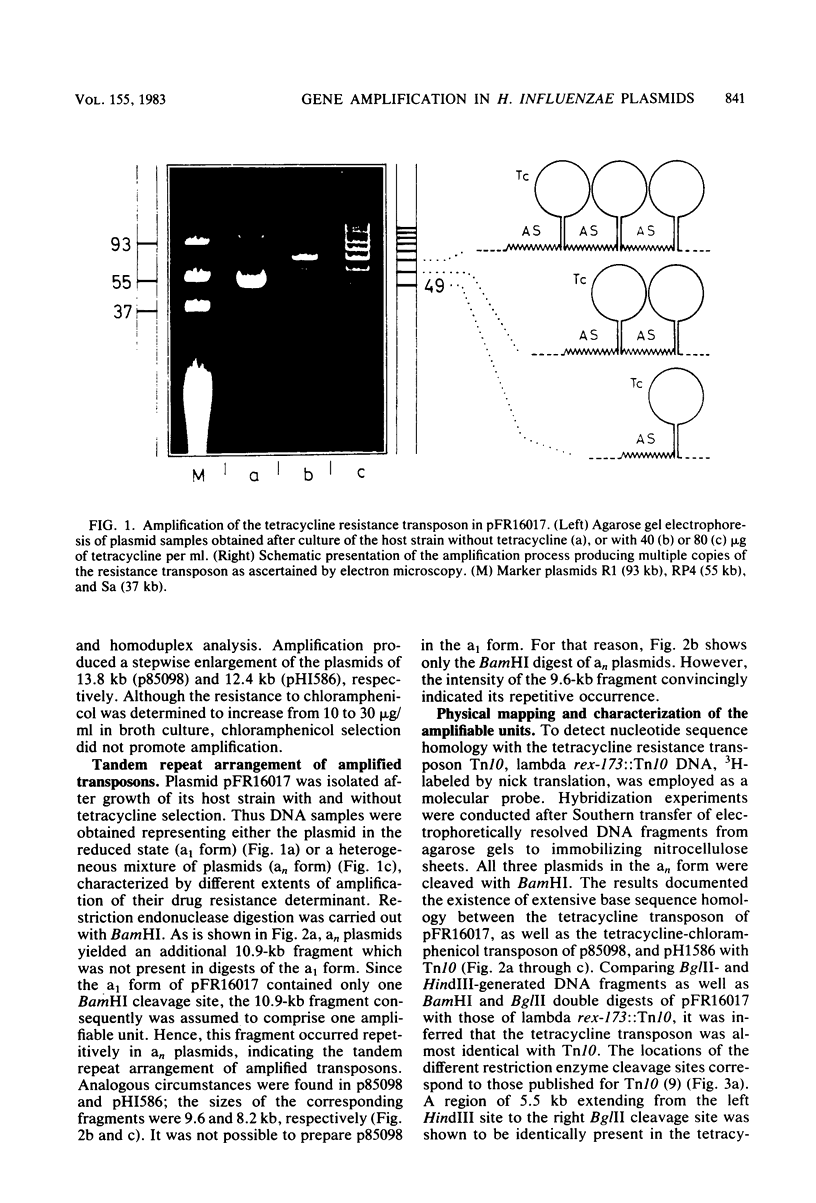
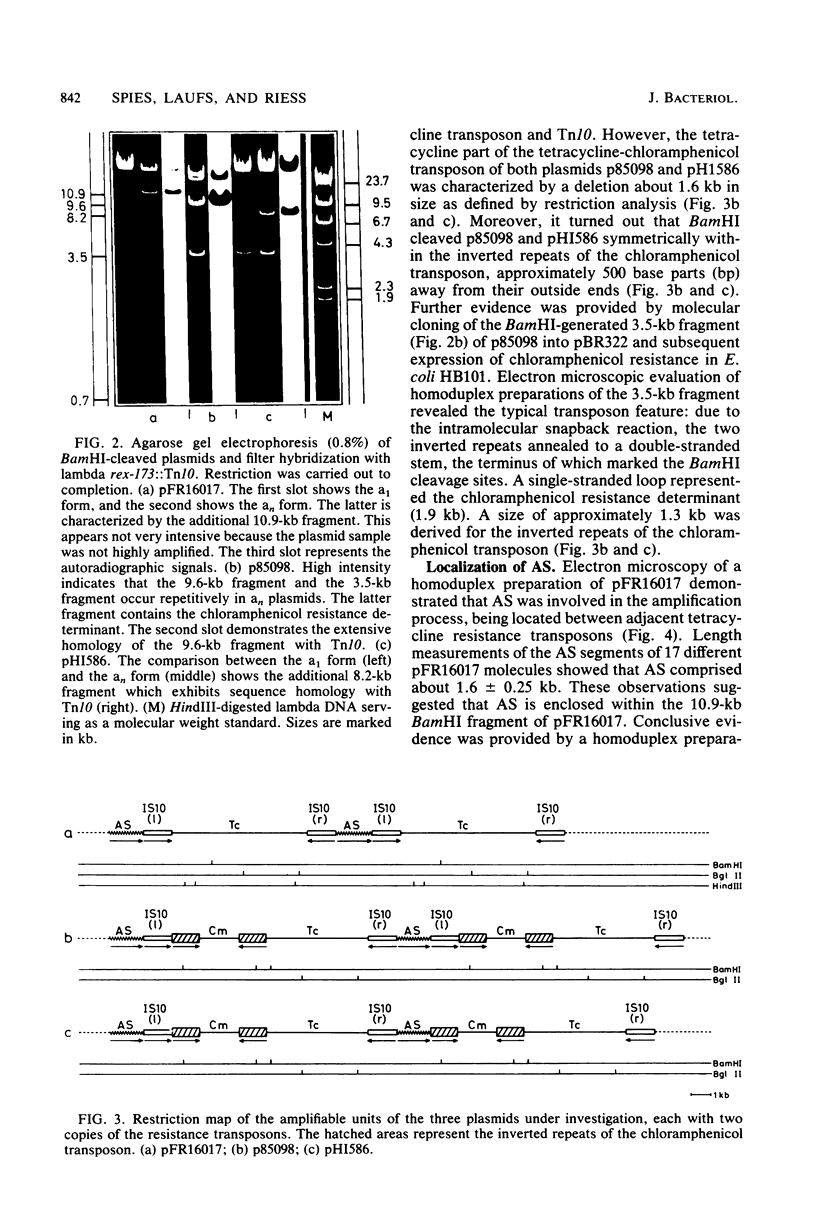
Intramolecular amplification produces tandem repeats of tetracycline and combined tetracycline-chloramphenicol resistance determinants in conjugative plasmids of Haemophilus influenzae. This process depends on host recombination pathways. Physical mapping revealed the tetracycline transposon involved in amplification to be almost identical with Tn10, including two IS10 insertion elements. The chloramphenicol resistance determinant of the combined transposon is 1.9 kilobases (kb) in size and is bound by two 1.3-kb inverted repeats. Insertion in the close vicinity of the inside end of the left-hand IS10 generates a deletion of a 1.6-kb Tn10 region. The amplifiable units were resolved to comprise not only the respective resistance transposons, but also an additional 1.6-kb sequence (designated AS) which was demonstrated to be identically present in the different amplification systems studied. AS separates amplified transposons from each other, thereby maintaining the same orientation. Moreover, AS is present at the left flank of the transposons, but is missing at the right one. It was shown that AS represents a general constituent of the H. influenzae plasmids of the 45-kb class. Evaluation of the results suggests that AS is responsible for the recombinational events involved in the gene amplification process.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson R. P., Roth J. R. Tandem genetic duplications in phage and bacteria. Annu Rev Microbiol. 1977;31:473–505. doi: 10.1146/annurev.mi.31.100177.002353. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Yagi Y., Bauer B. Plasmid-determined tetracycline resistance in Streptococcus faecalis: evidence for gene amplification during growth in presence of tetracycline. Proc Natl Acad Sci U S A. 1975 May;72(5):1720–1724. doi: 10.1073/pnas.72.5.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Hu S., Otsubo E., Davidson N., Saedler H. Electron microscope heteroduplex studies of sequence relations among bacterial plasmids: identification and mapping of the insertion sequences IS1 and IS2 in F and R plasmids. J Bacteriol. 1975 May;122(2):764–775. doi: 10.1128/jb.122.2.764-775.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys G. O., Willshaw G. A., Anderson E. S. A simple method for the preparation of large quantities of pure plasmid DNA. Biochim Biophys Acta. 1975 Apr 2;383(4):457–463. doi: 10.1016/0005-2787(75)90318-4. [DOI] [PubMed] [Google Scholar]
- Jahn G., Laufs R., Kaulfers P. M., Kolenda H. Molecular nature of two Haemophilus influenzae R factors containing resistances and the multiple integration of drug resistance transposons. J Bacteriol. 1979 May;138(2):584–597. doi: 10.1128/jb.138.2.584-597.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R. A., Berg D. E., Allet B., Reznikoff W. S. Restriction enzyme cleavage map of Tn10, a transposon which encodes tetracycline resistance. J Bacteriol. 1979 Jan;137(1):681–685. doi: 10.1128/jb.137.1.681-685.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAISER A. D., HOGNESS D. S. The transformation of Escherichia coli with deoxyribonucleic acid isolated from bacteriophage lambda-dg. J Mol Biol. 1960 Dec;2:392–415. doi: 10.1016/s0022-2836(60)80050-2. [DOI] [PubMed] [Google Scholar]
- Kaulfers P. M., Laufs R., Jahn G. Molecular properties of transmissible R factors of Haemophilus influenzae determing tetracycline resistance. J Gen Microbiol. 1978 Apr;105(2):243–252. doi: 10.1099/00221287-105-2-243. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Barker D. F., Ross D. G., Botstein D. Properties of the translocatable tetracycline-resistance element Tn10 in Escherichia coli and bacteriophage lambda. Genetics. 1978 Nov;90(3):427–461. doi: 10.1093/genetics/90.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Kooistra J., van Boxel T., Venema G. Characterization of a conditionally transformation-deficient mutant of Haemophilus influenzae that carries a mutation in the rec-1 gene region. J Bacteriol. 1983 Feb;153(2):852–860. doi: 10.1128/jb.153.2.852-860.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs R., Riess F. C., Jahn G., Fock R., Kaulfers P. M. Origin of Haemophilus influenzae R factors. J Bacteriol. 1981 Aug;147(2):563–568. doi: 10.1128/jb.147.2.563-568.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S. B., McMurry L. Detection of an inducible membrane protein associated with R-factor-mediated tetracycline resistance. Biochem Biophys Res Commun. 1974 Feb 27;56(4):1060–1068. doi: 10.1016/s0006-291x(74)80296-2. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Marko M. A., Chipperfield R., Birnboim H. C. A procedure for the large-scale isolation of highly purified plasmid DNA using alkaline extraction and binding to glass powder. Anal Biochem. 1982 Apr;121(2):382–387. doi: 10.1016/0003-2697(82)90497-3. [DOI] [PubMed] [Google Scholar]
- McCarthy D. Plasmid recombination in Haemophilus influenzae. J Mol Biol. 1982 Jun 5;157(4):577–596. doi: 10.1016/0022-2836(82)90500-9. [DOI] [PubMed] [Google Scholar]
- Meyer J., Iida S. Amplification of chloramphenicol resistance transposons carried by phage P1Cm in Escherichia coli. Mol Gen Genet. 1979 Oct 3;176(2):209–219. doi: 10.1007/BF00273215. [DOI] [PubMed] [Google Scholar]
- Ptashne K., Cohen S. N. Occurrence of insertion sequence (IS) regions on plasmid deoxyribonucleic acid as direct and inverted nucleotide sequence duplications. J Bacteriol. 1975 May;122(2):776–781. doi: 10.1128/jb.122.2.776-781.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rownd R., Mickel S. Dissociation and reassociation of RTF and r-determinants of the R-factor NR1 in Proteus mirabilis. Nat New Biol. 1971 Nov 10;234(45):40–43. doi: 10.1038/newbio234040a0. [DOI] [PubMed] [Google Scholar]
- Schmitt R., Bernhard E., Mattes R. Characterisation of Tn1721, a new transposon containing tetracycline resistance genes capable of amplification. Mol Gen Genet. 1979 Apr 17;172(1):53–65. doi: 10.1007/BF00276215. [DOI] [PubMed] [Google Scholar]
- Schöffl F., Arnold W., Pühler A., Altenbuchner J., Schmitt R. The tetracycline resistance transposons Tn1721 and Tn1771 have three 38-base-pair repeats and generate five-base-pair direct repeats. Mol Gen Genet. 1981;181(1):87–94. doi: 10.1007/BF00339010. [DOI] [PubMed] [Google Scholar]
- Setlow J. K., Boling M. E., Beattie K. L., Kimball R. F. A complex of recombination and repair genes in Haemophilus influenzae. J Mol Biol. 1972 Jul 21;68(2):361–378. doi: 10.1016/0022-2836(72)90218-5. [DOI] [PubMed] [Google Scholar]
- Setlow J. K., Brown D. C., Boling M. E., Mattingly A., Gordon M. P. Repair of deoxyribonucleic acid in Haemophilus influenzae. I. X-ray sensitivity of ultraviolet-sensitive mutants and their behavior as hosts to ultraviolet-irradiated bacteriophage and transforming deoxyribonucleic acid. J Bacteriol. 1968 Feb;95(2):546–558. doi: 10.1128/jb.95.2.546-558.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuy J. H. Mechanism of additive genetic transformation in Haemophilus influenzae. J Bacteriol. 1980 Dec;144(3):999–1002. doi: 10.1128/jb.144.3.999-1002.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin B. E., Nordström K. R plasmid gene dosage effects in Escherichia coli K-12: copy mutants of the R plasmic R1drd-19. Plasmid. 1977 Nov;1(1):1–7. doi: 10.1016/0147-619x(77)90003-8. [DOI] [PubMed] [Google Scholar]
- Wiebauer K., Schraml S., Shales S. W., Schmitt R. Tetracycline resistance transposon Tn1721: recA-dependent gene amplification and expression of tetracycline resistance. J Bacteriol. 1981 Sep;147(3):851–859. doi: 10.1128/jb.147.3.851-859.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y., Clewell D. B. Amplification of the tetracycline resistance determinant of plasmid pAM alpha 1 in Streptococcus faecalis: dependence on host recombination machinery. J Bacteriol. 1980 Aug;143(2):1070–1072. doi: 10.1128/jb.143.2.1070-1072.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y., Clewell D. B. Identification and characterization of a small sequence located at two sites on the amplifiable tetracycline resistance plasmid pAMalpha1 in Streptococcus faecalis. J Bacteriol. 1977 Jan;129(1):400–406. doi: 10.1128/jb.129.1.400-406.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Klingeren B., van Embden J. D., Dessens-Kroon M. Plasmid-mediated chloramphenicol resistance in Haemophilus influenzae. Antimicrob Agents Chemother. 1977 Mar;11(3):383–387. doi: 10.1128/aac.11.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]








