Abstract
TraR is an Agrobacterium transcriptional regulator whose activity requires the pheromone N-3-oxooctanoyl-l-homoserine lactone. TraR was purified as a complex with the pheromone and contained one pheromone molecule per protein monomer. TraR–pheromone complexes bound to a single DNA site and activated two promoters that flank this site. Promoter expression was elevated 30-fold by using a supercoiled template. Pheromone binding increased the affinity of TraR for this binding site. Pheromone also increased TraR abundance in vivo by causing a 20-fold decrease in TraR turnover rates.
Keywords: pheromone, transcription, Agrobacterium
Although it was recently thought that few bacteria were able to communicate by exchanging chemical signals, we now know that this behavior is widespread within many prokaryotic lineages (1). Many of these signaling systems use a protein resembling LuxI of Vibrio fisheri to generate an acyl-homoserine lactone that acts as a pheromone and a protein resembling LuxR that serves as a pheromone receptor and transcriptional activator. Most of these systems are found in bacteria that colonize plants or animals and are thought to provide a mechanism for bacteria to estimate their population densities, a process sometimes referred to as quorum sensing (2, 3). The plant pathogen Agrobacterium tumefaciens provides one of the best studied examples of quorum-responsive gene expression (4–7). This organism is widely known for its ability to transfer fragments of oncogenic DNA to host plants, resulting in crown gall tumors (8, 9). This DNA originates from a megaplasmid called the tumor-inducing (Ti) plasmid. In addition to mediating interkingdom DNA transfer, Ti plasmids are capable of conjugal DNA transfer between agrobacteria (10). Genes required for conjugation (tra genes) are positively regulated by the TraR protein, which is activated by the pheromone N-3-oxooctanoyl-l-homoserine lactone, sometimes referred to as Agrobacterium autoinducer, or AAI (4, 7). AAI is synthesized by the TraI protein (11). Both traR and traI are located on the Ti plasmid, and both are positively autoregulated by TraR–AAI complexes (6).
TraR is thought to contain an amino-terminal module (≈165 amino acid residues) that binds AAI and also mediates multimerization and a carboxyl-terminal module (≈65 amino acid residues) that binds particular DNA sites near target promoters and also makes stimulatory contacts with RNA polymerase (RNAP) (2). Genetic evidence suggests that TraR binds to a site centered 42 nucleotides upstream of transcription start sites (12). LuxR-type proteins have been the subject of extensive genetic research, but very few biochemical studies of LuxR-type proteins have been published (13–15).
MATERIALS AND METHODS
Overproduction and Purification of TraR and RNAP.
Strains and plasmids used are listed in Table 1. The traR coding sequence was fused to the φ10 promoter (22) of bacteriophage T7 by PCR amplification, using plasmid pCF222 (6) as template and oligonucleotides 5′-GCTCTAGACATATGCAGCACTGGCTGGACAAG-3′ and 5′-AATACGACTCACTATAG-3′ as primers. The resulting PCR product was digested with NdeI and PstI and was cloned into pRSETA (Invitrogen) digested with the same enzymes, resulting in plasmid pJZ358. Strain BL21/DE3(pJZ358) was cultured at 28°C in 1 liter of AT medium (4) containing 2 mg/ml of ampicillin, 10 μg/ml of thiamine, and 100 nM AAI to an OD600 of ≈0.4, was treated with isopropyl β-d-thiogalactoside to a final concentration of 50 μM, and was incubated at 28°C overnight. This very high concentration of ampicillin was used to ensure that plasmid-free bacteria did not accumulate. Cells were collected and resuspended in 10 ml of TEDG buffer (50 mM Tris⋅HCl, pH 7.9/0.5 mM EDTA/1 mM DTT/5% glycerol) plus 0.15 M NaCl and were disrupted by using French pressure cell (20,000 psi). Cell debris was removed by ultracentrifuge (150,000 × g for 0.5 hr), and the supernatant was chromatographed by using 30 ml of heparin agarose (2-cm-diameter column) equilibrated with TEDG plus 0.15 M of NaCl. Bound proteins were eluted by using a 100-ml linear gradient of NaCl from 0.15 to 1 M. Peak fractions containing TraR were pooled, precipitated by using ammonium sulfate (95% saturation), resuspended, dialyzed overnight against buffer TEDG plus 0.15 M NaCl and 0.05% Tween 20, and applied to a 1-ml Mono S column (Amersham Pharmacia) equilibrated with TEDG plus 0.15 M NaCl. TraR protein was eluted by using a 15-ml linear gradient NaCl from 0.15 to 1 M. To purify RNA polymerase from A. tumefaciens, strain KYC55 (17) was cultured in 2 liters of AT medium to OD600 = 0.6. This strain lacks a Ti plasmid, ensuring that the RNAP preparation will not contain TraR. RNAP was purified by Polymin P precipitation by using a protocol described previously (23).
Table 1.
Strains and plasmids
| Strain or plasmid | Relevant genotype | Reference |
|---|---|---|
| BL21/DE3 | E. coli B Plac-gene 1 of bacteriophage T7 | 16 |
| KYC55 | A. tumefaciens R10, no Ti plasmid, Knr | 17 |
| WCF47 | A. tumefaciens R10, ΔtraI | 18 |
| pCF372 | PtraI-lacZ cloned into broad host range plasmid pUCD2 | 18 |
| pCF218 | traRmc cloned into broad host range plasmid pSW213 | 6 |
| pUCD1002 | High copy cloning vector | 19 |
| pPZP201 | Broad host range cloning vector | 20 |
| pBluescript+ | Narrow host range cloning vector | Stratagene |
| pRESTA | T7 promoter cloning vector | Invitrogen |
| pBend3 | Derivative of pBluescript for assays of DNA bending | 21 |
| pJZ304 | traA-traC intergenic region cloned into pBend3 | This study |
| pJZ335 | Plac-traR cloned into pPZP201 | 25 |
| pJZ358 | traR cloned into pRSETA | This study |
| pJZ368 | traR cloned into pUCD1002 | This study |
| pJZ373 | traA-traC intergenic region cloned into pBluescript+ | This study |
| pJZ374 | Fill-in tra box mutation of pJZ373 | This study |
Gel Retardation Assays.
A 251-nt PCR product containing traA-traC intergenic region was created by PCR amplification using oligonucleotides 5′-GCATCTAGAGCCCGGTCTCACCGGGCCGAG-3′ and 5′-GCCGTCGACGATTTCTTCCCGGATTTTCGATGA-3′ and was cloned between the SalI and XbaI sites of plasmid pBend3 (21), resulting in plasmid pJZ304. The same PCR fragment was cloned into pBlusecript II SK(+) (Stratagene), resulting in pJZ373. pJZ373 was sequentially treated with BglII, T4 DNA polymerase, and T4 DNA ligase, creating plasmid pJZ374, which contains a “fill-in” tra box mutation. For gel retardation assays, pJZ304 was digested with HindIII and XbaI, and the resulting fragments were end-labeled by using [α-32P]dCTP and the Klenow fragment of DNA polymerase I. Binding reactions contained 10−12 M DNA and TraR protein in the concentrations indicated and contained or lacked 10 μM AAI, in a buffer of 10 mM Tris⋅HCl (pH 7.9), 1 mM EDTA, 1 mM DTT, 60 mM potassium glutamate, 30 μg/ml calf thymus DNA, 20 μg/ml BSA, and 10% glycerol. After 20 min of incubation at room temperature, samples were size-fractionated by using 5% polyacrylamide gels in 0.5× TAE buffer (20 mM Tris⋅acetate/1 mM EDTA, pH 8.5). Radioactivity of free DNA and TraR–DNA complexes was quantitated by using a Storm B840 PhosphorImager (Molecular Dynamics).
DNase I Footprinting Assays.
To label the traA coding strand of pJZ304, this plasmid was digested with XbaI, was end-labeled by using [α-32P]dNTP, and then was digested with HindIII, and the 360-nt fragment containing the tra box was size-fractionated by electrophoresis and was eluted from the gel by overnight incubation in a buffer containing 0.5 M ammonium acetate and 1 mM EDTA. Labeling of the traC coding strand was done in the same way except that the plasmid was digested sequentially by using SalI and EcoRI. DNase I digestions were carried out by using ≈0.1 pmol of labeled DNA fragment and 5 μM TraR protein, in a buffer containing 10 mM Tris⋅HCl (pH 7.9), 5 mM MgCl2, 1 mM CaCl2, 2 mM DTT, 2 μg/ml calf thymus DNA, 100 mM KCl, and 5% glycerol (final volume 100 μl). Where indicated, 10 μM AAI also was added. After incubation at room temperature for 30 min, 0.25 units of DNase I was added. The tubes were incubated at room temperature for 30 sec, and the reactions were stopped by addition of 0.25 M EDTA and 5 mg/ml yeast tRNA. DNA fragments were ethanol precipitated and size-fractionated by using denaturing 6% polyacrylamide gels. Positions of G + A residues were determined by using a published protocol (24).
In Vitro Transcription Reactions.
Transcription reaction mixes (20 μl) contained 50 mM Tris⋅HCl (pH 7.9), 6 mM MgCl2, 2 mM spermidine, 10 mM DTT, 0.5 mM each NTP, 0.5 μg of plasmid pJZ304 as template DNA, and 2 μg A. tumefaciens RNAP. Where indicated, 0.2 μM TraR and 10 μM AAI also were added. In some reactions, pJZ304 was converted to a linear form by digestion with EcoRI. Samples were incubated at 28°C for 30 min, at which time 0.5 unit of DNase I was added and were incubated for 5 min to digest template DNA. The samples were extracted twice with phenol/chloroform and were precipitated by using ethanol. The resulting RNA was subjected to S1 protection assays (25), using 5′-radiolabeled oligonucleotides complementary to traA (49 nt, from nucleotide −4 to +45), traC (44 nt, from nucleotide −4 to +40), or bla (44 nt, from nucleotide −4 to +40).
Analysis of AAI Content of TraR.
A sample of each fraction (100 μl) from the Mono S column eluate described above was treated with 1 μl of 20 mg/ml proteinase K at 37°C for 1 hr, and ethyl acetate was extracted twice. The ethyl acetate fractions were pooled, were evaporated to dryness, and were resuspended in 10 μl water. Aliquots (1 μl) were applied to C18 reversed-phase thin layer chromatography plates (Whatman) and were chromatographed by using 60% methanol and 40% water as described (26). After chromatography, the plates were dried and overlaid with 125 ml of agar containing AT medium, 40 μg/ml of 5-bromo-4-choloro-3-indolyl β-d-galactoside, and the reporter strain WCF47(pCF372)(pCF218) (18). TLC plates were incubated overnight at 28°C and were examined for 5-bromo-4-choloro-3-indolyl β-d-galactoside hydrolysis. To estimate the TraR:AAI mole ratio, TraR protein was purified by using the procedures described above except that AAI (100 nM) was provided in all buffers during purification. After allowing TraR to bind to the Mono S matrix, the protein was washed with 15 ml of a buffer containing 50 mM phosphate buffer (pH 7.9) plus 0.15 M NaCl and then was eluted by using a 15-ml linear gradient from 0.15 to 1 M NaCl. The resulting TraR sample was subjected to amino acid analysis by Cornell BioResource Center (Ithaca, NY) using a Waters PICO-TAG amino acid analyzer.
Measurements of TraR Turnover in Whole Cells.
Plasmid pJZ368 was created by ligation of HindIII-digested pJZ335 (a derivative of pPZP201 expressing Plac-traR; see ref. 25) with HindIII-digested pUCD1002, a high copy number cloning vector. Strain KYC55(pJZ368) was cultured in 120 ml of AT minimal medium in the absence or presence of 1 μM AAI to an OD600 of ≈0.5. [35S]Methionine was added to each culture to a final concentration of 5 μCi/ml. After an interval of 5 min, nonlabeled methionine was added to a final concentration of 2 mM. Samples (20 ml) were withdrawn at the various intervals. Cells were transferred to prechilled tubes and were centrifuged at 4°C, were washed once with cold buffer containing 0.05% Sarkosyl, 0.5 M NaCl, 50 mM Tris⋅HCl (pH 7.9), and 20 mM EDTA, were centrifuged, and were stored as cell pellets at −80°C. The pellets then were resuspended in 0.5 ml of a buffer containing 50 mM Tris⋅HCl (pH 7.9), 200 mM NaCl, 2 mM EDTA, 1 mM DTT, 1 mM β-mercaptoethanol, 1% Nonidet P-40, and 0.5 mM PMSF, and were disrupted by using a French Press. The resulting lysates were ultracentrifuged and subjected to immunoprecipitation by using affinity-purified TraR antibody and Protein-A agarose (27). To measure traR mRNA accumulation, strains KYC55(pJZ368) and KYC55(pPZP201) (both of which lack a Ti plasmid [17]) were cultured in AT broth containing or lacking 100 nM AAI. Total nucleic acid was purified from each strain and was quantitated by using radiolabeled oligonucleotide probes. Oligonucleotides 5′-GCCGGTGAAGCCGAAATGGTCGGCGATGTCCGCCAGCCCGACTC-3′ and 5′-GCGACTGCCCTGCTGAACATCGTTGCTGCTCCATCCAC-3′ were used to detect traR and aadA mRNA, respectively (aadA confers resistance to spectinomycin and is expressed by both plasmids). The 3′ end of each oligonucleotide contains four nucleotides that are not complementary to the mRNA (italicized). Quantitative S1 analysis was carried out by using a published protocol (25).
RESULTS
Binding of Purified TraR to Predicted tra Box DNA.
In an effort to overproduce the native TraR protein, we fused the traR coding sequence to the φ10 promoter of bacteriophage T7 (22), creating pJZ358. When Escherichia coli strain BL21/DE3(pJZ358) was cultured in growth medium supplemented with isopropyl β-d-thiogalactoside, it produced large amounts of TraR, but virtually none of this protein was soluble (data not shown). However, when the same strain was cultured in the presence of 100 nM AAI, ≈25% of the TraR protein remained in the soluble fraction, even after prolonged ultracentrifugation. The protein was purified to >95% homogeneity by chromatography using heparin agarose and Mono S resins.
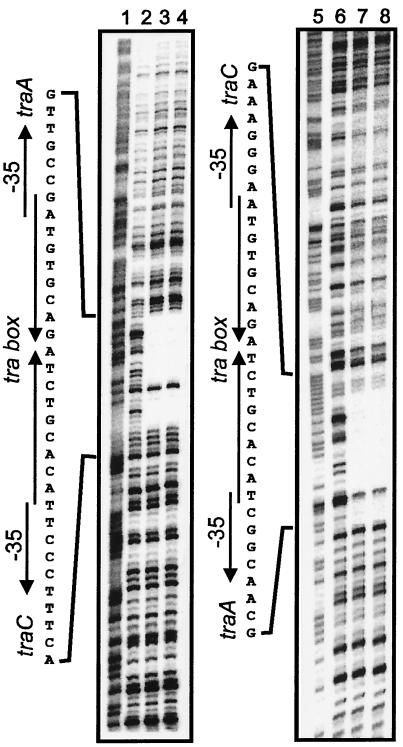
Purified TraR protein retarded the mobility of a DNA fragment containing a predicted TraR binding site (tra box), forming a single complex (Fig. 1A). The Kd for binding was 1 × 10−8 M, and the Hill coefficient was 1.2 (Fig. 1B), indicating that TraR bound this site with little, if any, cooperativity. DNase I footprinting indicated that TraR bound precisely to a predicted tra box (Fig. 2). The fact that one nucleotide of each strand within this protected region was DNase I-sensitive suggests that TraR bound to a single face of this site. This tra box sequence has a perfect dyad symmetry and contains a BglII site at its center. Addition of four nucleotides to this site (thus increasing the distance between the arms and placing them on opposite helical faces) abolished detectable TraR binding (data not shown). We also tested TraR for the ability to bend the DNA at its binding site by introducing the binding site into plasmid pBend3 (21). The resulting plasmid was digested with a variety of restriction endonucleases that create permuted DNA fragments. These fragments, either unbound or bound to TraR, did not exhibit position-dependent gel mobility, indicating that TraR does not detectably bend the DNA to which it binds (data not shown).
Figure 1.
TraR gel retardation assays. (A) Plasmid pJZ304 was digested with HindIII and XbaI and was end-labeled. Binding reactions contained 10−12 M DNA, and TraR (monomer) was added at 75 nM (lane 1), 37.5 nM (lane 2), 18.8 nM (lane 3), 9.4 nM (lane 4), 4.7 nM (lane 5), 2.4 nM (lane 6), 1.2 nM (lane 7), 0.6 nM (lane 8), and 0 nM (lane 9). (B) Hill plot of data obtained in A.
Figure 2.
DNase I protection of a predicted TraR binding site. A 360-nt fragment was end-labeled on the traA coding strand (lanes 1–4) or the traC coding strand (lanes 5–8) and was incubated without TraR (lanes 2 and 6), with 5 μM TraR (lanes 3 and 7), or with 5 μM TraR and 10 μM AAI (lanes 4 and 8). Lanes 1 and 5 show the positions of all G and A residues.
Purified TraR Activates Two tra Promoters on Supercoiled but not Linear DNA.
We wanted to test whether purified TraR can activate transcription of the divergent traA and traC promoters, which are activated by this protein in vivo (6). Because TraR is poorly active in whole E. coli cells, we used RNAP that was purified from A. tumefaciens. This preparation of RNAP efficiently transcribed a control consensus promoter in a runoff transcription assay (data not shown). However, the same preparation of RNAP, in combination with TraR, did not efficiently transcribe traA or traC in similar assays (data not shown). Addition of AAI did not stimulate transcription activity.
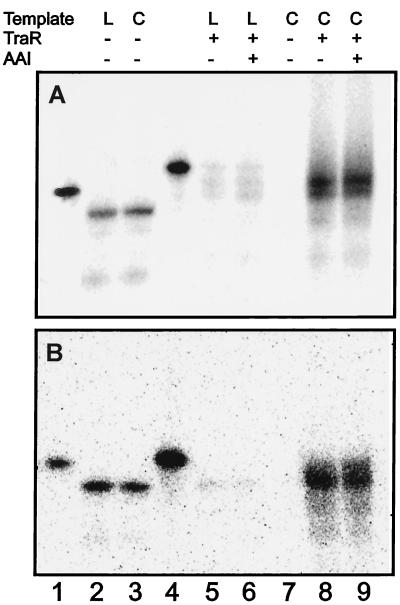
RNAP has been reported to transcribe several bacterial promoters more strongly on supercoiled templates than on linear templates (28). To determine whether this was true for the traA and traC promoters, we carried out transcription reactions with RNAP, TraR, a supercoiled template, and nonradiolabeled NTPs. The resulting nonlabeled RNA was hybridized with 5′-radiolabeled oligonucleotides complementary either to traA or to traC. Both promoters were efficiently transcribed under these conditions (Fig. 3 A, lane 8 and B, lane 8), and, in both cases, transcription was not detected in the absence of TraR (Fig. 3 A, lane 7 and B, lane 7). Similar transcription reactions were carried out in which the template DNA was digested by using a restriction endonuclease. In these reactions, transcription was reduced ≈30-fold (Fig. 3 A, lane 5 and B, lane 5). In contrast, linearization of the template DNA did not affect transcription levels of a control bla promoter (compare Fig. 3 A and B, lanes 2 and 3). We conclude that TraR and RNAP can activate traA and traC when these promoters are cloned on a supercoiled template but are largely inactive when the same template is converted to a linear form.
Figure 3.
In vitro transcription by purified TraR and A. tumefaciens RNA polymerase. (A) Plasmid pJZ304 was incubated with A. tumefaciens RNAP and nonlabeled NTPs to synthesize mRNA in vitro. Where indicated, TraR and AAI were added to each reaction (0.2 and 10 μM, respectively), and total nucleic acids from these reactions were hybridized by using probes complementary to traA or bla. Probes complementary to bla or traA are shown (lanes 1 and 4, respectively). (B) Same as A except that a probe hybridizing to traC was used in lanes 4–9. “L” and “C” denote linear DNA template and circular DNA template, respectively.
Copurification of TraR and the Pheromone AAI.
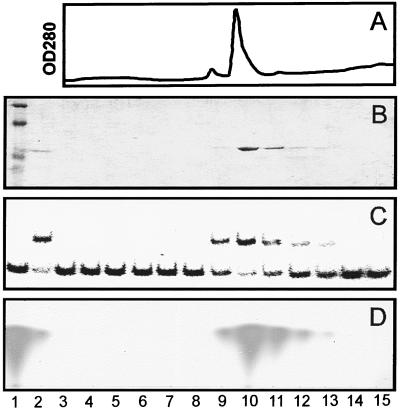
In all of the experiments described above, TraR activities were not stimulated by exogenous AAI. Because TraR was purified from cells that had been cultured in the presence of AAI, it seemed plausible that TraR had been purified as a complex with this pheromone. To test this idea, we repeated the purification procedure and assayed each fraction of the Mono S eluate for protein content (Fig. 4 A and B), gel retardation activity (Fig. 4C), and AAI content (Fig. 4D). AAI coeluted with TraR, indicating that AAI had remained tightly bound to TraR during purification. This in turn probably explains why TraR binding and transcription activities were not stimulated by exogenous AAI.
Figure 4.
Copurification of TraR and AAI. (A) OD280 of eluate from a Mono S column used to purify TraR. (B) Total protein content of each eluted fraction. Nonfractionated proteins (lane 2) and molecular mass standards (lane 1) also are shown. (C) Gel retardation activity of each fraction shown in B. Migration of DNA fragment in the absence of protein is shown (lane 1). (D) AAI content of each fraction shown in B. AAI was detected by proteolyzing a portion of each fraction, extracting with ethyl acetate, and chromatographing the products by thin layer chromatography (26). A synthetic AAI standard is shown (lane 1).
To estimate the TraR:AAI mole ratio, we purified TraR by using the same procedures except that 100 nM AAI was provided in all buffers during purification. After allowing TraR to bind to the Mono S chromatography matrix during the second purification step, the protein was washed with 15-column volumes of a buffer lacking AAI and then was gradient-eluted with buffers lacking AAI. The resulting TraR preparation was submitted to the Cornell BioResource Center for amino acid analysis. This procedure caused quantitative conversion of synthetic AAI to homoserine (as well as, presumably, 3-oxooctanoic acid). We were therefore able to quantitate the amount of released homoserine and the amount of each amino acid and to use these data to calculate the mole ratio of AAI to TraR monomers. Because the calculated ratio was 0.95 (Table 2), we conclude that each monomer of TraR binds one molecule of AAI.
Table 2.
Amino acid analysis of TraR-AAI complexes
| Amino acid | Concentration, μM | Residues per monomer | Concentration of each residue, μM |
|---|---|---|---|
| Ala | 843.8 | 28 | 30.1 |
| Arg | 561.6 | 10 | 56.1 |
| Asx | 349.6 | 21 | 16.6 |
| Cys | 48.1 | 1 | 48.1 |
| Glx | 428.9 | 18 | 23.8 |
| Gly | 391.8 | 11 | 35.6 |
| His | 308.4 | 11 | 28 |
| Ile | 554.1 | 19 | 29.1 |
| Leu | 530.5 | 16 | 33.1 |
| Lys | 486.4 | 16 | 30.4 |
| Met | 201.5 | 6 | 33.6 |
| Phe | 288.8 | 11 | 26.3 |
| Pro | 230.8 | 6 | 38.5 |
| Ser | 305 | 11 | 27.7 |
| Thr | 525.4 | 18 | 29.2 |
| Tyr | 223.1 | 7 | 31.8 |
| Val | 344.7 | 11 | 31.2 |
| Average* | 28.4 | ||
| Homoserine | 26.9 | (1) | 26.9 |
Calculated by regression analysis of number of amino acid residues plotted against concentration of each amino acid.
Removal of AAI from TraR–AAI Complexes by Prolonged Dialysis.
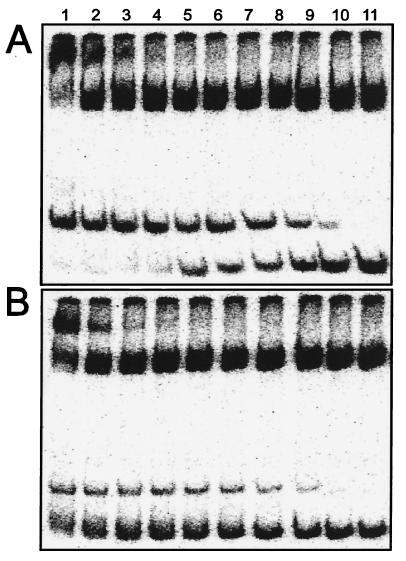
In an effort to understand how AAI alters the properties of TraR, we attempted to remove AAI by dialysis. In repeated trials, dialysis against buffers lacking detergents caused TraR to lose all DNA binding activity, even in the presence of exogenous AAI. We therefore dialyzed TraR against a buffer containing 3% Tween 20 and changed the dialysis buffer daily for 6 days. Each day, a portion of TraR was removed and assayed for AAI content. Approximately 90% of the AAI had been removed from the TraR within 3 days (data not shown). However, 10% of the AAI remained bound to TraR, and this residual AAI was not removed even after 3 additional days of dialysis. The concentration of soluble TraR did not change during the course of dialysis (data not shown). The resulting TraR preparation (designated apo-TraR) was tested in gel retardation assays in the presence and absence of exogenous AAI. In the presence of exogenous AAI, TraR bound to this fragment with an affinity of 2 × 10−8 M (Fig. 5A) and a Hill coefficient of 1.0 (data not shown). These properties are extremely similar to TraR–AAI complexes before dialysis (Fig. 1A). Apo-TraR showed a lower affinity for DNA at all protein concentrations (Fig. 5B), indicating that AAI stimulates binding affinity between TraR and its binding site. This effect was most pronounced at high TraR concentrations because even at the highest concentration tested, only ≈25% of the DNA was shifted. Binding showed negative cooperativity (Hill coefficient 0.5). We postulate that the observed DNA binding was caused by the residual AAI in our TraR preparation. Negative binding cooperativity could have been caused by apo-TraR interfering with binding.
Figure 5.
DNA binding affinity of apo-TraR in the presence and absence of AAI. TraR–AAI complexes were dialyzed for 7 days in a buffer containing Tween 20 (3%) and then were dialyzed once against a buffer containing a trace of Tween 20 (0.01%). The resulting preparation of TraR was tested for gel retardation activity by using the same DNA fragment as used in Fig. 2, in the presence (A) or absence (B) of 10 μM AAI. TraR was provided at the following concentrations: 600 nM (lane 1), 300 nM (lane 2), 150 nM (lane 3), 75 nM (lane 4), 37.5 nM (lane 5), 18.8 nM (lane 6), 9.4 nM (lane 7), 4.7 nM (lane 8), 2.3 nM (lane 9),1.8 nM (lane 10), and 0 nM (lane 11).
AAI Increases TraR Stability and Abundance in Vivo.
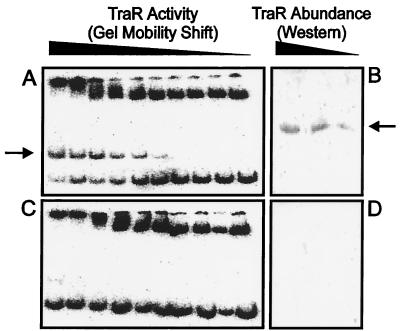
We attempted to purify TraR from A. tumefaciens by using a strain that expresses TraR from a Plac promoter, which is a moderately strong, constitutive promoter in A. tumefaciens (29). This strain also lacks a traI gene and therefore cannot synthesize AAI. When a strain containing this plasmid was cultured in the presence of exogenous AAI, the resulting cleared lysate contained amounts of TraR that were readily detectable by gel retardation assays (Fig. 6A) and by Western immunoblots (Fig. 6B). Surprisingly, when the same strain was cultured without exogenous AAI, TraR was completely undetectable by either assay (Fig. 6 C and D).
Figure 6.
TraR abundance is enhanced by AAI. A strain of A. tumefaciens that constitutively expresses TraR was cultured in the presence (A and B) or absence (C and D) of AAI and was concentrated and lysed. The cleared lysate was serially diluted in 2-fold increments and was used in gel retardation experiments by using pJZ304 DNA as described above (A and C) and for Western immunoblots by using rabbit polyclonal antiserum (B and D).
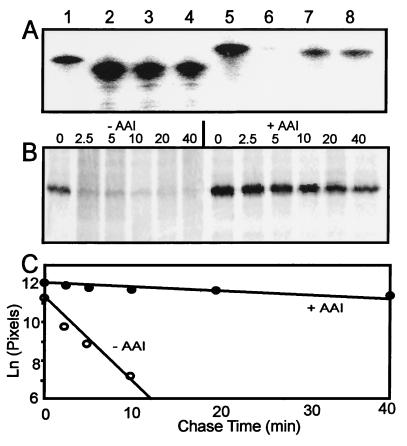
The finding that TraR protein failed to accumulate in the absence of AAI, even when expressed from a constitutive promoter, indicates that AAI stabilizes traR mRNA against cytoplasmic RNases, increases translation of this message, or stabilizes TraR protein against cytoplasmic proteases. We used a 5′-radiolabeled oligonucleotide probe to measure the abundance of traR mRNA in the presence or absence of AAI and found no difference (Fig. 7A, compare lanes 7 and 8). We therefore conducted pulse–chase experiments to measure TraR protein stability. Radiolabeled TraR was far more stable in the presence of AAI than in its absence (Fig. 7B). The half-life of TraR was estimated to be 2 min in the absence of AAI and 35 min in the presence of AAI (Fig. 7C). These data fully support the independent Western immunoblots and gel shift assays described above. We conclude that AAI dramatically decreases the rate of TraR turnover. This phenomenon may fully account for the AAI-dependent increase in TraR accumulation.
Figure 7.
Increase in TraR abundance by AAI by decreased protein turnover rate. (A) Cellular abundance of traR mRNA in a strain that contains a plasmid with a Plac-traR fusion. A radiolabeled oligonucleotide probe complimentary to traR (lane 5) or aadA (lane 1) was hybridized to cellular mRNA of strains KYC55(pJZ368) (lanes 3, 4, 7, and 8) or to KYC55(pPZP201) (lanes 2 and 6). The former strain previously was cultured in the presence (lanes 3 and 7) or absence (lanes 4 and 8) of 100 nM AAI. (B) Immunoprecipitated TraR protein that had been radiolabeled with [35S]methionine and then had been treated with excess unlabeled methionine for the time intervals indicated above each lane (in minutes). Strain KYC55(pJZ368) was cultured in the absence (left) or presence (right) of 1 μM AAI. (C) Natural log of radioactivity of each band shown in B, as determined by using a Storm PhosphorImager.
DISCUSSION
In this study, we have demonstrated that full length, native TraR protein binds to a site upstream of the divergent traA and traC promoters. This site is similar in sequence and position to a site upstream of the traI promoter and is also similar to a putative LuxR binding site (12). We also have reconstituted TraR-dependent transcription of the traA and traC promoters. We confirmed the prediction that TraR tightly binds AAI and demonstrated that one AAI molecule is bound per TraR monomer. The half-life of TraR in A. tumefaciens was found to be strongly increased by exogenous AAI. We propose that apo-TraR is highly sensitive to cytoplasmic proteolysis whereas AAI converts TraR to a more protease-resistant form. One possibility is that AAI converts TraR from protease-sensitive monomers to protease-resistant higher-order oligomers. Many bacterial proteins have been characterized as unstable to cytoplasmic proteases, and several have been found to be stabilized by binding to other proteins. However, this may be the first known example of a regulatory protein that is stabilized by binding of a low molecular mass ligand (S. Gottesman, personal communication).
Stabilization of TraR by AAI could have important consequences for regulation of target genes. It is already known that, in wild-type bacteria, the tra regulon is under positive autoregulation at several levels. First, transcription of traR is positively autoregulated (as well as being positively regulated by OccR–octopine complexes), increasing the pool size of TraR by increasing its production rate. Second, transcription of traI is also positively regulated by TraR–AAI complexes, thus increasing the pool of AAI. We now demonstrate that AAI further increases the pool size of TraR by decreasing its rate of destruction. These autoregulatory circuits may help to operate a molecular “switch” that attempts to maintain tra expression either at very low levels or at very high levels. Similar patterns of positive autoregulation are seen in most but not all other quorum-sensing systems.
Our finding that TraR–AAI complexes are extremely stable during purification suggests that they also may be highly stable in vivo. If so, then proteolysis may play an essential role in turning off the expression of the tra regulon. At low cell densities, AAI is thought to diffuse rapidly from the cytoplasm, such that newly synthesized TraR protein may fail to bind AAI. Our data indicate that this apo-TraR should be rapidly degraded. TraR–AAI complexes, though more stable than apo-TraR, also are degraded eventually, thereby releasing AAI, which diffuses from the bacterial cytoplasm. In a previous study, the pheromone 3-oxohexanoyl homoserine lactone diffused rapidly from V. fischeri cells that express the cognate receptor LuxR (30). It was concluded that LuxR–pheromone complexes rapidly disassociate. Although these data could reflect a difference between the LuxR and TraR proteins, it is also possible that proteolysis of LuxR could have played a significant role in release of the pheromone.
Acknowledgments
We thank Anatol Eberhard for the generous gift of AAI and John Helmann, Anatol Eberhard, Clay Fuqua, Pete Greenberg, and the members of our laboratory for helpful discussions. We are grateful to Jim Russell and his colleagues for help with cell disruptions and to Bob Sherwood for amino acid analysis. This study was supported by National Institutes of Health Grant GM42893 and the Cornell University College of Agriculture and Life Science and the Department of Microbiology.
ABBREVIATIONS
- Ti plasmid
tumor-inducing plasmid
- AAI
Agrobacterium autoinducer
- RNAP
RNA polymerase
References
- 1.Dworkin M. Microbial Cell-Cell Interactions. Washington, DC: Am. Soc. Microbiol.; 1991. [Google Scholar]
- 2.Fuqua C, Winans S C, Greenberg E P. Ann Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 3.Fuqua C, Winans S C, Greenberg E P. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L, Murphy P J, Kerr A, Tate M E. Nature (London) 1993;362:446–448. doi: 10.1038/362446a0. [DOI] [PubMed] [Google Scholar]
- 5.Piper K R, Beck von Bodman S, Farrand S K. Nature (London) 1993;362:448–450. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- 6.Fuqua W C, Winans S C. J Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang I, Li P-L, Zhang L, Piper K R, Cook D M, Tate M E, Farrand S K. Proc Natl Acad Sci USA. 1994;91:4639–4643. doi: 10.1073/pnas.91.11.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheng J, Citovsky V. Plant Cell. 1996;8:1699–1710. doi: 10.1105/tpc.8.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winans S C. Microbiol Rev. 1992;56:12–31. doi: 10.1128/mr.56.1.12-31.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrand S K. In: Bacterial Conjugation. Clewell D B, editor. New York: Plenum; 1993. pp. 255–291. [Google Scholar]
- 11.Moré M I, Finger L D, Stryker J L, Fuqua C, Eberhard A, Winans S C. Science. 1996;272:1655–1658. doi: 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- 12.Fuqua C, Winans S C. J Bacteriol. 1996;178:435–440. doi: 10.1128/jb.178.2.435-440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevens A M, Dolan K M, Greenberg E P. Proc Natl Acad Sci USA. 1994;91:12619–12623. doi: 10.1073/pnas.91.26.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.You Z, Fukushima J, Ishiwata T, Chang B, Kurata M, Kawamoto S, Williams P, Okuda K. FEMS Microbiol Lett. 1996;142:301–307. doi: 10.1111/j.1574-6968.1996.tb08447.x. [DOI] [PubMed] [Google Scholar]
- 15.Nasser W, Bouillant M L, Salmond G, Reverchon S. Mol Microbiol. 1998;29:1391–1405. doi: 10.1046/j.1365-2958.1998.01022.x. [DOI] [PubMed] [Google Scholar]
- 16.Studier F W, Rosenberg A H, Dunn J J, Dunbendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 17.Cho K, Fuqua C, Winans S C. J Bacteriol. 1997;179:1–8. doi: 10.1128/jb.179.1.1-8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu J, Beaber J W, Moré M I, Fuqua C, Eberhard A, Winans S C. J Bacteriol. 1998;180:5398–5405. doi: 10.1128/jb.180.20.5398-5405.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallie D R, Novak S, Kado C I. Plasmid. 1985;14:171–175. doi: 10.1016/0147-619x(85)90078-2. [DOI] [PubMed] [Google Scholar]
- 20.Hajdukiewicz P, Svab Z, Maliga P. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- 21.Zwieb C, Brown R S. Nucleic Acids Res. 1990;18:583–587. doi: 10.1093/nar/18.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoepfer R. Gene. 1993;124:83–85. doi: 10.1016/0378-1119(93)90764-t. [DOI] [PubMed] [Google Scholar]
- 23.Burgess R R, Jendrisak J J. Biochemistry. 1975;14:4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- 24.Eckert R L. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. New York: Wiley; 1987. pp. 7.5.1–7.5.9. [Google Scholar]
- 25.Zhu J, Winans S C. Mol Microbiol. 1998;27:289–297. doi: 10.1046/j.1365-2958.1998.00672.x. [DOI] [PubMed] [Google Scholar]
- 26.Shaw P D, Ping G, Daly S L, Cha C, Cronan J E, Rinehart K L, Farrand S K. Proc Natl Acad Sci USA. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 421–470. [Google Scholar]
- 28.Choy H E, Adhya S. Proc Natl Acad Sci USA. 1993;90:472–476. doi: 10.1073/pnas.90.2.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen C-Y, Winans S C. J Bacteriol. 1991;173:1139–1144. doi: 10.1128/jb.173.3.1139-1144.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaplan H B, Greenberg E P. J Bacteriol. 1985;163:1210–1214. doi: 10.1128/jb.163.3.1210-1214.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]