Abstract
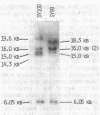
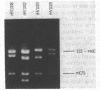
In this work, the previously described inversion in the lactose region of the Escherichia coli K-12 chromosome (D. J. Savić, J. Bacteriol. 140:311-319, 1979) is analyzed in greater detail. The results presented indicate that the inversion most likely occurred by a homologous recombination between alpha 3 beta 3 and beta 5 alpha 5 IS3 elements.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Molecular consequences of deletion formation mediated by the transposon Tn9. Nature. 1980 May 1;285(5759):38–41. doi: 10.1038/285038a0. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Deonier R. C., Hadley R. G., Hu M. Enumeration and identification of IS3 elements in Escherichia coli strains. J Bacteriol. 1979 Mar;137(3):1421–1424. doi: 10.1128/jb.137.3.1421-1424.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Sherratt D. J. Sequence analysis at IS1 insertion sites: models for transposition. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1257–1261. doi: 10.1101/sqb.1979.043.01.142. [DOI] [PubMed] [Google Scholar]
- Hadley R. G., Deonier R. C. Specificity in formation of type II F' plasmids. J Bacteriol. 1979 Sep;139(3):961–976. doi: 10.1128/jb.139.3.961-976.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn B., Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980 Nov;11(3-4):291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- Lloyd R. G., Low B., Godson G. N., Birge E. A. Isolation and characterization of an Escherichia coli K-12 mutant with a temperature-sensitive recA- phenotype. J Bacteriol. 1974 Oct;120(1):407–415. doi: 10.1128/jb.120.1.407-415.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low B. Formation of merodiploids in matings with a class of Rec- recipient strains of Escherichia coli K12. Proc Natl Acad Sci U S A. 1968 May;60(1):160–167. doi: 10.1073/pnas.60.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niaudet B., Ehrlich S. D. In vitro genetic labeling of Bacillus subtilis cryptic plasmid pHV400. Plasmid. 1979 Jan;2(1):48–58. doi: 10.1016/0147-619x(79)90005-2. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sommer H., Cullum J., Saedler H. Integration of IS3 into IS2 generates a short sequence duplication. Mol Gen Genet. 1979;177(1):85–89. doi: 10.1007/BF00267256. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Youngs D. A., Smith K. C. Evidence for the control by exrA and polA genes of two branches of the uvr gene-dependent excision repair pathway in Escherichia coli K-12. J Bacteriol. 1973 Oct;116(1):175–182. doi: 10.1128/jb.116.1.175-182.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]