Abstract
Smad3 and Smad4 are sequence-specific DNA-binding factors that bind to their consensus DNA-binding sites in response to transforming growth factor β (TGFβ) and activate transcription. Recent evidence implicates Smad3 and Smad4 in the transcriptional activation of consensus AP-1 DNA-binding sites that do not interact with Smads directly. Here, we report that Smad3 and Smad4 can physically interact with AP-1 family members. In vitro binding studies demonstrate that both Smad3 and Smad4 bind all three Jun family members: JunB, cJun, and JunD. The Smad interacting region of JunB maps to a C-terminal 20-amino acid sequence that is partially conserved in cJun and JunD. We show that Smad3 and Smad4 also associate with an endogenous form of cJun that is rapidly phosphorylated in response to TGFβ. Providing evidence for the importance of this interaction between Smad and Jun proteins, we demonstrate that Smad3 is required for the activation of concatamerized AP-1 sites in a reporter construct that has previously been characterized as unable to bind Smad proteins directly. Together, these data suggest that TGFβ-mediated transcriptional activation through AP-1 sites may involve a regulated interaction between Smads and AP-1 transcription factors.
Keywords: Smad3, Smad4, cJun, JunB, transforming growth factor
Transforming growth factor β (TGFβ) is a multipotent cytokine that regulates a variety of cellular activities, such as cell proliferation, differentiation, and extracellular matrix (ECM) formation. The combined actions of these cellular responses are likely to mediate more global effects of TGFβ including its role in development, wound healing, immune responses, and the pathogenesis of cancer (1–3). The identification of genes transcriptionally regulated by TGFβ and the elucidation of the molecular mechanisms responsible for this transcriptional regulation will help define how TGFβ exerts its cellular effects and its role in resulting physiological processes. Although progress has been made in the identification of TGFβ target genes, including the cyclin-dependent kinase inhibitors p21 and p15 (1, 2) and the ECM component plasminogen activator inhibitor-1 (PAI-1) (3), which has subsequently contributed toward our understanding of TGFβ-mediated growth inhibition and ECM deposition, the mechanisms by which TGFβ controls gene expression remain largely unknown.
Numerous studies have characterized the differential expression of specific genes in response to TGFβ, revealing a common link in the ability of TGFβ to regulate many of these genes through the functions of the AP-1 family of transcription factors. This protein family, which includes the Fos and Jun proteins, binds a specific DNA sequence and facilitates transcriptional regulation (4). The ability of TGFβ to induce the expression of several genes, including PAI-1, clusterin, monocyte chemoattractant protein-1 (JE/MCP-1), type I collagen, and TGFβ1 itself depends on specific AP-1 DNA-binding sites in the promoter regions of these genes (3, 5–10). Furthermore, TGFβ-mediated transcriptional activation of several of these genes requires AP-1 proteins (5, 8–10). Intriguingly, the expression of many AP-1 proteins themselves is induced as an early response to TGFβ in a cell type-specific manner (11–14). It has been demonstrated that this induced expression of particular AP-1 family members is involved in TGFβ-mediated regulation of subsequent target genes (10). In addition, genetic studies of TGFβ signaling in Drosophila melanogaster reveal a direct overlap between AP-1 and TGFβ signaling and suggest an evolutionarily conserved convergence of these pathways (15). Together, these studies demonstrate a link between TGFβ signaling and AP-1 in the TGFβ-regulated expression of various genes. The molecular mechanisms responsible for the TGFβ-mediated transcriptional activation of these genes are just beginning to be elucidated.
Insight into the mechanism of TGFβ-regulated gene expression has come about with the discovery of the Smad family of proteins. The Smads are phosphorylated by the activated type I receptor in response to ligand (16). Specifically, Smad2 and Smad3 were shown to be inducibly phosphorylated in response to TGFβ (17–19). Smad phosphorylation results in heteromerization of either Smad2 or Smad3 with Smad4 (20–23). Smad4-containing heteromers then enter the nucleus where they can activate transcription of specific genes (24, 25). Current research is focused on elucidating the role of Smads in TGFβ-induced transcriptional activation.
Through attempts made at understanding the mechanism of Smad-mediated transcriptional activation, two distinct roles for Smads have emerged: Smads as DNA-binding factors and Smads as transcription factor-binding proteins. Several lines of evidence suggest that Smads activate transcription by binding directly to DNA. For instance, transcription of a reporter plasmid containing the concatamerized consensus Smad-binding site is induced by TGFβ in a Smad4-dependent manner (26). Smad3 and Smad4 were recently shown to form a complex on similar DNA sequences derived from the PAI-1 promoter (27). Mutation of these sequences in the PAI-1 promoter reduced TGFβ responsiveness. Furthermore, Gal4 fusions with the C-terminal domains of Smad1 and Smad4 activate transcription from concatamerized Gal4 DNA-binding sites (28).
Other evidence suggests that Smads can activate transcription by binding to other transcription factors. For example, the interaction between Smad2/Smad4 heteromers and the transcription factor FAST-1 is critical for the formation of the activin responsive factor (ARF), an activin-inducible DNA-binding complex in Xenopus (24, 29). Overexpression of the Smad-binding domain of FAST-1 blocked ARF formation and transcriptional induction of an activin-inducible early response gene. Together, these data indicate that although Smads bind DNA directly, association with other transcription factors may play a crucial role in Smad-mediated transcriptional activation.
In an attempt to identify transcription factors involved in Smad-mediated transcriptional activation, we performed a yeast two-hybrid screen using Smad3 as a bait. Two interacting cDNAs encoding two different clones of the AP-1 family member, JunB, were isolated, indicating that Smads may bind to AP-1 members directly. Supporting a direct interaction between Smads and AP-1, we show that Smad3 and Smad4 bind all known members of the Jun family of proteins in vitro. Furthermore, we demonstrate that Smad3 is critical for the ability of TGFβ to activate AP-1 sites independent of Smad DNA binding. These data, therefore, provide insight into a possible mechanism by which TGFβ activates AP-1-mediated transcription through the induction of Smad/AP-1 complex formation.
MATERIALS AND METHODS
Materials.
TGFβ1 was a generous gift of Amgen Biologicals. Human keratinocyte cells (HaCaT) were the generous gift of P. Baukamp and N. Fusenig. A HaCaT cDNA library in the pACT2 expression vector was the generous gift of Y. Xiong. The full length cDNAs for murine Jun family members, FosB, cFos, Fra2, and human Fra1, were the generous gifts of R. Wisdom. Smad3 polyclonal antibody was generated against amino acids 200–219 of Smad3 and affinity purified in this laboratory. Antibodies used included JunB polyclonal N-17 (Santa Cruz Biotechnology), cJun monoclonal KM-1 (Santa Cruz Biotechnology), cJun polyclonal no. 9162 (NEB, Beverly, MA), cJun polyclonal no. 06–828 (Upstate Biotechnology, Lake Placid, NY) and JunD-329 polyclonal antibody (Santa Cruz Biotechnology).
Cell Culture.
COS cells were maintained in DMEM with 10% FBS. HaCaT cells were maintained in MEM with 10% FBS. Primary fibroblasts were prepared from day 14 embryos by mechanical dissociation of whole embryos by passage through an 18-gauge needle and plating onto gelatin-coated 10-cm tissue culture plates in DMEM with the inclusion of 20% FBS. Cells were grown to confluence and carried in DMEM with 10% FBS. All experiments were performed on littermate fibroblasts at the same passage number.
Plasmid Construction.
The BamHI fragment containing full length human Smad3 cDNA was subcloned from pGEX-3X into pGBT9 (CLONTECH) (30). cDNAs encoding each Jun and Fos family member were subcloned into pCMV5 and pCMV6 expression vectors (CMV, cytomegalovirus). Full length JunB was PCR amplified with the following primers: CGGGATCCCGATGTGCACGAAAATGG (5′ primer) and GGATCCTCAGAAGGCGTGTCC (3′ primer). Full length cJun was PCR amplified with the following primers: CGGGATCCCGATGACTGCAAAGATGGAAACG (5′ primer) and CGGGATCCCGTCAAAACGTTTGCAACTGC (3′ primer). The cDNAs were completely sequenced and subcloned into pACT2 (CLONTECH). Construction of 4xSBSMT and 4xAP1MT reporter plasmids was previously described (30).
Yeast Two-Hybrid Assay.
The yeast strain Hf7c was transformed with Smad3/pGBT9, and expression of the appropriate-size fusion protein was confirmed by Western blotting by using GAL4 DBD monoclonal antibody (Santa Cruz Biotechnology). Bait-expressing yeast were transformed with a HaCaT cDNA library in the pACT2 expression vector. Individual cDNAs (5 × 106) were screened. Transformants (484) grew on media lacking tryptophan, leucine, and histidine that contained 5 mM 3-aminotriazole. Transformants (242) were positive for β-galactosidase activity, which was measured by the appearance of blue color on colony filter lifts incubated in the presence of 5-bromo-4-chloro-3-indolyl-β-d-galactosidase (X-Gal). Bait dependence for each positive transformant was established similarly.
Binding Studies.
Full length JunB in pGEM4 was digested with BspHI, BssHII, or DraI (NEB). The full length construct and the digested DNAs were used as templates for in vitro transcription and translation (TNT) with [35S]methionine in rabbit reticulocyte lysates (Promega). The TNT–JunB lysates were incubated with an equal amount of bacterially purified glutathione S-transferase (GST), or GST-Smad3 or GST-Smad4 (30) in B/P (150 mM NaCl/50 mM Tris, pH 7.5/0.1% Tween/1 mM DTT) for 2.5 hours at 4°C. The GST reactions were washed three times in TBS (500 mM NaCl/25 mM Tris, pH 7.5/0.1% Tween-20/1 mM DTT). Samples were resolved by SDS/PAGE. The gels were treated with 10% sodium salicylate, dried, and exposed to film. Whole-cell COS lysates overexpressing each AP-1 member were lysed as described (30) and incubated with the GST fusions as described above. The binding reactions were washed three times with B/P and separated by SDS/PAGE.
For endogenous protein interactions, HaCaT cells were treated with 100 pM TGFβ1 in DMEM/10% FBS for 15, 30, or 60 min. Cells were then lysed and either whole-cell or nuclear extracts were prepared (30, 31). Four hundred fifty μg of each whole-cell lysate was incubated with an equal amount of bacterially purified GST-Smad3 or GST-Smad4 normalized for protein by Coomassie blue and for volume of glutathione-Sepharose added to each binding reaction. After 2 hr at 4°C, the reactions were washed three times and separated by SDS/PAGE. One hundred μg of each nuclear lysate was diluted to 150 mM NaCl with buffer A and incubated with GST, GST-Smad3, or GST-Smad4, as above. For phosphatase treatment, HaCaT cells were treated for 15 min with 100 pM TGFβ1 or DMEM containing 10% FBS, penicillin, streptomycin, and 0.5 M sorbitol and whole-cell extracts were prepared as described (30). Briefly, the lysates were treated with potato acid phosphatase (0.034 units) and calf intestinal phosphatase (2 units) for 30 min at 37°C.
Western Blot Analysis.
Electrophoresed proteins were transferred to Immobilon (Millipore) and treated as previously described, except that the blots were blocked and blotted in PBS/0.1% Tween-20/5% milk (30).
Luciferase Assays.
Transfections were performed by using a standard DEAE-Dextran protocol (32). Primary fibroblasts were allowed to recover from glycerol shock for 20 hr before treating with 100 pM TGFβ1 in DMEM/0.2% FBS. Luciferase assays were performed as previously described (33). All transfections were normalized to β-galactosidase activity by cotransfection of 0.5 μg of CMV-β-galactosidase expression vector.
RESULTS
Smad3 and JunB Associate in Yeast.
To identify Smad3-binding proteins, we performed a yeast two-hybrid screen using the Gal4 DNA-binding domain fused to Smad3 as a bait. Of the five million yeast transformants screened for Smad3 binding, 242 transformants were positive for growth in the absence of histidine and for the appearance of blue color on staining with 5-bromo-4-chloro-3-indolyl-β-d-galactosidase (X-Gal). Two of the clones sequenced contained two different cDNA fragments encoding the AP-1 member, JunB (Fig. 1). Clone 44 lacked the N-terminal 126 amino acids of JunB indicating that these residues are not required for binding in yeast. Fusions of the Gal4 activation domain with cDNAs encoding JunB and cJun also tested positive for interaction with the Smad3 bait protein (data not shown).
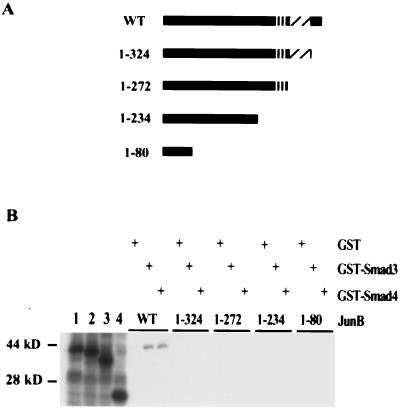
Figure 1.
Isolation of JunB from a Gal4-Smad3 screen of a human keratinocyte library (HaCaT) in yeast. Schematic diagram of two Smad3-interacting clones encoding JunB. Amino acids associated with particular functional domains of JunB are shown.
In Vitro Binding of Smads and AP-1.
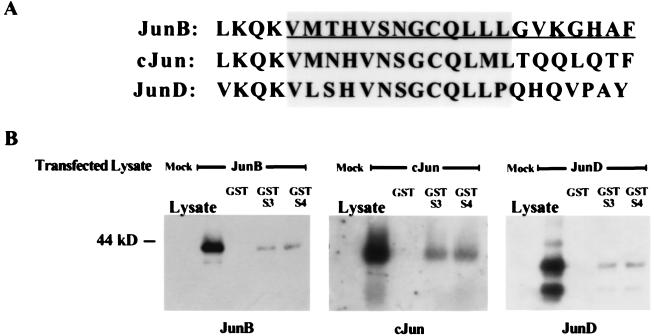
To determine whether this interaction occurs in solution with recombinant proteins, GST pulldown experiments were performed. Bacterially produced GST-Smad3, but not GST alone, bound TNT-JunB (Fig. 2B). In addition, GST-Smad4-bound TNT–JunB, indicating that AP-1 binding is not exclusive to Smad3. Similar studies with GST-Smad1, GST-Smad2, and GST-Smad5 showed that these proteins also bind JunB TNT products but with lower affinity than that observed with GST-Smad3 and GST-Smad4 (data not shown). To map the Smad interaction domain, GST-Smad fusion proteins were used to pull down various TNT–JunB deletion products (Fig. 2 A and B). Deletion of only 20 amino acids from the C terminus of JunB abrogated the interaction between GST-Smad3 and GST-Smad4 and JunB (Fig. 2). Thirteen of these 20 amino acids are conserved among Jun family members, including cJun and JunD (Fig. 3A). To determine whether these proteins also interact with Smads, we tested whether GST-Smad3 and GST-Smad4 could associate with AP-1 members from transfected COS cell lysates overexpressing each Jun member. As shown in Fig. 3B, all three Jun family members associated with both GST-Smad3 and GST-Smad4 to a similar extent. Furthermore, GST-Smad3 and GST-Smad4 bound full length TNT–cJun and deletion of 20 amino acids from the C terminus of TNT–cJun also reduced association with GST-Smad3 and GST-Smad4 (data not shown). Conversely, in studies using lysates from transfected cells overexpressing each Fos family member, no association with GST-Smad3 or GST-Smad4 was observed (data not shown). Consistent with these findings, the amino acids required for Jun binding to GST-Smads in vitro are not conserved among Fos family members.
Figure 2.
In vitro association of Smad3 and Smad4 with JunB. (A) Schematic diagram of JunB deletion mutants used in the binding study shown in B. (B) In vitro association of JunB and JunB deletion mutant TNT products with GST, GST-Smad3, or GST-Smad4. Bacterially produced GST, GST-Smad3, and GST-Smad4 proteins were coupled to glutathione-Sepharose, incubated with the indicated TNT product, centrifuged, and the Sepharose-bound proteins visualized by Coomassie stained SDS/PAGE (data not shown). The amount of each GST protein and the volume of glutathione-Sepharose used in each binding reaction were normalized. Each reticulocyte lysate was produced as described in Materials and Methods. Domains depicted in A refer to the same domains depicted in Fig. 1. Five percent of each reticulocyte lysate input was run in lanes 1–4: lane 1, JunB full length; lane 2, 1–324; lane 3, 1–272; lane 4, 1–80.
Figure 3.
Association of Smad3 and Smad4 with Jun family members. (A) The Smad-binding site on JunB is conserved among Jun family members. The very C-terminal amino acids of the Jun proteins are aligned. The leucine at the beginning of the JunB and cJun sequences is the most C-terminal leucine of the leucine zipper domain. Amino acids that were deleted in the JunB 1–324 mutant are underlined. Conserved amino acids that may play a role in Smad binding are shaded. (B) In vitro association of overexpressed JunB, cJun, and JunD from COS cell lysates with GST, GST-Smad3, or GST-Smad4. COS cells transfected with JunB/pCMV, cJun/pCMV, or JunD/pCMV were lysed, and these extracts were treated with GST fusion proteins prepared as described in Materials and Methods.
Smad3 and Smad4 Associate with An Inducibly Phosphorylated Form of Endogenous cJun.
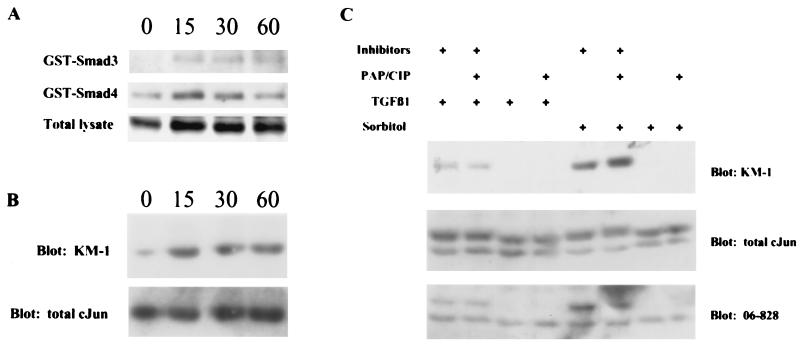
To determine whether GST-Smad3 and GST-Smad4 can associate with endogenous cJun, we incubated GST-Smad3 and GST-Smad4 with TGFβ-treated HaCaT whole-cell extracts. Western blot analysis of these binding reactions with a cJun-specific antibody raised against a phosphopeptide containing phosphorylated Ser-63 of cJun (KM-1 from Santa Cruz Biotechnology) showed that GST-Smad3 and GST-Smad4 bind this form of cJun (Fig. 4A). We noticed that the level of cJun recognized by the KM-1 antibody appeared to increase with TGFβ treatment. To more clearly determine whether the level of cJun was induced by TGFβ, nuclear lysates were analyzed by using the KM-1 antibody and a different cJun antibody raised against a nonphosphorylated N-terminal portion of cJun (no. 9162, NEB). As shown in Fig. 4B, the form of cJun recognized by the KM-1 antibody is clearly induced by TGFβ treatment, whereas the levels of total cJun recognized by the NEB antibody remain relatively unchanged. This result suggests that cJun is rapidly phosphorylated, at least on the residue of Ser-63, in response to TGFβ. Subsequent binding studies with these nuclear lysates and GST, GST-Smad3, and GST-Smad4 revealed that GST-Smad3 and GST-Smad4 bound equally well to cJun recognized by either antibody (data not shown). To confirm that the TGFβ-induced increase in the cJun species detected by the KM-1 antibody was indeed the phosphorylated form of cJun, we performed a phosphatase assay using HaCaT lysates treated with TGFβ (Fig. 4C). As a positive control, HaCaT cells were treated with 0.5M sorbitol, which has been shown to induce cJun N-terminal kinase (JNK) kinase activity (34), and similar results were obtained with lysates treated with 50 μg/ml anisomycin. Under phosphatase treatment conditions, the TGFβ-induced form of cJun recognized by the KM-1 antibody was lost, whereas total cJun detected by the no. 9162 antibody did not change. The specificity of the KM-1 antibody for the phosphorylated form of cJun was further confirmed by using another antibody (no. 06–828, Upstate Biotechnology) raised against a cJun peptide also containing phosphorylated Ser-63.
Figure 4.
GST-Smad3 and GST-Smad4 associate with an inducibly phosphorylated form of endogenous cJun. (A) HaCaT cells were treated with 100 pM TGFβ1 for 15, 30, or 60 min and lysed. GST-Smad3 and GST-Smad4 were incubated with 450 μg of whole-cell HaCaT lysates, and the binding reactions were analyzed by Western blot by using a cJun specific monoclonal antibody raised against a phosphopeptide containing phosphorylated Ser-63 of cJun, KM-1 from Santa Cruz. The lower row shows 60 μg total lysate blotted with the same antibody as the upper rows. (B) Nuclear lysates (60 μg) from HaCaT cells treated as in A were analyzed with the KM-1 antibody. This blot was stripped and reprobed with antibody no. 9162 from NEB raised against a fusion protein containing the unphosphorylated N-terminal portion of cJun. (C) HaCaT cells were treated with 100 pM TGFβ1 or 0.5 M sorbitol for 15 min, lysed in the absence or presence of phosphatase inhibitors, and treated with or without potato acid phosphatase and calf intestinal phosphatase. The reactions were analyzed by SDS/PAGE and Western blotting. The upper row shows 60 μg of each lysate blotted with the KM-1 antibody. This blot was stripped and reprobed with no. 9162 antibody, shown in the middle row. This blot was stripped again and reprobed with no. 06–828 antibody (Upstate Biotechnology). The bands in the KM-1 blot align with the upper bands in the no. 9162 antibody and the no. 06–828 antibody blots.
Smad3 Is Required for TGFβ-Mediated Activation of AP-1 Sites Independent of Smad DNA-Binding Activity.
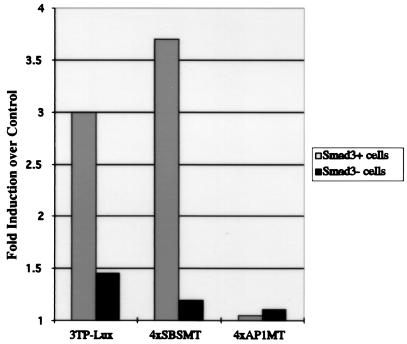
In an attempt to determine whether the interaction between AP-1 and Smads contributes to the ability of TGFβ to activate the transcription of AP-1 sites, we transfected Smad3 heterozygous and Smad3 homozygous null primary mouse embryonic fibroblasts with AP-1 site-containing reporter constructs previously described (Fig. 5) (30). Intriguingly, 4xSBSMT, which contains consensus AP-1 sites adjacent to mutated Smad DNA-binding sites incapable of Smad protein binding, requires Smad3 for transcriptional activation by TGFβ. TGFβ is unable to activate transcription, however, in the analogous reporter, 4xAP1MT, in which the Smad-binding sites are intact and the AP-1 sites are mutated. Thus, within the context of the 4xSBSMT reporter, the AP-1 sites are required for transcriptional activation by TGFβ. Activation of these AP-1 sites requires Smad3 but is independent of Smad DNA binding, suggesting that the ability of Smad3 to act through the AP-1 sites in this reporter is required for transcriptional activation by TGFβ.
Figure 5.
Smad3 is required for TGFβ-mediated transcriptional activation of 4xSBSMT. Smad3 homozygous null or Smad3 heterozygous mouse primary fibroblasts were transfected with 3 μg of 3TP-Lux, 4xSBSMT, or 4xAP1MT reporter constructs. Twenty hours after transfection, vehicle or 100 pM TGFβ1 was added. Twenty-four hours after treatment, luciferase activity was quantified. The averages of duplicate transfections were used to determine the fold induction by TGFβ over the vehicle control. The 3TP-Lux data is shown as a positive control for the experiment (35).
DISCUSSION
Over the past year, a model for the functional role of Smads in TGFβ-mediated transcriptional regulation has emerged. Here, we provide evidence supporting a role for Smads as transcriptional coactivators, in addition to their role as DNA binding-dependent activators of transcription. Smads may thus transduce the TGFβ signal to the promoter level and activate transcription through direct physical interaction with DNA-bound AP-1 proteins.
The potential role of Smads as transcriptional coactivators of AP-1 is supported by a previous study in which we reported that TGFβ as well as Smad3/Smad4 cooverexpression could activate transcription of 4xWT, a luciferase reporter containing a concatamerized TGFβ-responsive element derived from 3TP-Lux that contains both consensus AP-1 binding sites and Smad4-binding sites (30). Interestingly, a mutation in this sequence that inhibited Smad association had no effect on the ability of TGFβ or Smad3/Smad4 cooverexpression to induce transcription of this reporter. In contrast, a mutation in the adjacent AP-1 site that inhibited the association of a constitutively bound, AP-1-containing complex, abrogated the ability of TGFβ or Smad overexpression to activate transcription of the 4xWT reporter. These data suggest that within the context of this reporter, TGFβ-mediated transcriptional activation does not depend on the DNA- binding function of Smads but rather the ability of Smads to act through the AP-1 sites. In light of the interaction studies described here, Smads may act through these AP-1 sites by binding directly to AP-1 proteins. In studies of Smad3 wild-type and homozygous null fibroblasts, it was demonstrated that TGFβ-mediated transcriptional activation of 3TP-Lux (35) and the cJun promoter (14), which contain both Smad and AP-1 DNA-binding sites, was lost in fibroblasts lacking Smad3. Transfection of Smad3 into the null fibroblasts rescued transcriptional activation by TGFβ. Here, we further show that activation of the AP-1 sites in a reporter containing mutant Smad-binding sites similarly requires Smad3. Taken together, the interaction and transcriptional activation data suggest that the regulated association between Smads and AP-1 may be necessary for the TGFβ-mediated transcriptional activation of AP-1 sites. Consistent with these findings, recently published data demonstrate that cotransfection of Smad3 and Smad4 and cJun facilitates mild synergistic transcriptional activation of AP-1-site containing reporters (36).
We demonstrate that Smad3 and Smad4 bind JunB directly in vitro, and that the interaction involves a stretch of 20 C-terminal amino acids of JunB. It is possible that deletion of these 20 amino acids that abrogated Smad binding may alter the conformation of JunB, rendering it incapable of binding Smads. However, we do show that both cJun and JunD, which contain conserved identity in 13 of these 20 C-terminal amino acids, also bind GST-Smad3 and GST-Smad4, whereas Fos family members that lack this conserved amino acid sequence do not bind GST-Smad3 or GST-Smad4. Therefore, we postulate that Smad3 and Smad4 binding requires specific residues within the C-terminal 20 amino acids of JunB. Furthermore, direct association with Smads may be a conserved function of the Jun family of transcription factors.
Because Smads translocate to the nucleus in response to TGFβ (20–23), the interaction between Smads and Jun family members may be regulated largely by TGFβ-induced alteration of Smad subcellular localization. Intriguingly, we found that endogenous cJun was rapidly phosphorylated in response to TGFβ, most likely at Ser-63. Phosphorylation at this site peaks by 15 min and is maintained over the course of an hour of TGFβ treatment, a time course that overlaps TGFβ-induced Smad entry into the nucleus (17, 21, 22, 25). This inducibly phosphorylated form of cJun binds both GST-Smad3 and GST-Smad4. Although this phosphorylation does not appear to alter the association of Smads and AP-1 in vitro, we suspect this phosphorylation may contribute to TGFβ-mediated transcription in vivo. This notion is supported by evidence showing that JNK is activated in response to TGFβ (37, 38), and that this kinase is known to phosphorylate cJun at Ser-63 and -73, thereby enhancing the ability of cJun to activate transcription (39). These data, in combination with previously discussed results, suggest that TGFβ treatment may initiate two simultaneous signaling pathways that converge on AP-1 complexes in the nucleus: a Smad-mediated pathway and a JNK-mediated pathway. The combined result of these pathways may be stronger interactions between Smads and AP-1 and as a result, a more robust induction of transcription. Although cJun phosphorylation may indeed enhance association with Smads under physiological conditions, we do not have evidence to suggest that Jun phosphorylation is required for the Smad/AP-1 interaction, since we observe the interaction under in vitro conditions in which JNK-mediated phosphorylation would not occur and also detect a constitutive interaction of GST-Smads with endogenous cJun in the absence of TGFβ treatment. Furthermore, JunB, which is not a JNK substrate, also binds to Smads (40). Future work is necessary to define the role of Jun phosphorylation in TGFβ-mediated transcription in vivo.
The data presented here may provide a plausible explanation for the specificity of TGFβ-mediated induction of specific responsive promoters that contain AP-1 DNA-binding sites. On TGFβ treatment, Smad3 and Smad4 heteromerize and enter the nucleus, where they can associate with TGFβ-responsive promoters by binding a discreet DNA sequence and/or AP-1 members bound to AP-1 sites on the same promoter. Thus, Smads in response to TGFβ act as the signaling intermediates to initiate transcription from specific promoters by recruiting required factors to form an active transcriptional complex. The transcriptional adapter molecule p300/CBP, which binds directly to AP-1 and serves as a coactivator of AP-1-mediated transcription, has recently been shown to associate directly with Smads in response to TGFβ (41–44). TGFβ-induced Jun modification may promote the stability of these interactions, thereby facilitating complex formation. Given that AP-1 complex composition depends on differential expression of specific family members, the distinct constitution of AP-1 complexes in different cell types may contribute to promoter targeting specificity by TGFβ. This possibility is supported by the observation that under conditions where Smads bind Jun proteins, Smads are unable to associate with Fos family members. Thus, it is possible that AP-1 complexes containing Fos members may have a lower affinity for Smads than AP-1 complexes containing Jun–Jun dimers, and that promoters associated with these specific AP-1 complexes would be favored for Smad binding and TGFβ-mediated activation. The induced interaction of Smads with particular AP-1 complexes in vivo may determine the ability of TGFβ to initiate transcription from specific AP-1 site-containing promoters.
Acknowledgments
We are grateful to Alison Meloni and Michael Nichols for technical assistance with the yeast two-hybrid screen. We thank Gary Reuther for his help with the in vitro interaction studies and Jonathan Yingling for providing reagents, members of the Wang lab for helpful discussions on this project, and Yong Yu for technical assistance. This work was supported by grants CA75368 from the National Institutes of Health and DAMD17-94-J-4065 from the Department of the Army. N.T.L. was supported by a National Science Foundation Predoctoral Fellowship; J.P.F. was supported by a predoctoral fellowship from the Department of the Army.
ABBREVIATIONS
- TGFβ
transforming growth factor β
- HaCaT
human keratinocyte cells
- JNK
cJun N-terminal kinase
- CMV
cytomegalovirus
- GST
glutathione S-transferase
- TNT
transcription and translation
References
- 1.Datto M B, Li Y, Panus J, Howe D J, Xiong Y, Wang X-Y. Proc Natl Acad Sci USA. 1995;92:5545–5549. doi: 10.1073/pnas.92.12.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hannon G J, Beach D. Nature (London) 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 3.Keeton M R, Curriden S A, van Zonneveld A-J, Loskutoff D J. J Biol Chem. 1991;266:23048–23052. [PubMed] [Google Scholar]
- 4.Karin M, Liu Z, Zandi E. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 5.Jin G, Howe P H. J Biol Chem. 1997;272:26620–26626. doi: 10.1074/jbc.272.42.26620. [DOI] [PubMed] [Google Scholar]
- 6.Armendariz-Borunda J, Simkevich C P, Roy N, Raghow R, Kang A H, Seyer J M. Biochem J. 1994;304:817–824. doi: 10.1042/bj3040817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang E, Goldberg H. J Biol Chem. 1995;270:4473–4477. doi: 10.1074/jbc.270.9.4473. [DOI] [PubMed] [Google Scholar]
- 8.Kim S J, Angel P, Lafyatis R, Hattori K, Kim K Y, Sporn M B, Karin M, Roberts A B. Mol Cell Biol. 1990;10:1492–1497. doi: 10.1128/mcb.10.4.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeshita A, Chen Y, Watanabe A, Kitano S, Hanazawa S. J Immunol. 1995;155:419–426. [PubMed] [Google Scholar]
- 10.Mauviel A, Chung K Y, Agarwal A, Tamai K, Uitto J. J Biol Chem. 1996;271:10917–10923. doi: 10.1074/jbc.271.18.10917. [DOI] [PubMed] [Google Scholar]
- 11.Beauchamp R D, Sheng H M, Ishizuka J, Townsend C M J, Thompson J C. Ann Surg. 1992;216:300–307. doi: 10.1097/00000658-199209000-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blatti S P, Scott R E. Cell Growth Differ. 1992;3:429–434. [PubMed] [Google Scholar]
- 13.Pertovaara L, Sistonen L, Bos T J, Vogt P K, Keski-Oja J, Alitalo K. Mol Cell Biol. 1989;9:1255–1262. doi: 10.1128/mcb.9.3.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong C, Rougier-Chapman E M, Frederick J P, Datto M B, Liberati N T, Li J-M, Wang X-F. Mol Cell Biol. 1999;19:1821–1830. doi: 10.1128/mcb.19.3.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riesgo-Escovar J R, Hafen E. Science. 1997;278:669–672. doi: 10.1126/science.278.5338.669. [DOI] [PubMed] [Google Scholar]
- 16.Abdollah S, Macias-Silva M, Tsukazaki T, Hayashi H, Attisano L, Wrana J L. J Biol Chem. 1997;272:27678–27685. doi: 10.1074/jbc.272.44.27678. [DOI] [PubMed] [Google Scholar]
- 17.Liu X, Sun Y, Constantinescu S N, Karam E, Weinberg R A, Lodish H F. Proc Natl Acad Sci USA. 1997;94:10669–10674. doi: 10.1073/pnas.94.20.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin C H, Miyazono K, et al. EMBO J. 1997;16:5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macias-Silva M, Abdollah S, Hoodless P, Pirone R, Attisano L, Wrana J. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 20.Wu R Y, Zhang Y, Feng X H, Derynck R. Mol Cell Biol. 1997;17:2521–2528. doi: 10.1128/mcb.17.5.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagna G, Hata A, Hemmati-Brivanlou A, Massague J. Nature (London) 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Feng X-H, Wu R-Y, Derynck R. Nature (London) 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Musci T, Derynck R. Curr Biol. 1997;7:270–276. doi: 10.1016/s0960-9822(06)00123-0. [DOI] [PubMed] [Google Scholar]
- 24.Liu F, Pouponnot C, Massague J. Genes Dev. 1997;11:3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Souchelnytskyi S, Tamaki K, Engstrom U, Wernstedt C, ten Dijke P, Heldin C H. J Biol Chem. 1997;272:28107–28115. doi: 10.1074/jbc.272.44.28107. [DOI] [PubMed] [Google Scholar]
- 26.Zawel L, Dai J L, Buckhaults P, Zhou S, Kinzler K W, Vogelstein B, Kern S E. Mol Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 27.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier J M. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu F, Hata A, Baker J, Goody J, Carcamo J, Harland R, Massague J. Nature (London) 1996;381:620–623. doi: 10.1038/381620a0. [DOI] [PubMed] [Google Scholar]
- 29.Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Nature (London) 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- 30.Yingling J M, Datto M B, Wong C, Frederick J P, Liberati N T, Wang X- F. Mol Cell Biol. 1997;17:7019–7028. doi: 10.1128/mcb.17.12.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schreiber E, Matthias P, Müller M, Schaffner W. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wrana J L, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang X F, Massague J. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 33.Datto M B, Yu Y, Wang X-F. J Biol Chem. 1995;270:28623–28628. doi: 10.1074/jbc.270.48.28623. [DOI] [PubMed] [Google Scholar]
- 34.Bogoyevitch M A, Ketterman A J, Sugden P H. J Biol Chem. 1995;270:29710–29717. doi: 10.1074/jbc.270.50.29710. [DOI] [PubMed] [Google Scholar]
- 35.Datto M B, Frederick J P, Pan L, Borton A J, Zhuang Y, Wang X-F. Mol Cell Biol. 1999;19:2495–2504. doi: 10.1128/mcb.19.4.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Feng X-H, Derynk R. Nature (London) 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- 37.Atfi A, Djelloul S, Chastre E, Davis R, Gespach C. J Biol Chem. 1997;272:1429–1432. doi: 10.1074/jbc.272.3.1429. [DOI] [PubMed] [Google Scholar]
- 38.Wang W, Zhou G, Hu M C-T, Yao Z, Tan T-H. J Biol Chem. 1997;272:22771–22775. doi: 10.1074/jbc.272.36.22771. [DOI] [PubMed] [Google Scholar]
- 39.Dérijard B, Hibi M, Wu I-H, Barrett T, Su B, Deng T, Karin M, Davis R J. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 40.Kallunki T, Deng T, Hibi M, Karin M. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- 41.Shen X, Hu P-C, Liberati N T, Datto M B, Frederick J P, Wang X-F. Mol Biol Cell. 1998;9:3309–3319. doi: 10.1091/mbc.9.12.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Topper J N, DiChiara M R, Brown J D, Williams A J, Falb D, Collins T, Gimbrone M A J. Proc Natl Acad Sci USA. 1998;95:9506–9511. doi: 10.1073/pnas.95.16.9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janknecht R, Wells N J, Hunter T. Genes Dev. 1998;12:2114–2119. doi: 10.1101/gad.12.14.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng X H, Zhang Y, Wu R Y, Derynck R. Genes Dev. 1998;12:2153–2163. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]