Abstract
RNA editing in higher plant plastids alters mRNA sequences by C-to-U conversions at highly specific sites through an unknown mechanism. To elucidate how the cytidine residues to be edited are specifically recognized and distinguished from other cytidines in close proximity, we have changed in vivo the distances of two plastid RNA-editing sites from their essential upstream cis-acting sequence element. Analysis of RNA editing in transgenic chloroplasts revealed that reduction of this distance by 1 nt entirely abolishes RNA editing. Surprisingly, deletions or combinations of deletional and point mutations that shift a heterologous cytidine residue in the same distance from the upstream cis-element as the editing site in the wild type result in transfer of the RNA-editing activity to the heterologous cytidine whereas the wild-type site remains unedited. Our results suggest that the molecular identity of at least some editing sites in the chloroplast genome is defined by their distance from an essential upstream sequence element.
Posttranscriptional alterations of single nucleotides within an mRNA are referred to as RNA editing and have been described for a variety of genetic systems (1), including higher-plant mitochondria (2–4) and chloroplasts (refs. 5 and 6; for review, see, e.g., ref. 7). It appears useful to formally distinguish between two major types of RNA editing: insertional/deletional and conversional editing. The insertional/deletional type of editing is best known from kinetoplasts of trypanosomes, where uridine residues are inserted or deleted (ref. 8; for review, see, e.g., ref. 9). Editing in mammalian nuclei and plant organelles is of the conversional type and typically involves purine-to-purine or pyrimidine-to-pyrimidine transitions. The editing processes in different genetic systems employ widely different mechanisms, implying that editing activities may have evolved several times independently.
The editing systems in higher-plant mitochondria and chloroplasts share many similar features and, thus, may have originated from common evolutionary roots. RNA editing in both organelles proceeds mainly by C-to-U conversions with the exception of few reverse events. Editing is an early posttranscriptional event and an essential processing step in the maturation of organellar transcripts: the nucleotide conversions usually alter the coding properties of the mRNA, thereby facilitating the synthesis of functional proteins (10).
A central question surrounding plant organellar RNA editing is how to explain the extraordinarily high specificity with which the editing apparatus selects individual cytosine residues for modification. The sequences flanking editing sites lack any apparent conserved, consensus sequence-like elements at the primary or at the secondary structure level. A number of in vivo studies in transgenic chloroplasts have demonstrated that mRNA sequences flanking the editing site are involved directly in plastid RNA editing (11–14). However, the molecular mechanism by which RNA-editing sites are recognized with high specificity as well as how the editing machinery distinguishes between the cytidines to be edited and other cytidines in close proximity are completely unknown. The absence of consensus motifs at the mRNA level may indicate that editing sites in plant organelles are selected by a molecular mechanism different from the recognition of primary or secondary structural features at the editing site itself.
Here we provide evidence from transgenic in vivo studies that the position of a cytidine residue in relation to an essential upstream cis-acting sequence element determines whether or not this cytidine can be edited. Our results suggest a model for editing site recognition in which the editing machinery binds to an upstream sequence element and recognizes the editing site as being a downstream cytidine in a defined distance.
MATERIALS AND METHODS
Plant Material.
Tobacco plants (Nicotiana tabacum cv Petit Havana) were grown under sterile conditions on agar-solidified MS medium (15) containing 30 g/liter sucrose. Homoplasmic transformed lines were rooted and propagated on the same medium. The previously generated transplastomic tobacco line Nt-pRB59 (12) was kept under identical conditions. For seed assays and tests of maternal transgene inheritance, wild-type and transformed plants were transferred to soil and grown to maturity under standard greenhouse conditions.
List of Oligonucleotides.
Oligonucleotides included: P3, 5′-CAGTTGGAAGAATTTGTCC-3′; P10, 5′-AACCTCCTATAGACTAGGC-3′; P11, 5′-AGCGAAATGTAGTGCTTACG-3′; P16, 5′-TTTTTCTAGACGCTCATATTCATTACCGTA-3′; P28, 5′-TAGCACCCTCTTGATAGAAC-3′; P29, 5′-CGCTATGGAACTCGCCGCC-3′; P30, 5′-TTTTGGATCCTACGTCAGGAGTCCATTGATGAGAAGGGCTGGGGA-3′; P31, 5′-TTTTGGATCCTACGTCAGGAGTCCATTGATGAGAAGGCTGGGGA-3′; P32, 5′-TTTTGGATCCTACGTCAGGAGTCCATTGATGAGAAGGGGCTGGGGGAAAGC-3′; P33, 5′-TTTTGGATCCTACGTCAGGAGTCCATTGATAGGAAGGGCTGGGGA-3′; 7355, 5′-GACTATAGATCGAACCTATCC-3′; 1020, 5′-CAAGATCCATTACGTGTCCAAGG-3′.
Construction of Plastid Transformation Vectors.
Chimeric genes containing mutated sequences with the ndhB editing sites IV and V were constructed by using the previously generated plastid transformation vector pRB51, which contains a minilinker between the aadA coding region and the psbA 3′ untranslated region (12). Editing sites IV and V- containing ndhB fragments for insertion into pRB51 were prepared by PCR amplification of the corresponding plastid sequences (16) from −42, with respect to editing site IV, to +22, with respect to editing site V. A 5′ XbaI restriction site, a 3′ BamHI site, and the desired (insertional or deletional) mutations were introduced with the primer sequences. After digestion with XbaI and BamHI, the fragments were cloned into the similarly cut pRB51 and the correctness of mutagenesis and cloning were verified by DNA sequencing with primer P3. In this way, the following transformation vectors were generated: pRB67-Δ1 (ndhB insertion amplified with the primer pair P16/P30), pRB68-Δ2 (primer pair P16/P31), pRB69-i1 (primer pair P16/P32), and pRB73-Δ1/cm (compensatory mutation; primer pair P16/P33).
Plastid Transformation and Selection of Transplastomic Tobacco Lines.
Young leaves were harvested from sterile plants and bombarded with plasmid-coated tungsten particles by using the DuPont PDS1000He biolistic gun (17). Primary spectinomycin-resistant lines were selected on RMOP regeneration medium containing 500 mg/liter spectinomycin dihydrochloride (18). PCR, using the chimeric aadA gene-specific primer pair P10/P11, eliminated spontaneous, spectinomycin-resistant mutants and identified true plastid transformants. Correct integration of the constructs into the chloroplast genome was verified by PCR with the primer pair P11/1020 (12). For each construct, three independent, transplastomic lines were subjected to four additional rounds of regeneration on RMOP/spectinomycin to obtain homoplasmic tissue. Homoplasmy was verified by a highly sensitive PCR assay (12).
Isolation of Nucleic Acids.
Total plant nucleic acids were extracted according to a rapid miniprep procedure described by Doyle and Doyle (19). Total cellular RNA was isolated by using the TRIzol reagent (GIBCO/BRL). For cDNA synthesis, an aliquot of the RNA preparation was treated with DNase I. Vector DNA for biolistic transformation and templates for plasmid sequencing were prepared by using the Qiagen column-purification system.
cDNA Synthesis and PCR.
Reverse transcription was primed with a random hexanucleotide primer mixture for 10 min at room temperature. The elongation reaction was carried out with SuperScript II reverse transcriptase (GIBCO/BRL) at 42°C following the manufacturer’s instructions. DNA and cDNA templates were amplified according to standard PCR protocols.
DNA Sequencing.
Plasmid DNA was sequenced by cycle sequencing, using the fluorescence-labeled oligonucleotide P3 as sequencing primer. Primer pairs P11/P28 or P29/P28 were used to generate the substrate for direct sequencing of transgene-derived PCR products. Amplification products were purified for sequencing by electrophoresis on 2% agarose gels and subsequent extraction from excised gel slices by using the Qiaex II kit (Qiagen). Sequence determination was carried out by a modified chain-termination method (20). Oligonucleotide P28 served as sequencing primer for the PCR products. RNA-editing efficiencies were quantitated by using a PhosphorImager and a quantitation procedure developed earlier (12).
RESULTS
Integration of Mutated Sequences Containing Two ndhB Editing Sites into the Tobacco Plastid Genome.
The plastid ndhB gene encodes a subunit of a putative chloroplast NADH dehydrogenase (21). The ndhB mRNA was shown previously to undergo several base changes by RNA editing (22, 23). It contains nine editing sites in tobacco (23), six of which are grouped in three pairs with two closely spaced sites each.
We recently have defined a minimum sequence context that is necessary and sufficient to direct editing at ndhB sites IV and V in vivo (12). In these analyses, a sequence element 5′ upstream of the two editing sites (in the −2 to −12 region with respect to site IV) was identified that is absolutely required for eliciting the editing reaction at both sites. Most of the nucleotides in the small, 8-nt spacer in between sites IV and V turned out to have little or no influence on editing efficiencies (14). Both editing sites are embedded in sequences with numerous other cytidine residues in close proximity (Fig. 1), and it is not clear how the plastid RNA-editing machinery distinguishes between these cytidines and recognizes the editing sites with high specificity.
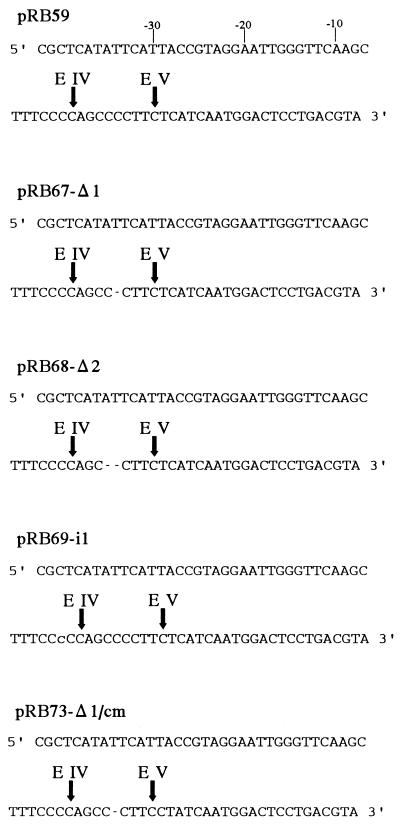
Figure 1.
Sequences of the ndhB segment insertions in the chloroplast transformation vectors used in this study. Plasmid pRB59 contains the wild-type sequence (12) and spans an ndhB segment from −42, with respect to the 5′ editing site (site IV), to +22, with respect to the 3′ editing site (site V). Vector pRB67-Δ1 carries a deletion of a single nucleotide in the 8-nt spacer separating sites IV and V. pRB68-Δ2 has a deletion of 2 nt in the spacer, and in pRB69-i1, a single C residue was inserted into a tetracytidine motif harboring editing site IV (denoted by a lowercase C at an arbitrarily chosen position). Vector pRB73-Δ1/cm is identical to pRB67-Δ1 but carries an additional compensatory point mutation expected to restore editing at the 3′ site. See text for details. Editing sites as in the wild type are marked by vertical arrows. Deleted nucleotides are indicated by dashes, and inserted nucleotides are shown as lowercase letters. The nucleotides upstream of site IV (harboring the essential cis-acting elements for the editing of both sites in the –2/−12 region; ref. 12) are numbered in the pRB59 sequence, with editing site IV as nucleotide “0.”
The requirement for a cis-acting element in proximity to, but spatially separated from, the editing sites raises an attractive possibility of how the cytidines to be edited are specifically recognized: the editing apparatus could act only on those cytidines that are in an exactly defined distance from the essential upstream sequence element. To test this hypothesis, we have constructed a series of chloroplast transformation vectors in which this distance was altered. For this purpose, we chose the ndhB editing sites IV and V as a model system because they are well characterized with respect to the sequence requirements for editing.
The ndhB sequence manipulations were carried out in a −42/+22 segment spanning the two editing sites IV and V. Incorporated into a chimeric context, this segment was shown to yield 95% editing at site IV and 75% at site V (12, 14). In the final transformation vectors, the mutated ndhB sequence segment is linked to and cotranscribed with a selectable marker gene (aadA) conferring resistance to spectinomycin (12, 17). The flanking regions of homology to sequences of the tobacco plastid genome target the chimeric aadA/ndhB transgenes to the intergenic spacer region between the psbE operon and the petA gene, which is known to be a suitable target site for the uptake of transgenes (10, 12). In this way, the transformation vectors with the ndhB insertions shown in Fig. 1 were constructed.
The chimeric aadA/ndhB genes were integrated into the tobacco plastid genome by using the biolistic process. Sterile tobacco leaves were bombarded with plasmid DNA-coated tungsten particles and subsequently subjected to selection for spectinomycin resistance on a plant-regeneration medium (18). Primary plastid transformants appeared after 1–2 months. Correct integration of the transgene into the chloroplast genome was confirmed by PCR-based assays. Subsequently, homoplasmic transplastomic lines were purified by repeated plant regeneration under selective conditions.
Deletion of a Single Nucleotide in the Spacer Between Editing Sites IV and V Selectively Abolishes Editing at Site V.
In a first experiment toward determining the influence of nucleotide phasing on editing-site recognition, we left the 5′ editing site (site IV) and its distance from the upstream cis-element unchanged and merely altered the distance of the downstream site V by 1 nt. Analysis of partially edited cDNA clones recently has established that the two editing sites are edited independently and not in a 3′ → 5′ or 5′ → 3′ polar fashion. Consequently, loss of editing at one site is not expected to abolish editing at the other (14). Moreover, using a scanning-point mutagenesis approach, the four cytidine residues in the 8-nt spacer between the two editing sites (Fig. 1) were shown not to be involved in editing-site recognition (14). Thus, this C4 motif seemed to be a suitable site at which to introduce the desired mutations. We first deleted one of the cytidine residues from the spacer between editing sites IV and V (pRB67-Δ1, Fig. 1) and generated transgenic tobacco plants carrying this mutated ndhB segment in their chloroplast genome.
Sequencing of the cDNA population derived from the chimeric aadA/ndhB gene construct revealed that editing at the 5′ site (IV) is not affected by the single nucleotide deletion (Fig. 2A). This finding is consistent with the idea that the essential elements for editing-site recognition reside upstream of both sites (12). Editing at site V, however, turned out to be completely abolished in the Nt-pRB67-Δ1 transplastomic tobacco lines. Because the nucleotide deleted in pRB67-Δ1 can be changed by point mutagenesis without any effect on editing of site V (13), loss of site V editing in the Nt-pRB67-Δ1 lines could indicate that, indeed, the distance of the editing site from the upstream cis-element is critical for editing-site selection.
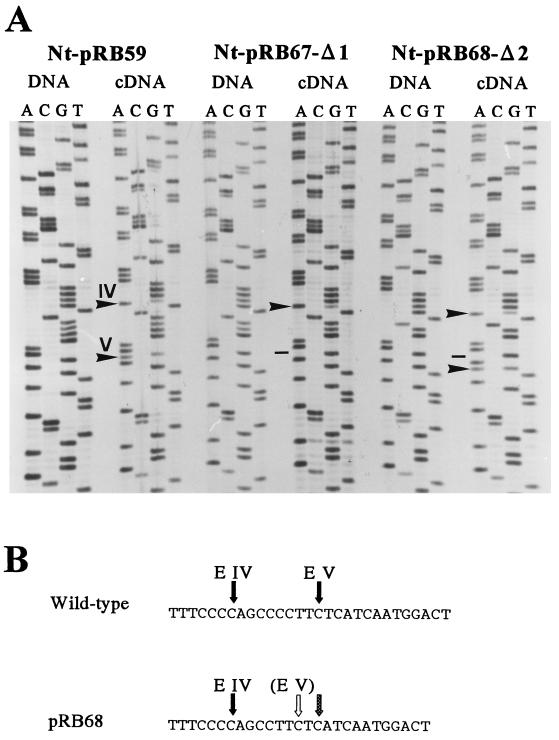
Figure 2.
RNA editing in the Nt-pRB67-Δ1 and Nt-pRB68-Δ2 transplastomic tobacco plants. (A) Sequence analysis to test for transgene mRNA editing in the Nt-pRB67-Δ1 and Nt-pRB68-Δ2 transplastomic lines in comparison with editing of the wild-type ndhB sequence placed in an identical transgenic context (line Nt-pRB59). DNA and cDNA were amplified with primer pair P11/P28 and sequenced directly with oligonucleotide P28. Owing to the polarity of this primer, the sequence ladders reflect the DNA strand complementary to the mRNA sequence. Arrowheads point to the editing positions in the cDNA lanes (G in DNA; A in cDNA), dashes denote lack of editing, and roman numerals indicate editing sites IV and V (see Fig. 1). Note the lack of editing at site V in Nt-pRB67-Δ1 (dash) and induction of editing at a downstream cytidine (arrowhead) in Nt-pRB68-Δ2. (B) Shift of the ndhB site V editing activity to a heterologous site in Nt-pRB68-Δ2 transplastomic tobacco lines. Deletion of two cytidine residues from the spacer sequence between editing sites IV and V results in the transfer of a downstream cytidine (stippled arrow) in a distance from site IV (solid arrow) identical to that of what was previously site V (solid arrow in the wild-type sequence; open arrow in the pRB68-Δ2 sequence). Whereas wild-type site V remains unedited, editing is induced at the cytidine, which is now 9 nt from site IV.
Deletion of Two Nucleotides from the Spacer Between Editing Sites IV and V Induces Editing at a Novel Site.
We then deleted two of the four cytidines in the spacer between editing sites IV and V (pRB68-Δ2; Figs. 1 and 2B). This deletion is different from the above-described mutation in that it shifts a downstream cytidine in place of the wild-type editing site V (Figs. 1 and 2B). If the distance from the essential upstream cis-element indeed were the major determinant for editing-site recognition, then the editing machinery would now find a cytidine residue in the correct phase with the cis-element and possibly would be able to act on this heterologous substrate cytidine.
As expected, editing at the 5′ site IV also was not affected by the 2-nt deletion present in the Nt-pRB68-Δ2 plants. Also, as in the Nt-pRB67-Δ1 lines, editing at site V was completely lost in the Nt-pRB68-Δ2 plants. However, the novel “in-phase” cytidine now undergoes editing with efficiency (65%) similar to that in site V in the Nt-pRB59 control lines (75%, Fig. 2). This transfer of the editing activity to a heterologous site may suggest that this editing site indeed may be recognized as a cytidine being in a defined distance from an upstream cis-acting sequence element.
A Single-Nucleotide Insertion Upstream of Site IV Affects Editing at Both Sites.
We next wanted to test whether the identity of the upstream editing site IV is determined similarly by its distance from the upstream cis-element, as is shown for site V. Site IV is part of a tetracytidine motif, with the 3′ most cytidine as the editing position (Fig. 1). We inserted an additional cytidine into this motif. If the distance from the 5′ cis-element was also the critical determinant for site IV, then the fourth of the now five cytidines should undergo editing. Alternatively, if the position of the editing site in the surrounding sequence context was the recognition principle, then, as in the wild type, the 3′ most (i.e., the fifth) cytidine should be recognized. Simultaneously, this cytidine insertion changes the distance of editing site V from the upstream cis-element. In contrast to the Nt-pRB67-Δ1 and Nt-pRB68-Δ2 transplastomic lines, where this distance is reduced by 1 or 2 nt, respectively, it now is increased by 1 nt in transformation vector pRB69-i1 (Fig. 1).
Analysis of site IV editing in the Nt-pRB69-i1 transplastomic lines revealed that only the fourth cytidine in the pentacytidine motif was edited (Fig. 3). No editing was detected at the 3′ most cytidine edited in the wild type, suggesting that, for ndhB editing site IV as well, the position of the cytidine in relation to the upstream cis-acting element determines the identity of the editing site. In the Nt-pRB69-i1 lines, cytidines are both the 5′ and the 3′ neighboring nucleotides of the edited cytidine. Our finding that, nonetheless, only the cytidine in the correct distance from the upstream cis-element undergoes editing is indicative of a remarkably high accuracy with which this recognition mechanism operates.
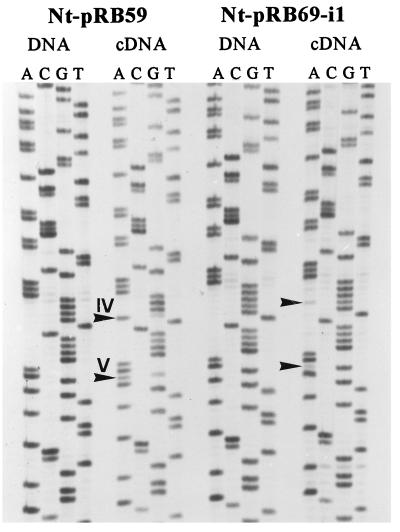
Figure 3.
Sequence analysis to test the effect of a single C nucleotide inserted into a tetracytidine motif containing editing site IV as the 3′ most cytidine residue. For comparison, editing in the wild-type ndhB sequence as contained in line Nt-pRB59 also is shown. DNA and cDNA samples were amplified with primer pair P11/P28 and sequenced directly with primer P28. Because of the polarity of this primer, the autoradiograph shows the sequences of the DNA strand complementary to the mRNA. Arrowheads mark the editing positions in the cDNA lanes, and roman numerals indicate editing-site numbers.
The editing efficiency at the fourth cytidine is significantly lower (approximately 20%) than that at site IV in the Nt-pRB59 control lines (95%). This most probably is caused by the presence of a different 3′ neighboring nucleotide of the editing site. This explanation is in accordance with the earlier findings that the nucleotides immediately adjacent to the editing position contribute significantly to the efficiency of the editing reaction (12–14).
As expected from the results with the nucleotide deletions in the spacer region (as in pRB67-Δ1 and pRB68-Δ2), the insertion of the cytidine in the Nt-pRB69-i1 lines also exerts a negative effect on RNA editing at downstream site V. However, we reproducibly detected a residual editing activity of approximately 10% (Fig. 3). Thus, in contrast to the deletion of 1 nt, which leads to a complete loss of editing, insertion of 1 nt does not entirely abolish editing. From an evolutionary point of view, such a slightly relaxed specificity would be tolerable because the nucleotides 5′ and 3′ of editing site V are not cytidines. However, in the case of site IV, which is flanked by other cytidines, the editing machinery seems to measure the distance from the upstream cis-element with perfect accuracy, thereby preventing misediting of neighboring cytidines, which potentially would result in the synthesis of nonfunctional proteins.
A Compensatory Point Mutation Restores RNA Editing in the Single-Nucleotide Deletion Mutant.
As described above, deletion of a single nucleotide in the spacer region between editing sites IV and V led to a complete loss of editing at site V (Nt-pRB67-Δ1 lines, Fig. 2). To provide additional evidence for the distance being the critical determinant for editing-site selection, we have attempted to restore editing in these mutants by the introduction of a compensatory point mutation that creates a cytidine residue in the “correct” distance from the upstream cis-element. Construction of this double mutant was accomplished by changing the T nucleotide, which was in the potentially editable position in vector pRB67-Δ1 (Fig. 1), into a C. The original sequence context (i.e., the 3′ neighboring nucleotide known to influence the efficiency of the editing reaction) was maintained partially by an additional C-to-T change immediately downstream, which restores the 3′ thymidine flanking editing site V in the wild-type sequence (Fig. 1). The resulting transformation vector was termed pRB73-Δ1/cm (Fig. 1). If the distance from the upstream sequence element determines the editing-site selection, then the prediction is that editing now should take place at the newly created cytidine downstream of editing site V (Figs. 1 and 4B).
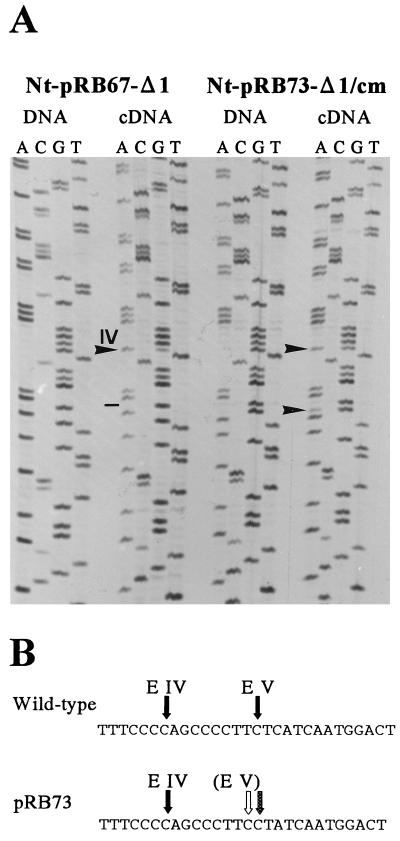
Figure 4.
Restoration of site V editing in the single-nucleotide-deletion mutant (construct pRB67-Δ1) by introduction of a compensatory point mutation, creating an editable C the correct distance from the upstream cis-element (construct pRB73-Δ1/cm). (A) Sequence analysis to test for transgene mRNA editing in the Nt-pRB73-Δ1/cm transplastomic lines. For comparison, editing in the Nt-pRB67-Δ1 lines also is shown. PCR and sequencing primers are as in Fig. 3. Arrowheads point to the editing positions in the cDNA lanes (G in DNA; A in cDNA), a dash indicates lack of editing, and roman numerals mark editing site IV. Note the lack of editing at site V in Nt-pRB67-Δ1 (dash) and restoration of editing after conversion of the downstream nucleotide into a cytidine in Nt-pRB73-Δ1/cm (arrowhead). (B) Restoration of editing in the Nt-pRB73-Δ1/cm transplastomic lines. The single-nucleotide deletion in the spacer, abolishing editing at site V in Nt-pRB67-Δ1 (solid arrow in the wild-type sequence; open arrow in the pRB73-Δ1/cm sequence), is compensated by mutational creation of an “in-phase” cytidine immediately 3′ of editing site V (stippled arrow).
Analysis of the transgene-derived cDNA population from the generated Nt-pRB73-Δ1/cm transplastomic tobacco lines revealed that, as predicted, the introduced compensatory point mutation restores RNA editing in the single-nucleotide deletion mutant (Fig. 4). The editing efficiency (calculated to be approximately 35%) is lower compared with that at site V in the pRB59 control lines (75%). This most probably is due to the presence of a different neighboring nucleotide 5′ of the editing site (T in the wild type and C in the Nt-pRB73-Δ1/cm lines, Fig. 4B). This 5′ neighboring position was shown earlier to exert a significant influence on the efficiency of the editing reaction (12–14).
DISCUSSION
Chloroplast transformation, though being a laborious and time-consuming technology, is currently the method of choice for the study of RNA editing in higher-plant plastids. Earlier studies attempted to define the cis-acting sequence requirements for RNA-editing-site selection and to define minimum substrates for the plastid editing machinery (12, 13). Using two well characterized ndhB editing sites from tobacco, the scope of this study was to test whether the distance of plastid editing sites from an upstream cis-element could be a determinant for editing-site selection. This could explain how the editing apparatus selectively recognizes the editing site and distinguishes between the cytidine to be edited and other cytidines in the immediate neighborhood.
We report here that the recognition of the two ndhB RNA-editing sites is critically dependent on their distance from an upstream essential cis-acting element. Apparently, only those cytidine residues that are the correct distance from this upstream element can be recognized by the editing apparatus. Small changes of this distance can abolish editing completely, indicating that this distance is “measured” by the editing apparatus with high accuracy. We have shown that heterologous cytidine residues can undergo editing when placed the correct distance downstream of the cis-element, suggesting that this distance is a major determinant for selection of the correct cytidine for modification by the editing machinery.
C-to-U editing at a single position in the mammalian apolipoprotein B (apoB) mRNA was demonstrated to involve an essential RNA sequence element in close proximity to the editing site. This cis-acting element was termed the “mooring sequence” and is believed to mediate both substrate recognition and editosome assembly (for review, see, e.g., ref. 24). In this system, the editosome will edit any cytidine that is located in a 3- to 5-nt distance 5′ from the mooring sequence. In addition to the much higher number of editing sites the editing machineries in plant organelles have to deal with, plastid editing—at least in the case of the ndhB sites examined here—differs in two aspects from mammalian apoB editing. First, the mooring sequence-like, essential cis-acting element resides upstream of the editing site for plastid ndhB editing but downstream of the editing site in the case of apoB editing. Second, whereas there is a larger window for the distance of the editing site from the mooring sequence in apoB editing, this distance seems to be more precisely defined in plastid RNA editing. At present, we can only speculate about a possible evolutionary relationship between mammalian C-to-U editing and the editing systems in plant organelles. Clarification of this point would require a thorough comparison of the factors involved in editing in both systems. However, whereas the editing enzyme for apoB editing meanwhile is well characterized, the trans-factors involved in plant organellar RNA editing still await their molecular identification.
There is now compelling evidence for the participation of at least two factors in the editing reactions in plastids: an essential cis-acting element at the mRNA level (12, 13) and a site-specific trans-acting factor of unknown molecular identity (11, 25). By analogy to apoB editing, the essential upstream sequence element could serve as mooring sequence, allowing for binding of the editing apparatus to its RNA substrate mediated by the site-specific trans-factor. For ndhB editing sites IV and V, this model implies the existence of two specificity factors (one site IV-specific and one site V-specific) accounting for the “measuring” of two distinct distances from the essential upstream cis-element. This upstream sequence element harbors either a single mooring sequence employed by both specificity factors or two distinct, but largely overlapping mooring sequences (evidenced by deletion of this region, which abolishes editing at both sites; ref. 12). The existence of separate specificity factors for sites IV and V is also in agreement with the earlier finding that the two sites are edited independently (14). The data presented here support the idea that plastid editing sites are recognized specifically by a sophisticated interplay of a cis-acting element and a site-specific trans-acting factor. In addition to these qualitative determinants, other factors are known to influence the efficiency of the editing reaction in a quantitative fashion: (i) the identity of the nucleotides immediately adjacent to the editing site (13, 14) and (ii) upstream as well as downstream sequence elements outside the minimum sequence context (12, 13) that have not yet been characterized in detail.
It remains to be determined whether the presence of an upstream cis-element and the distance of the editing site from it are also the major determinants for the recognition of other plastid or even plant mitochondrial editing sites. Given the laborious and time-consuming procedures involved in the generation of homoplasmic plants with transgenic chloroplasts, a major obstacle in this respect is posed by the lack of efficient in vitro systems for plant organellar RNA editing (26, 27). Therefore, the development of faithful in vitro assays for plant RNA editing represents one of the major challenges for the future.
Acknowledgments
We thank Drs. Pal Maliga and Zora Svab (Rutgers University, Piscataway, NJ) for providing the plastid aadA gene. We thank Dr. Jörg Kudla (University of Ulm, Germany) for helpful suggestions for the preparation of this manuscript. This research was supported by grants from the Deutsche Forschungsgemeinschaft to R.B. (Bo 1482/3–2).
ABBREVIATION
- cm
compensatory mutation
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Smith H C, Gott J M, Hanson M R. RNA. 1997;3:1105–1123. [PMC free article] [PubMed] [Google Scholar]
- 2.Covello P S, Gray M W. Nature (London) 1989;341:662–666. doi: 10.1038/341662a0. [DOI] [PubMed] [Google Scholar]
- 3.Gualberto J M, Lamattina L, Bonnard G, Weil J-H, Grienenberger J M. Nature (London) 1989;341:660–662. doi: 10.1038/341660a0. [DOI] [PubMed] [Google Scholar]
- 4.Hiesel R, Wissinger B, Schuster W, Brennicke A. Science. 1989;246:1632–1634. doi: 10.1126/science.2480644. [DOI] [PubMed] [Google Scholar]
- 5.Hoch B, Maier R M, Appel K, Igloi G L, Kössel H. Nature (London) 1991;353:178–180. doi: 10.1038/353178a0. [DOI] [PubMed] [Google Scholar]
- 6.Kudla J, Igloi G L, Metzlaff M, Hagemann R, Kössel H. EMBO J. 1992;11:1099–1103. doi: 10.1002/j.1460-2075.1992.tb05149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maier R M, Zeltz P, Kössel H, Bonnard G, Gualberto J M, Grienenberger J M. Plant Mol Biol. 1996;32:343–365. doi: 10.1007/BF00039390. [DOI] [PubMed] [Google Scholar]
- 8.Benne R, Van den Burg J, Brakenhoff J P J, Sloof P, Van Boom J H, Tromp M C. Cell. 1986;46:819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- 9.Kable M L, Heidmann S, Stuart K D. Trends Biochem Sci. 1997;22:162–166. doi: 10.1016/s0968-0004(97)01041-4. [DOI] [PubMed] [Google Scholar]
- 10.Bock R, Kössel H, Maliga P. EMBO J. 1994;13:4623–4628. doi: 10.1002/j.1460-2075.1994.tb06784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaudhuri S, Carrer H, Maliga P. EMBO J. 1995;14:2951–2957. doi: 10.1002/j.1460-2075.1995.tb07295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bock R, Hermann M, Kössel H. EMBO J. 1996;15:5052–5059. [PMC free article] [PubMed] [Google Scholar]
- 13.Chaudhuri S, Maliga P. EMBO J. 1996;15:5958–5964. [PMC free article] [PubMed] [Google Scholar]
- 14.Bock R, Hermann M, Fuchs M. RNA. 1997;3:1194–1200. [PMC free article] [PubMed] [Google Scholar]
- 15.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 16.Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, et al. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Svab Z, Maliga P. Proc Natl Acad Sci USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svab Z, Hajdukiewicz P, Malgia P. Proc Natl Acad Sci USA. 1990;87:8526–8530. doi: 10.1073/pnas.87.21.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doyle J J, Doyle J L. Focus. 1990;12:13–15. [Google Scholar]
- 20.Bachmann B, Lüke W, Hunsmann G. Nucleic Acids Res. 1990;18:1309. doi: 10.1093/nar/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burrows P A, Sazanov L A, Svab Z, Maliga P, Nixon P J. EMBO J. 1998;17:868–876. doi: 10.1093/emboj/17.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maier R M, Neckermann K, Hoch B, Akhmedov N B, Kössel H. Nucleic Acids Res. 1992;20:6189–6194. doi: 10.1093/nar/20.23.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freyer R, López C, Maier R M, Martin M, Sabater B, Kössel H. Plant Mol Biol. 1995;29:679–684. doi: 10.1007/BF00041158. [DOI] [PubMed] [Google Scholar]
- 24.Chan L, Chang B H-J, Nakamuta M, Li W-H, Smith L C. Biochim Biophys Acta. 1997;1345:11–26. doi: 10.1016/s0005-2760(96)00156-7. [DOI] [PubMed] [Google Scholar]
- 25.Bock R, Koop H-U. EMBO J. 1997;16:3282–3288. doi: 10.1093/emboj/16.11.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Araya A, Domec C, Begu D, Litvak S. Proc Natl Acad Sci USA. 1991;89:1040–1044. doi: 10.1073/pnas.89.3.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu W, Schuster W. J Biol Chem. 1995;31:18227–18233. doi: 10.1074/jbc.270.31.18227. [DOI] [PubMed] [Google Scholar]