Abstract
Gene expression is in part controlled by chromatin remodeling factors and the acetylation state of nucleosomal histones. The latter process is regulated by histone acetyltransferases and histone deacetylases (HDACs). Previously, three human and five yeast HDAC enzymes had been identified. These can be categorized into two classes: the first class represented by yeast Rpd3-like proteins and the second by yeast Hda1-like proteins. Human HDAC1, HDAC2, and HDAC3 proteins are members of the first class, whereas no class II human HDAC proteins had been identified. The amino acid sequence of Hda1p was used to search the GenBank/expressed sequence tag databases to identify partial sequences from three putative class II human HDAC proteins. The corresponding full-length cDNAs were cloned and defined as HDAC4, HDAC5, and HDAC6. These proteins possess certain features present in the conserved catalytic domains of class I human HDACs, but also contain additional sequence domains. Interestingly, HDAC6 contains an internal duplication of two catalytic domains, which appear to function independently of each other. These class II HDAC proteins have differential mRNA expression in human tissues and possess in vitro HDAC activity that is inhibited by trichostatin A. Coimmunoprecipitation experiments indicate that these HDAC proteins are not components of the previously identified HDAC1 and HDAC2 NRD and mSin3A complexes. However, HDAC4 and HDAC5 associate with HDAC3 in vivo. This finding suggests that the human class II HDAC enzymes may function in cellular processes distinct from those of HDAC1 and HDAC2.
DNA is packaged in eukaryotic cells in the form of nucleosomal arrays. Each nucleosome core consists of 145 bp of DNA wound around an octamer of H2A, H2B, H3, and H4 histone proteins. These nucleosome cores then are packaged into higher-order structures with additional factors to form chromatin (1).
The incorporation of DNA into chromatin creates a repressive environment that has been implicated in transcriptional silencing. Two cellular processes serve to alter chromatin structure (2). Chromatin remodeling factors such as SWI/SNF, RSC, NURF, and NRD (reviewed in refs. 3–6) have been shown to increase the accessibility of the DNA, presumably by modifications of the nucleosomal structure. A second cellular mechanism involves alterations of the acetylation state of nucleosomal histones. Hypoacetylated chromatin often is associated with silent genes, whereas hyperacetylation is correlated with actively transcribed genes. However, this rule is not absolute. Acetylation of K12 on histone H4 is observed in silent heterochromatin regions in Drosophila and yeast (reviewed in ref. 7). Furthermore, there is increasing evidence for regulation of nonhistone proteins by acetylation, which may function in activation as well as repression of transcription (8–10). The acetylation state of histones and perhaps nonhistone proteins is regulated by a dynamic interaction of histone acetyltransferase and histone deacetylase (HDAC) enzymes.
Previously, three human HDACs (11–14) and five yeast HDACs (15, 16) had been identified, and several of these were biochemically characterized. These HDACs, together with the prokaryotic enzymes acetylspermine deacetylase (ASD) and acetoin utilization protein (acuC), comprise a deacetylase superfamily. In yeast, members of this superfamily can be subdivided into two classes based on size and sequence considerations, as well as the observation that Rpd3p and Hda1p function in biochemically distinct complexes. The first class (I) consists of Rpd3p, Hos1p, and Hos2p, and the second class contains Hda1p. Similarly in mammals, HDAC1, HDAC2, and HDAC3 conform to class I criteria, whereas no human class II HDAC proteins have been identified previously. The National Center for Biotechnology Information (NCBI) database blast program was used to search human expressed sequence tags (ESTs) for amino acid sequence similarity to yeast Hda1p. This search revealed a number of ESTs that appeared to derive from human class II HDACs. A combination of cDNA library screening and PCR-based strategies facilitated the cloning of three human class II HDAC enzymes, HDAC4, HDAC5, and HDAC6. The expression levels of these HDAC proteins in human tissues are more variable than the human class I HDACs. Class II HDACs deacetylate all four core histones in vitro and are sensitive to the potent HDAC inhibitor trichostatin A. Based on coimmunoprecipitation experiments, these HDACs are not associated with the previously identified NRD and mSin3A complexes that contain HDAC1 and HDAC2, and therefore are likely to be components of distinct complexes that perform alternate functions.
MATERIALS AND METHODS
Cloning of HDAC4, HDAC5, and HDAC6.
The amino acid sequence of yeast Hda1p was used in a tblastn search of the NCBI databases to identify human homologs of Hda1p. A cDNA clone for HDAC4 was obtained from the Kazusa DNA Research Institute, Kisarazu, Japan (gene name KIAA0288, GenBank accession no. 3024889). The full-length clone was 8,459 bp, with a predicted ORF of 2,903 bp. A comparison of the HDAC4 clone with the HDAC5 sequence, however, revealed that the putative 5′ untranslated region of HDAC4 was highly homologous to the N-terminal coding sequence of HDAC5. A truncation in the HDAC4 ORF seems to have been caused by a C-to-T point mutation in the putative 5′ untranslated region of the HDAC4 clone, resulting in the conversion of a glutamate codon to a stop codon. This hypothesis was confirmed by obtaining the remaining N-terminal 352 bp by PCR from a 5′Stretch cDNA Leukemia Library (CLONTECH) and sequencing the product. A C-terminal FLAG epitope tag was incorporated into the complete HDAC4 ORF of 3,255 bp, which then was inserted into the NotI–EcoRI sites of a mammalian expression vector (pBJ5).
An EST (GenBank accession no. R64669) homologous to yHda1p was identified and obtained from the I.M.A.G.E. Consortium (Lawrence Livermore National Laboratory, Berkeley, CA). This sequence was used to generate a random primed probe (Boehringer Mannheim) to screen a Lambda ZAP II Jurkat cDNA library (Stratagene). A 2.3-kb cDNA clone was identified that contained a partial ORF of HDAC5. A blastn search of the NCBI database facilitated the assembly of the full-length cDNA sequence as a combination of a second clone from the Kazusa DNA Research Institute (gene name KIAA0600, GenBank accession no. 3043724) and the cDNA clones containing the 11-js mRNA sequence (GenBank accession no. AF039241), kindly provided by Jeff Swensen (University of Utah). An ORF of 3,369 bp was identified and assembled into a C-terminal FLAG construct by subcloning into the NotI–XhoI sites of a pBJ5 vector.
A second EST homologous to yHDA1p was identified and used to screen the Lambda ZAP II Jurkat cDNA library. The sequence of the 2.5-kb clone produced was used in a blastn search to reveal the full-length sequence of HDAC6 in the jm-21 mRNA sequence (GenBank accession no. AJ011972). This information was used to obtain the remaining 1.5 kb by nested PCR from a U957 cDNA library, kindly provided by Don Ayer (University of Utah), to produce the full-length ORF of 3,648 bp. A C-terminal FLAG epitope tag was incorporated into this clone, which was inserted into the NotI–SpeI sites of pBJ5.
Northern Blot Analysis.
Multiple human tissue Northern blots were obtained from CLONTECH. Probes were generated, and blots were stripped by using Strip-EZ DNA probe synthesis and removal kit (Ambion). Prehybridization and hybridization was carried out according to the manufacturer’s instructions by using ExpressHyb solution (CLONTECH). For HDAC4 expression analysis, a 12-lane tissue blot was probed with the 895-bp SalI–SacI fragment of the HDAC4 gene. For HDAC5 expression analysis, an eight-lane tissue blot was probed with the 993-bp SacI–SacII fragment of the HDAC5 gene. This blot was stripped and reprobed for HDAC6 expression by using the 667-bp SphI–AvrII fragment of the HDAC6 gene. Blots were stripped and reprobed with β-actin cDNA as a control (CLONTECH).
Antibodies, Immunoprecipitation, and Western Blotting.
Antibodies against HDAC1 (11), HDAC3 (17), and RbAp48 (17) have been described. Antibodies against mSin3A were kindly provided by Don Ayer (18), antibodies to the PHD domain of CHD4 were provided by Weidong Wang (National Institute on Aging) (6), and antibodies to the N-terminal domain of metastasis-associated factors (MTA) were provided by Yasushi Toh (Kyushi University, Fukuoka, Japan) (19, 20).
Forty-eight hours after transfection, cells were lysed in JLB (50 mM Tris⋅HCl, pH 8/150 mM NaCl/10% glycerol/0.5% Triton X-100) containing a complete protease inhibitor cocktail (Boehringer-Mannheim). Lysis proceeded for 10 min at 4°C, after which the cellular debris was pelleted by centrifugation at 14 K for 5 min. Recombinant proteins were immunoprecipitated from the supernatant by incubation with α-FLAG M2 agarose affinity gel (Sigma) for 2 hr at 4°C. For Western blot analysis, the beads were washed three times for 5 min at room temperature with MSWB (50 mM Tris⋅HCl, pH 8/150 mM NaCl/1 mM EDTA/0.1% NP-40), and the proteins were separated by SDS/PAGE. For enzyme activity assays, the beads were washed three times with JLB at 4°C.
HDAC Assays.
[3H]Acetate-incorporated histones were isolated from butyrate-treated HeLa cells by hydroxyapitite chromatography as described (4). Immunoprecipitates were incubated with 1.4 μg (10,000 dpm) histones for 3 hr at 37°C. HDAC activity was determined by scintillation counting of the ethyl-acetate-soluble [3H]acetic acid (11). Inhibition of enzyme activity by trichostatin (TSA) was performed by incubating samples with 300 nM TSA (Wako) for 10 min before addition of the labeled histones.
Histone Isolation and Substrate Specificity Determination.
For deacetylase assays, 6 μg of histones was incubated with immunoprecipitated recombinant enzyme or negative control (RbAp48 transfected) for 3 hr at 37°C in HD buffer (50 mM Tris, pH 8.0/150 mM NaCl/10% glycerol). Reactions were stopped with SDS loading buffer, and proteins were separated by 20% SDS/PAGE.
HDAC6 Mutagenesis.
The H216A and H611A mutations were produced by PCR overlap extension. For each mutant, internal primers to both strands with the corresponding CAC-to-GCC mismatches were synthesized and used to amplify two overlapping fragments containing the mutation in the common region. These fragments then were used as templates in a second PCR with the same flanking external primers. The H216A fragment was ligated into the NotI and DraIII sites of the wild-type HDAC6-pBJ5 construct. The H611A fragment was ligated into the DraIII and BstEII sites of the wild-type HDAC6-pBJ5. The H216/611A double mutant was constructed by ligating the H216A fragment into the NotI and DraIII sites of the HDAC6-H611A-pBJ5 clone. Plasmids were sequenced to ensure the incorporation of the mutations.
Cell Culture and Transfections.
TAg-Jurkat cells were transfected by electroporation with 5 μg of FLAG-epitope-tagged pBJ5 constructs for expression of recombinant proteins. Cells were mock-transfected without DNA or with an untagged RbAp48 construct in pBJ5 as a negative control. Cells were harvested 48 hr posttransfection.
RESULTS
Identification of Three Class II Human HDAC Enzymes.
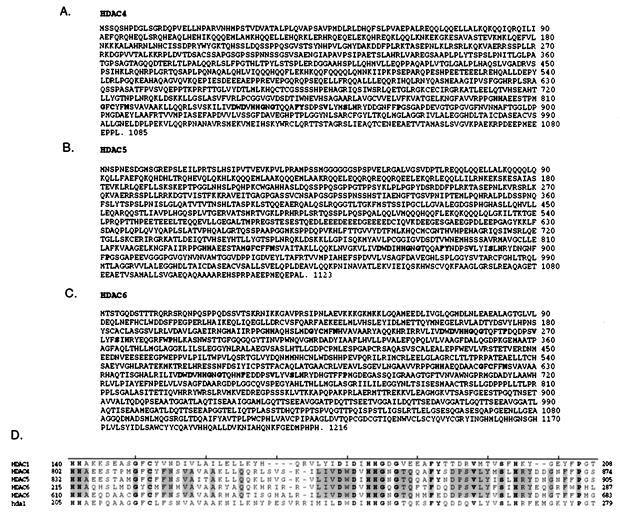
The NCBI database was screened with the yeast Hda1p amino acid sequence to identify human homologs. The complete ORFs of HDAC4, HDAC5, and HDAC6 were constructed as described in Materials and Methods. HDAC4 consists of 1,085 aa, with a putative catalytic region beginning at amino acid 802 (Fig. 1A). HDAC5 is highly homologous to HDAC4 (51% identity, 63% similarity), with 1,123 aa and a catalytic region beginning at amino acid 832 (Fig. 1B). HDAC6 is the largest HDAC protein yet identified in humans, with 1,216 aa. It is also unique in that it consists of an apparent internal dimer containing two catalytic domains, with the first beginning at amino acid 215 and the second at amino acid 610 (Fig. 1C). The two catalytic regions in HDAC6 are highly homologous to each other (47% identity, 64% similarity) and therefore the protein may have arisen evolutionarily from an in-frame gene duplication event. All of the catalytic domains of these three proteins are highly conserved with respect to each other and previously identified HDAC proteins (Fig. 1D). There are 15 invariant residues in this region between HDAC1, HDAC4, HDAC5, HDAC6 proteins and Hda1p, and a total of 37 invariant residues within the four catalytic domains of the three additional HDAC proteins. This level of sequence conservation strongly suggests that these additional proteins have deacetylase activity.
Figure 1.
Predicted amino acid sequences of human class II HDACs. The conserved residues of the catalytic domains are highlighted. (A) HDAC4, (B) HDAC5, and (C) HDAC6 predicted amino acid sequences. Note that there are two putative catalytic domains in HDAC6. (D) Alignment of catalytic domains of yeast HDA1p, human HDAC1, HDAC4, and HDAC5 with both catalytic regions of HDAC6. The residues that are conserved in these HDACs as well as in acuC (B. subtilis, GenBank accession no. 348052) and ASD (M. ramosa, GenBank accession no. 3023317) are in bold type, and those residues that are conserved within the class II human HDAC enzymes are boxed.
Differential Expression of Class II HDACs in Human Tissues.
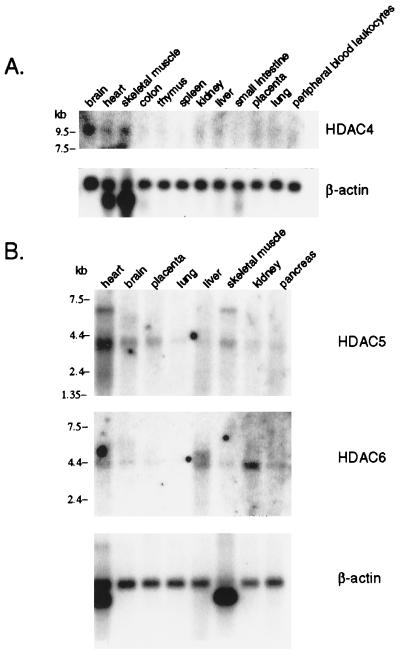
Northern blot analyses indicate differential tissue expression of the human class II HDACs (Fig. 2). HDAC4 is detectable as a 9.6-kb transcript in brain, heart, and skeletal muscle tissues. The signal is very weak, however, which may have been caused by the fact that the transcript is at the upper size limit for mRNA samples in this particular blot. It is possible that HDAC4 is expressed in other tissues, but the quantity of transcript in these samples may be too low to detect. This finding is consistent with the small number of ESTs corresponding to this cDNA present in the database. HDAC5 expression partially overlaps that of HDAC4 and is observed primarily in brain, heart, skeletal muscle, and placental tissues as a 3.7-kb transcript. HDAC6, which is present as a 5-kb transcript, has the highest expression levels in heart, liver, kidney, and pancreas. The differences in tissue expression may reflect a tissue specific function of these enzymes.
Figure 2.
Expression analysis of human class II HDAC family members. Multiple human tissue Northern blots were probed to determine mRNA expression of HDAC4, HDAC5, and HDAC6. Blots were stripped and reprobed with β-actin cDNA to normalize for total mRNA. The position of molecular size markers is indicated on the left.
In Vitro HDAC Activity of Class II HDACs.
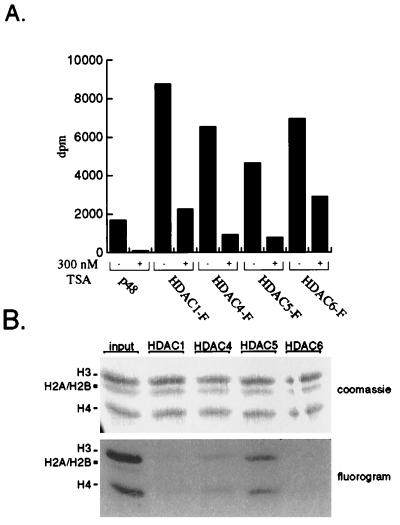
To determine whether HDAC4, HDAC5, and HDAC6 possess HDAC activity, the recombinant proteins were assayed for enzymatic activity in vitro. Epitope-tagged recombinant HDACs 1, 4, 5, and 6 were expressed in Tag-Jurkat cells and immunoprecipitated. The immunoprecipitates were incubated with 3H-acetate-labeled histones, and the subsequent release of 3H-acetate was quantified by scintillation counting. All four HDAC enzymes exhibit deacetylation activity that is at least 2-fold above background levels (Fig. 3A). In each case, activity is greatly reduced by the presence of 300 nM trichostatin, a potent HDAC inhibitor. HDAC1 and HDAC6 possess comparable activity, whereas that of HDAC4 and HDAC5 is somewhat reduced. This reduced activity is most likely caused by lower expression levels for these two recombinant proteins rather than inherently weaker HDAC activity. Furthermore, coimmunoprecipitation experiments demonstrate the association of HDAC4 and HDAC5 with HDAC3. It is possible that the observed HDAC activity is caused by HDAC3. However, sequence analysis suggests that HDAC4 and HDAC5 possess functional catalytic domains, and therefore should contribute to the activity. Therefore, in vitro, all three human class II HDACs can deacetylate histones in a trichostatin-sensitive manner.
Figure 3.
Class II HDAC enzymes deacetylate all four core histones in vitro. Recombinant FLAG-tagged HDACs were immunoprecipitated from transfected Jurkat cell extracts by using α-FLAG antibody (Sigma). Immunopurified enzymes were incubated with radiolabeled core histones as described in Materials and Methods. (A) The HDAC activity was measured by scintillation counting of the released [3H]acetic acid. Where indicated, immunoprecipitates were preincubated with trichostatin A (Wako) before addition of histones. Each assay was performed in duplicate and averaged. (B) Substrate specificity of class II HDACs. Deacetylase reactions were separated by 20% SDS/PAGE and stained with Coomassie (Upper). The gel was treated with EnHance (National Diagnostics), dried, and exposed to film (Lower). The identities of the core histones are indicated on the left. RbAp48 was transfected as a negative control.
Previously, Hda1p was shown to preferentially deacetylate histone H3 in vitro (16). To determine whether HDAC4, HDAC5, and HDAC6 display similar preferences, the immunopurified recombinant proteins were incubated with 3H-acetate-labeled histones, and the deacetylase reactions were separated by SDS/PAGE to identify the different histone isotypes. The gel then was exposed to autoradiography to determine the relative amount of acetylated histones remaining in each case. HDAC1, HDAC4, HDAC5, and HDAC6 deacetylate all four core histones equally well, though again deacetylation by HDAC4 and HDAC5 is incomplete (Fig. 3B).
Independently Active Catalytic Domains of HDAC6.
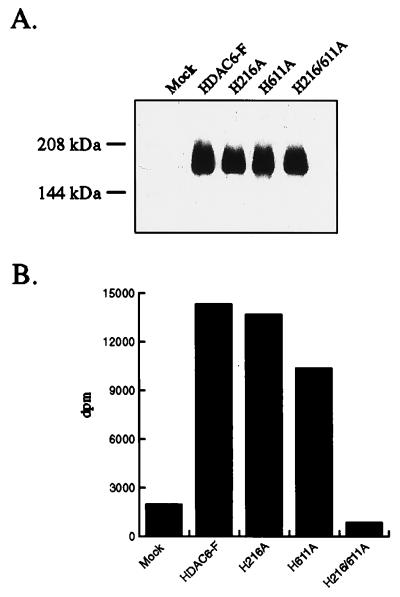
HDAC6 possesses two separate putative catalytic domains. Site-directed mutagenesis was performed to determine whether either domain required the other for catalytic activity. Histidine 141 of HDAC1 was shown previously to be critical for deacetylase activity (18). The corresponding histidine residues in each catalytic domain of HDAC6 were mutated to alanine, to produce the H216A and H611A single mutants and the H216/611A double mutant. The mutant HDAC6 proteins were expressed and assayed for in vitro deacetylation of histones. Mutation of either H216 or H611 to alanine results in a slight reduction of HDAC activity, and simultaneous mutation of both sites abrogates this activity completely (Fig. 4 A and B). Furthermore, a truncation of HDAC6 containing the N-terminal 460 aa, and therefore only the first catalytic domain, is still catalytically active (data not shown). Therefore, both catalytic domains of HDAC6 are fully functional HDACs and contribute independently to the overall activity of the wild-type HDAC6 protein.
Figure 4.
The catalytic domains of HDAC6 function independently. The histidine residues homologous to H141 of HDAC1 in each of the catalytic domains (H216 and H611) were mutated to alanine by PCR overlap extension. The single and double mutants were FLAG-tagged and expressed in Tag-Jurkat cells. The enzymes were immunoprecipitated by using α-FLAG antibodies (Sigma), and expression levels were compared by Western blotting (A). The mutant enzymes then were assayed for HDAC activity as before (B).
Expression and Coimmunoprecipitation of Class II HDACs.
Recombinant, FLAG epitope-tagged proteins were subjected to Western blot analysis to address possible protein–protein interactions. HDAC1 migrates as a band slightly above its expected size of 55 kDa in SDS/PAGE (Fig. 5A). Recombinant HDAC4 and HDAC6 appeared above their theoretical molecular masses of 119 kDa and 131 kDa, respectively. The expression level of HDAC5 is significantly lower than the others, and the protein appears as a doublet, both in the lysate (data not shown) and in the immunoprecipitate (Fig. 5A). This doublet may be the result of posttranslational modifications or partial proteolytic degradation. The high molecular weight diffuse signal apparent in the blot is most likely the result of crossreaction of the secondary mouse antibody with contaminating FLAG antibody used for the immunoprecipitation. This signal is partially masked by the comigration of the recombinant HDAC4, HDAC5, and HDAC6 proteins.
Figure 5.
Class II HDAC enzymes and HDAC1 are in different complexes in vivo. Recombinant FLAG-tagged HDACs were precipitated from transfected Jurkat cell extracts by using α-FLAG antibody (Sigma), separated by SDS/PAGE, and subjected to Western blot analysis. Blots were probed with (A) α-FLAG antibody (Sigma) to determine expression levels and (B) α-CHD4, -mSin3A, -MTA, -HDAC1, -HDAC3, and -Rbp48 antibodies to determine whether these proteins coimmunoprecipitated with the class II HDAC enzymes.
HDAC1 has been shown to be associated with a variety of transcription-related proteins, including the CHD chromodomain proteins, MTA, the corepressor mSin3A, and the histone-binding protein RbAp48. To determine whether HDAC4, HDAC5, and HDAC6 associated with the same proteins in vivo, a series of coimmunoprecipitation experiments was performed. Immunoprecipitates were probed with α-CHD4, α-mSin3A, α-MTA, α-RbAp48, α-HDAC1, and α-HDAC3 antibodies (Fig. 5B). The HDAC1 sample contains bands corresponding to all proteins with the exception of HDAC3, as anticipated. There is a band in the mSin3A blot of the HDAC4 immunoprecipitate, which appears at a lower molecular weight than expected for mSin3A. The nature of this band is unclear, because it does not correspond to any of the previously observed forms of mSin3A. HDAC4 coimmunoprecipitates with Rbp48 and HDAC3, while none of the other proteins were apparent. HDAC5 associates only with HDAC3, though it is possible that the expression levels were too low to detect other associated factors. HDAC6 does not appear to interact with any of these proteins, despite robust expression, nor did HDAC1 or HDAC2 (data not shown) coimmunoprecipitate with the class II HDACs. This analysis suggests that these class II HDAC proteins are biochemically distinct from HDAC1 in vivo.
DISCUSSION
Analysis of HDAC activity in Saccharomyces cerevisiae revealed the presence of two HDAC complexes, containing either Hda1p or Rpd3p (15). These two HDAC proteins are quite different in molecular mass and primary structure, though both contain conserved deacetylase catalytic domains. The three previously identified human HDAC proteins display greater homology to Rpd3p than to Hda1p. This observation suggested that mammalian cells might contain an uncharacterized class of biochemically distinct Hda1p-like proteins. A search of the NCBI database identified three putative class II human HDAC proteins. Biochemical characterization of the recombinant enzymes confirms that these proteins possess HDAC activity and are not associated with previously identified HDAC1 or HDAC2 complexes. Furthermore, the evolutionary conservation of these enzymes suggests that they possess critical biological functions.
Origin of the Class II HDACs.
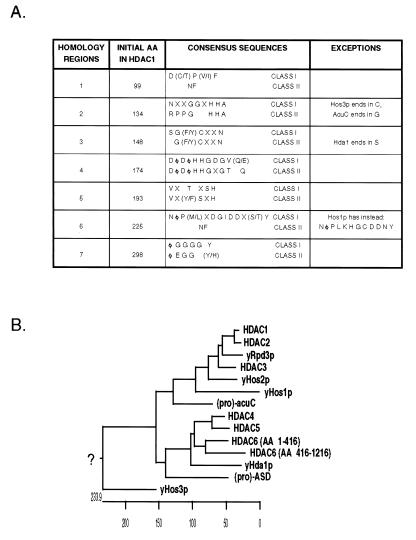
Previously, three human HDACs (11–14) and five yeast HDACs (15, 16) had been identified. These can be divided into two classes based on primary structure and biochemical characteristics. The first class contains the human HDAC1, HDAC2, HDAC3, and yeast Rpd3p, Hos1p, and Hos2p proteins, which range in size from 49 to 55 kDa in molecular mass. The second class contains yeast Hda1p and human HDAC4, HDAC5, and HDAC6. These proteins are significantly larger in size, ranging from 80 to 131 kDa. There are also seven clearly conserved sequence motifs, five of which are present in both groups, albeit with features distinct between the two classes (Fig. 6A). However, the homology between yHda1p and the human class II HDACs is limited to the catalytic regions (Fig. 1D) and does not extend to the N-terminal domains. The role of the extensive amino-terminal domains present in HDAC4 and HDAC5 is currently unknown. Although the N-terminal 100 aa of HDAC4 are not required for in vitro HDAC activity (data not shown), these regions may contribute to protein–protein or enzyme-substrate interactions. One exception to the above classification scheme is yeast Hos3p, which contains features of both families and cannot be classified into either category.
Figure 6.
Sequence analysis suggests that HDAC enzymes have diverged into two classes. (A) Alignment of human HDAC enzymes 1–6 with yeast Rpd3p, Hos1p, Hos2p, Hos3p, M. ramosa ASD, and B. subtilis acuC reveals the presence of seven conserved regions, whose consensus sequences differ between the two classes. Amino acids are represented by single letter codes; X represents any amino acid while Φ indicates a hydrophobic residue. NF, not found. (B) A phylogenetic analysis suggests that the HDAC enzymes diverged from a common prokaryotic ancestor to form two classes of HDAC proteins. Proteins from three different phyla were examined. Prokaryotic proteins are preceded by (pro), yeast proteins are preceded by y, and human proteins are capitalized. Note that yHos3p does not correlate well with either HDAC class.
The class II HDACs have been well conserved in higher eukaryotes. In Caenorhabditis elegans, an HDAC4 and HDAC5 homolog has been identified (C10E2.3, GenBank accession no. AF026202), as well as an HDAC6 homolog (F41H10.6, GenBank accession no. U61954), which contains the analogous two catalytic domains. Similarly, mouse HDAC5 (mHDA1) and HDAC6 (mHDA2) homologs recently have been cloned, though the mHDA1 sequence is shorter than the human HDAC5, which is possibly the result of a nonsense mutation in the 5′ region of the ORF (21). The evolutionary conservation of these proteins from nematode to human suggests that these enzymes perform critical functions.
The divergence of the two classes of HDACs appears to have occurred relatively early in evolution. A comparison of the human and yeast HDAC sequences with those of the eubacterium Bacillus subtilis acuC and the proteobacterium Mycoplana ramosa ASD reveal that these proteins contain features present primarily in class I and class II HDACs, respectively. Both enzymes are postulated to catalyze deacetylation of their substrates (22–25), further strengthening the evolutionary and functional link between these proteins and HDACs. This finding suggests that ASD and acuC may have diverged from a common ancestor and subsequently evolved to give rise to the two classes of eukaryotic HDACs that comprise the HDAC superfamily (Fig. 6B).
Biological Role of the Class II HDACs.
All three class II HDACs can deacetylate the four core histones in vitro. However, because HDAC4 and HDAC5 associate with HDAC3 in vivo, it is unclear whether both enzymes contribute to the deacetylase activity, though sequence analysis suggests that HDAC4 and HDAC5 have active catalytic domains. Furthermore, this assay does not confirm that histones are the substrates for these enzymes in vivo. It is possible that these HDAC proteins are responsible for deacetylation of nonhistone proteins, such as p53 or HMG I(Y), or small-molecule acetylpolyamines.
Some understanding of the biological role of the HDACs has been gained by the study of their associated factors. Previously, HDAC1 and HDAC2 have been shown to associate with two specific complexes. The NRD complex (4–6) contains a human SWI/SNF homolog, CHD4, which functions in ATP-dependent chromatin remodeling. Other members of this complex include MTA-1 and its homologs (19, 20). A second HDAC1/2 complex contains the corepressor mSin3A and has been shown to mediate repression of a variety of DNA-binding transcription factors (26). Interestingly, HDAC3 is not detected in either of these complexes, though in some cell types it does associate with HDAC1 and HDAC2 (C.A.H. and S.L.S. unpublished observations). These data suggest that the HDAC enzymes may perform specific functions by forming homo- and heterocomplexes with each other.
Coimmunoprecipitation data suggest that the human class II HDAC enzymes do not associate with the NRD or mSin3A complexes, nor do they interact with HDAC1 or HDAC2 (data not shown) in this cell line. However, HDAC3 does associate with HDAC4 and HDAC5. It appears that HDAC1 and HDAC2 preferentially associated with each other, whereas HDAC4 and HDAC5 complex with HDAC3. HDAC6 does not seem to associate with any class I HDAC protein. However, HDAC6 possesses an internal dimer of two functional catalytic domains, and therefore may not have need for an additional HDAC protein partner. This evidence for regulation of HDAC function by HDAC-protein pairing is further strengthened by the observation that the ASD protein exists as a homodimer (22), which suggests that this process is conserved throughout evolution.
RbAp48 is a component of both the NRD and mSin3A complexes, as well as a variety of histone metabolizing complexes, including histone acetyltransferase and chromatin assembly complexes. Furthermore, a direct interaction between RbAp48 and histone H4 and H2A has been demonstrated (27, 28). Coimmunoprecipitation revealed an association between RbAp48 and HDAC4. An interaction between RbAp48 and HDAC5 may not have been detected because of the low expression of this protein. However, RbAp48 does not associate with HDAC6, which may be the result of disruption of the protein–protein interaction by the antibody used in the immunoprecipitation. Alternatively, because RbAp48 frequently is found in complexes that contain histones, its absence suggests that HDAC6 may not interact with histones in vivo, but rather may deacetylate other substrates.
The identification of three additional human class II HDAC enzymes provides the means to explore the regulation and function of these HDAC enzymes. Our results also suggest that the function of HDAC complexes is controlled in part by altering the combination of HDAC proteins and their associated cofactors. Furthermore, the class II HDAC enzymes may deacetylate nonhistone substrates and therefore be involved in pathways distinct from the known HDAC1 and HDAC2 mSin3A-corepressor and NRD complexes.
Acknowledgments
We thank Jeff Tong, Don Ayer, Gavin Schniztler, and Brian Dynlacht for critical reading of the manuscript. This work was supported by National Institute of General Medical Sciences Grant GM38627 (to S.L.S.). C.A.H is supported by a National Institutes of Health predoctoral training grant, and C.M.G. is supported by a National Science Foundation predoctoral training grant. S.L.S. is an Investigator at the Howard Hughes Medical Institute.
ABBREVIATIONS
- HDAC
histone deacetylase
- EST
expressed sequence tag
- ASD
acetylspermine deacetylase
- acuC
acetoin utilization protein
- NCBI
National Center for Biotechnology Information
- MTA
metastasis-associated factors
Footnotes
References
- 1.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Nature (London) 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Workman J L, Kingston R E. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 3.Varga-Weisz P D, Becker P B. Curr Opin Cell Biol. 1998;10:346–353. doi: 10.1016/s0955-0674(98)80010-0. [DOI] [PubMed] [Google Scholar]
- 4.Tong J K, Hassig C A, Schnitzler G R, Kingston R E, Schreiber S L. Nature (London) 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, LeRoy G, Seelig H P, Lane W S, Reinberg D. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 6.Xue Y, Wong J, Moreno G T, Young M K, Cote J, Wang W. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 7.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 8.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 9.Gu W, Roeder R G. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 10.Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Mol Cell. 1998;2:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 11.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 12.Yang W M, Yao Y L, Sun J M, Davie J R, Seto E. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 13.Emiliani S, Fischle W, Van Lint C, Al-Abed Y, Verdin E. Proc Natl Acad Sci USA. 1998;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dangond F, Hafler D A, Tong J K, Randall J, Kojima R, Utku N, Gullans S R. Biochem Biophys Res Commun. 1998;242:648–652. doi: 10.1006/bbrc.1997.8033. [DOI] [PubMed] [Google Scholar]
- 15.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carmen A A, Rundlett S E, Grunstein M. J Biol Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- 17.Hassig C A, Tong J K, Fleischer T C, Owa T, Grable P G, Ayer D E, Schreiber S L. Proc Natl Acad Sci USA. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 19.Toh Y, Pencil S D, Nicolson G L. J Biol Chem. 1994;269:22958–22963. [PubMed] [Google Scholar]
- 20.Toh Y, Oki E, Oda S, Tokunaga E, Ohno S, Maehara Y, Nicolson G L, Sugimachi K. Int J Cancer. 1997;74:459–463. doi: 10.1002/(sici)1097-0215(19970822)74:4<459::aid-ijc18>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 21.Verdel A, Khochbin S. J Biol Chem. 1999;274:2440–2445. doi: 10.1074/jbc.274.4.2440. [DOI] [PubMed] [Google Scholar]
- 22.Fujishiro K, Ando M, Uwajima T. Biochem Biophys Res Commun. 1988;157:1169–1174. doi: 10.1016/s0006-291x(88)80997-5. [DOI] [PubMed] [Google Scholar]
- 23.Sakurada K, Ohta T, Fujishiro K, Hasegawa M, Aisaka K. J Bacteriol. 1996;178:5781–5786. doi: 10.1128/jb.178.19.5781-5786.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez J, Fortinagel P. Biochim Biophys Acta. 1972;279:554–560. doi: 10.1016/0304-4165(72)90177-8. [DOI] [PubMed] [Google Scholar]
- 25.Grundy F J, Waters D A, Takova T Y, Henkin T M. Mol Microbiol. 1993;10:259–271. doi: 10.1111/j.1365-2958.1993.tb01952.x. [DOI] [PubMed] [Google Scholar]
- 26.Pazin M J, Kadonaga J T. Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 27.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 28.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]