Abstract
Combined inhibition of HIV-1 reverse transcriptase and protease has significantly improved the treatment of AIDS. Nevertheless, resistance to these drugs occurs rapidly because of viral mutations, emphasizing the importance of identifying novel retroviral targets to develop new drug combinations. The critical role played by the nucleocapsid protein NCp7 of HIV-1 at different steps of the retrovirus life cycle makes it an attractive target for the development of new antiviral agents. NCp7 contains two highly conserved zinc fingers and is characterized by a three-dimensional structure that cannot be modified without a complete loss of infectivity of mutated viruses. Based on these structural data, we report that RB 2121, a cyclic peptide designed to mimic several essential biological determinants of NCp7, displays antiviral activity by inhibiting HIV-1 replication in CEM-4 cells infected by HIV-1. In vitro, RB 2121 does not interfere with HIV-1 cell entry and viral enzymes but is able to inhibit the annealing activities of NCp7 by recognizing nucleic acids. Analysis of proviral DNA synthesis by means of PCR has shown that RB 2121 acts at an early step of the retrovirus life cycle by inducing a dose-dependent reduction in transcribed DNA levels through inhibition of NCp7–reverse transcriptase interaction. Because of its original mechanism of action, RB 2121 provides an interesting lead for the rational development of new anti-HIV-1 agents that could be associated advantageously with enzyme inhibitors to counteract rapid virus mutations and resistance problems observed in tritherapies.
The association of HIV-1 reverse transcriptase (RT) and protease inhibitors led to an improvement in the treatment of AIDS. However, emergence of resistance to these polytherapies remains a critical problem. This problem points out the need to develop combination therapies associating new antiviral agents interfering with nonenzymatic and highly conserved viral proteins with the aim to discard the cross-resistance observed with the currently used drugs (1–4).
An attractive target could be the nucleocapsid protein NCp7 (Fig. 1) of HIV-1 found in tight association with the dimeric RNA genome in the retroviral core (5). NCp7 is a highly basic protein characterized by the presence of two well conserved and structured zinc-finger domains of the CX2CX4HX4C type (6). NCp7 plays a major role in viral infectivity during the early and late stages of the retroviral life cycle (7). In vivo, NCp7 is required for the protection of genome against cellular nucleases and, as a component of Gag polyprotein precursor, is involved in genomic RNA packaging and morphogenesis of virus particles (8–10). In vitro, NCp7 participates in the dimerization of viral genome (5) and in the annealing of the tRNA3Lys primer at the initiation site of reverse transcription (11–12), and enhances the global processivity of the RT by activating strand transfers and by improving the RNase H activity of RT (13–19).
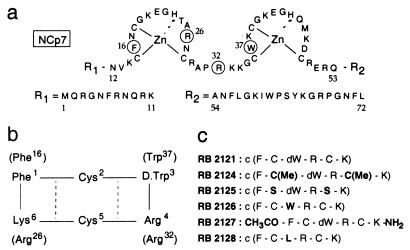
Figure 1.
Structures of NCp7 and RB 2121. (a) Sequence of HIV-1 Lai NCp7. The biological determinants (Phe-16, Arg-26, Arg-32, and Trp-37) used for designing NCp7 mimics are numbered. (b) Sequence of the cyclic hexapeptide mimic of NCp7, RB 2121. NCp7-targeted residues are in brackets. Hydrogen bonds evidenced by NMR are indicated by dotted lines. (c) RB 2121 and its analogues.
The NCp7 structure shows that the zinc fingers are in spatial proximity, whereas the N- and C-terminal sequences remain flexible (20–22). These structural characteristics seem to be crucial for NCp7 activities, because all mutations disrupting the conformation of the finger domain led to a complete loss of infectivity (8, 21, 23–25). The mutants are characterized by limited defects in RNA packaging and virus morphology, suggesting that other steps involving nucleic acids recognition and provirus DNA synthesis could be affected by modification of the NCp7 structure (18, 23, 26). The importance of the central fingered domain has been shown by the nuclear magnetic resonance (NMR) structure resolution of the complex between fragment (12–53)NCp7, encompassing the zinc-finger domain, and the single-stranded pentanucleotide d(ACGCC), which belongs to the ψ-packaging domain of viral RNA (27). The complex is stabilized by hydrophobic contacts of residues belonging to both zinc fingers and ionic interactions of Arg-26 and Arg-32 with the ribose phosphate backbone. Moreover, the Trp-37 residue was found stacked on a guanine ring, and, recently, this recognition process directing the protein–nucleic acid interaction also was found in the complex between NCp7 and the SL3 RNA domain (28). The biological relevance of this interaction is supported by the replacement of Trp-37 by Leu, which inhibits the annealing activities of NCp7, a critical step in RT-dependent synthesis of the provirus (29). Interestingly, no large differences were found in the structure of the free and complexed NCp7 (20, 27, 28).
In attempting to account for these results, we tried to inhibit NCp7 activities by designing small molecules that mimic a part of the three-dimensional structure of NCp7 and that should compete with NCp7 for the recognition of its targets. Therefore, a simplified scaffold deduced from the NCp7 structure was developed with the main biological determinants of the protein introduced on a cyclic peptide, RB 2121, enabling them to adopt a relative spatial orientation close to that found in NCp7.
RB 2121 showed dose-dependent antiviral properties by inhibiting HIV-1 Lai replication in a CEM-4 assay. By monitoring PCR-amplified early and late reverse transcription sequences, we establish that RB 2121 acts at an early step of the retroviral life cycle by inducing a dose-dependent reduction in transcribed DNA levels. This reduction seems to be caused by impairing the formation of a functional complex involving RT, NCp7, and nucleic acid (29, 30). RB 2121 behaves, therefore, as an interesting lead for the rational design of new antiretroviral agents with an original mechanism of action, completely different from that of NCp7 zinc ejectors (31, 32), RT, and protease blocking agents.
MATERIALS AND METHODS
Materials.
The peptides were synthesized by solid-phase methods by using the fluorenylmethoxycarbonyl strategy (12). Cyclization was carried out at high dilution (2 × 10−3 M) with benzotriazolyloxy-tris(dimethylamino)phosphonium hexafluorophosphate. Final deprotection was achieved with trifluoroacetic acid. Purification by high-pressure liquid chromatography yielded peptides that were >98% pure as determined by mass spectroscopy and 1H NMR spectroscopy (C.Z.D., B.P.R., and M.C.F.-Z., unpublished work). Cell lines CEM-4, C8166, and U1 were obtained from the National Institute for Biological Standards and Control (Hertfordshire, U.K.). The Lai virus strain was from the Institut Pasteur (Paris). HIV-1 RNA(77–257) and [33P]tRNA3Lys (a generous gift from J. L. Darlix, Ecole Normale Supérieure, Lyon, France) were purified as described (29).
NMR Spectroscopy and Calculations.
The conformational properties of RB 2121 in DMSO/H2O (70:30, vol/vol) solution, known to mimic the intracellular medium, were determined by 1H two-dimensional NMR techniques (Bruker AMX 600, Billerica, MA) by using totally correlated spectroscopy, nuclear Overhauser effect (NOE) spectroscopy (τm = 80 ms), and rotating-frame Overhauser effect spectroscopy experiments. The NH chemical-shift temperature dependencies were measured from 293 K to 333 K. A restrained molecular dynamics simulation was performed in vacuo on RB 2121 by using the NOEs as interproton distance restraints.
The CFF 91 force field in the Insight/Discover package (Biosym Technologies, Orsay, France) with a dielectric constant of 4 was used, and 100 structures were generated by using an annealing/energy-minimization algorithm: initialization was done at 1,000 K during 5,000 steps of 1 fs; dynamic study was continued for 5,000 additional steps by cooling by 100-K steps until reaching 200 K, after which the structure was energy-minimized by 2,000 steps of conjugate gradient. A representative low-energy conformation was selected and used as a starting point for energy minimization by using a standard template-forcing protocol to target Phe-16, Trp-37, Arg-26, and Arg-32 in their NCp7 solution conformation (33).
Annealing of tRNA3Lys to RNA(77–257): Direct Action of RB 2121 or Inhibition of (12–53)NCp7 Promotion.
HIV-1 RNA(77–257) and [33P]tRNA3Lys were heat-denatured for 2 min at 95°C and chilled at 0°C. An equimolar mixture of both RNAs (5 × 10−12 mol per assay) was incubated with RB 2121 alone from 0 μM to 500 μM or in the presence of 13 μM (12–53)NCp7 in 20 μl of a buffer containing 25 mM Tris⋅HCl (pH 7.5), 80 mM NaCl, and 0.1 mM MgCl2 for 15 min at 37°C. The reaction was stopped by addition of 1 μl of 20% (vol/vol) glycerol containing 0.01% bromophenol blue. RNA-protein complexes were analyzed by electrophoresis and visualized by autoradiography (29).
NCp7–RT Interaction; Inhibition by RB 2121.
HIV-1 p66/p51 RT (2 μg; Worthington) were immobilized on a nitrocellulose membrane (Amersham Pharmacia) in 100 μl of TBS buffer (25 mM Tris/100 mM NaCl/0.1 mM DTT/3 mM KCl, pH 7.4) for 3 h at room temperature. After treatment with Superblock blocking buffer (Pierce) for 3 h at room temperature, the membrane was incubated with RB 2121 (20–200 μM) or an inactive analog (200 μM) for 1 h at 37°C in 4 ml of 5% (vol/vol) dry milk TBS-T (TBS + 0.1% Tween 20) and then overnight at 4°C with NCp7 (8.5 × 10−7 M). Blotting was performed by incubation with 1/15,000 anti-NCp7 monoclonal antibody (34), followed by treatment with peroxidase-conjugated anti-mouse antibody and enhanced-chemiluminescence (Amersham Pharmacia) detection. These antibodies were unable to cross-react with RT, RB 2121, or its analogues. The inhibition was quantified on a Bio-profile imager (Vilber-Lourmat, Marne La Vallée, France).
Inhibition of HIV Replication in Infected CEM-4 Cells.
Acute CEM-4 infection was achieved in quadruplicate with HIV-1 Lai (550 ng of p24 per 106 cells; ref. 35). Various concentrations of inhibitors (up to 100 μM) were preincubated with cells for 3 h before or after virus infection. After 6 days, culture supernatants were tested for RT activity by a scintillation proximity assay (RPNQ100 Amersham Pharmacia SPA kit), and the enzyme activity was expressed as cpm per milliliter, corresponding to the amount of [3H]thymidine triphosphate incorporated per milliliter of culture fluid (36). Cell viability was determined by using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide procedure (35). Cytotoxicity was calculated as the decrease of cell viability observed in treated mock-infected cultures. Inhibition of the cytopathic effect was evaluated by using the ratio of cell viability in infected treated cultures over infected untreated controls. The ability of virions released in treated CEM-4 supernatants to infect fresh CEM-4 cells was assessed by using 2-fold serial dilutions of these supernatants. After 6 days, infectious titer was defined as the reverse of the last dilution that gave 1 μg/ml of p24 in supernatants.
Stimulated HIV-1 Production in Chronically Infected U1 Cells.
Chronically HIV-1-infected U1 cells (105 cells per ml) were pretreated with the inhibitors (up to 100 μM) 5 h before stimulation by 10−7 M phorbol 12-myristate 13-acetate (Sigma) or 10 units/ml tumor necrosis factor-α (Genzyme; ref. 36). Aliquots of culture supernatants were harvested 3 days after stimulation and tested for the presence of RT activity and cell viability.
Virus-Binding and Syncytia-Formation Assays.
The interaction between recombinant soluble CD4 and HIV-1 envelope glycoprotein gp120 (Repligen) was analyzed by using the NEMQuest HIV-1 CD4/gp120 recognition EIA kit (DuPont; ref. 37) in the presence of RB 2121 (up to 100 μM). A syncytia-formation assay was performed by mixing chronically infected CEM-4/Lai cells (≈105 cells per well) with noninfected indicator C8166 cells (ratio 1:2.5) and RB 2121 (up to 100 μM). At 24 h, the presence of syncytia was evaluated by microscopic examination (35). Cell viability was assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide. Aurintricarboxylic acid was used as a reference in both assays (1.2 μM and 4.4 μM, respectively).
In Vitro Assays of Viral Enzymes.
Inhibitory effects of RB 2121 on retroviral-enzyme activity were assessed as described (35). The exogenous RT assay was performed in the presence of RB 2121 by measuring the incorporation rate of labeled dGTP induced by recombinant RT (1 nM) on poly(C) or poly(dC) templates from (dG)12–18 primer. HIV-1 integrase assays were carried out by analyzing the autointegration of 32P-labeled HIV-1 U5-end oligonucleotide substrate promoted by recombinant HIV-1 integrase. The experiment for HIV-1 protease inhibition was achieved by measuring Gag processing (35).
PCR Detection of HIV-1 Proviral DNA Synthesis.
CEM-4 cells (n = 2 × 105) preincubated or not for 3 h with RB 2121 (from 4.7 μM up to 150 μM) were infected with HIV-1 Lai (5.50 μg/ml p24) for 2 h. Cells were washed twice. After a 16-h and a 24-h culture, total DNA was extracted. PCR analysis was performed with 0.5 μg of DNA for 35 cycles (38). Early RT-produced DNA sequences corresponding to the R-U5 domain were amplified by using primer 1 (nucleotides 498–518 in HIV Lai sequence; 5′-GGCTAACTAGGGAACCCACT-3′) and primer 2 (nucleotides 617–637; 5′-CTGCTAGAGATTTTCCACACT-3′), respectively, and late RT-produced DNA sequences corresponding to the R-Gag domain were amplified with primer 3 (nucleotides 528–548; 5′-CAATAAAGCTTGCCTTGAGTG-3′) and primer 4 (nucleotides 800–820; 5′-CCGCTTAATACTGACGCTCTC-3′, respectively. PCR products were run on 3% agarose gels, and bands, stained with ethidium bromide, were quantified by using a Bio-Rad Image analysis system. As a control for cellular DNA, the human β-globin sequence was amplified similarly.
Inhibition of NCp7–RT Interaction by Cyclic Peptides: Structure-Activity Relationships.
HIV-1 RT was covalently linked to CNBr-activated Sepharose 4B Beads (Amersham Pharmacia; ref. 30); 20 μl of beads (4.6 μg of RT; 3.9 × 10−11 mol) were equilibrated in TBS-T buffer with 0.1% Nonidet P40. Cyclic peptides (43 μM) were preincubated for 90 min at room temperature with immobilized RT in 100 μl of previously mentioned buffer before the addition of NCp7 (8.5 × 10−7 M). After 5 h at room temperature, beads were centrifuged and washed twice with cold TBS buffer. Bound NCp7 was recovered by heating at 80°C in Laemmli buffer [50 mM Tris/10% (vol/vol) glycerol/2% (vol/vol) SDS/0.05% bromophenol blue/200 mM DTT]. Preincubation of NCp7 before addition of cyclic peptide was also tested. Collected samples were loaded on an SDS/20% PAGE, transferred onto nitrocellulose, and analyzed by Western blotting by using monoclonal antibody (34) to identify NCp7.
RESULTS AND DISCUSSION
Rational Design of NCp7 Three-Dimensional Structure Mimics.
The nucleocapsid protein NCp7 was shown to possess a well defined three-dimensional structure with a spatial proximity between Phe-16 and Trp-37 aromatic residues and positively charged Arg-26 and Arg-32 amino acids (21–22). These water-accessible residues were shown by chemical and genetic manipulations to be essential for in vitro and in vivo functions of NCp7, suggesting that they are involved in the interaction of this protein with its biological targets. In a first approach, cyclic hexapeptides containing the four selected residues, Phe, Trp, Lys, and Arg, were designed by solid-phase synthesis to mimic the three-dimensional arrangement of their counterparts in NCp7, Phe-16, Trp-37, Arg-26, and Arg-32 (Fig. 1a; refs. 20 and 27). Among them, RB 2121 (Fig. 1b) was the most promising compound. In addition, to facilitate the convenient spatial orientation of the selected residues, peptide cyclization increases the resistance to peptidases, whereas cysteines allow different substitutions to be done. Different analogues of RB 2121 (Fig. 1c) were synthesized to study the importance of each residue. Cysteines were replaced by S-methylated cysteines (RB 2124) or serines (RB 2125). The d-tryptophan was mutated in l-tryptophan (RB 2126) or in leucine (RB 2128). RB 2127 is the linear analogue of RB 2121.
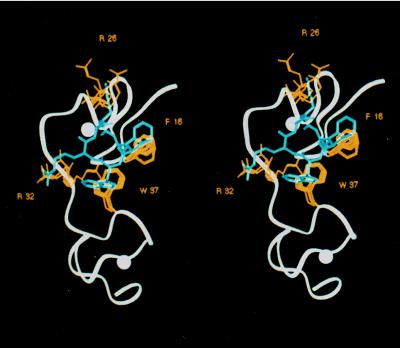
Structural analysis of RB 2121 was performed by 1H NMR spectroscopy and molecular modeling. The 1H NMR conformation of RB 2121 showed the classical structural parameter of a cyclic hexapeptide: hydrogen bonds NH(Cys-2)⋅⋅⋅CO(Cys-5) and NH(Cys-5)⋅⋅⋅CO(Cys-2), evidenced by the low temperature dependency Δδ/Δt of the concerned NH strong interresidue NOEs N1/N2 and N4/N5 and medium interresidue NOEs N1/N6, N5/α3, and N2/β5. These data indicated the existence of a well defined cyclic backbone, with side chains extended at the periphery. A molecular-dynamics simulation was performed by using the observed NOEs as interproton distance restraints. Superimposition of the 20 lowest-energy conformations showed a complete overlap of their backbones (rms = 0.2 Å) and a relatively restricted space for the side-chain orientations (Fig. 2). These conformations were relaxed by energy minimization and removal of the distance restraints. All converged toward a similar structure. A superimposition of this conformation with a set of seven low-energy conformations obtained for NCp7 from two-dimensional NMR studies shows that the cyclic structure provides an excellent template to overlap Phe-1, d-Trp-3, Arg-4, and Lys-6 side chains with the mean position of their respective analogues in NCp7, Phe-16, Trp-37, Arg-32, and Arg-26. Because of the free rotation around the indole bond, a similar volume could be fitted by the Trp ring in RB 2121 and NCp7.
Figure 2.
Stereoview of RB 2121 superimposed on (12–53)NCp7. Superposition of the NMR-derived structure of (12–53)NCp7 (in white), with the preferential conformation of RB 2121 (in blue) showing Phe-16, Arg-26, Arg-32, and Trp-37 side chains (in yellow) overlapping those of the corresponding amino acids in the cyclic peptide.
RB 2121 Competes with NCp7 for in Vitro Recognition of Its Nucleic Acids and Protein Targets.
The addition of dithiobisbenzamide-1 into an Acr-37–NCp7 solution, in which Trp-37 has been replaced by a 9-acridinylalanine (39), induces an irreversible time-dependent increase of fluorescence induced by zinc ejection from the second zinc finger (32). In contrast, RB 2121 had no effect on Acr-37–NCp7 fluorescence (data not shown), indicating that it did not function as a zinc ejector and was also unable to bind NCp7 as shown by NMR and surface plasmon resonance experiments (data not shown).
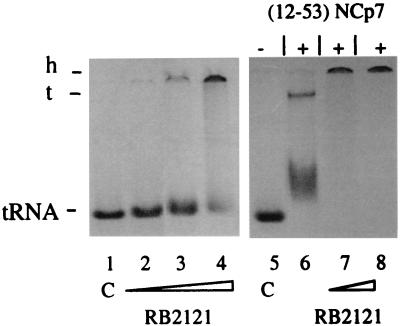
The nucleocapsid protein is involved critically in early steps of the retroviral life cycle by enhancing the global processivity of the reverse transcription of the HIV-1 RNA genome into double-stranded proviral DNA. It seems that a well structured zinc-finger domain and the presence of Trp-37 are essential determinants for NCp7 annealing activities (29) and RT recognition (30), accounting for the complete loss of infectivity of viruses in which mutations disrupting NCp7 conformation or replacement of Trp-37 by Leu have been introduced (23). Because RB 2121 was designed to mimic these structural characteristics, the effect of RB 2121 on the annealing of [33P]tRNA3Lys to RNA(77–257), which contains the PBS, was measured in the presence or absence of (12–53)NCp7, which contains the major structural elements shown in vivo to be essential for its biological functions, but did not induce nucleic acid aggregation through nonspecific electrostatic interactions (12, 29). After coincubation of both RNAs and peptides for 15 min at 37°C, the samples were analyzed by native PAGE and visualized by autoradiography. As depicted in Fig. 3, lane 6, 80 mol equivalents of (12–53)NCp7 are sufficient to induce the formation of the tRNA–RNA hybrid complex coexisting with complexed forms between (12–53)NCp7 and both types of RNA. When equimolar amounts of tRNA and RNA were coincubated with increasing concentrations of RB 2121 in the absence of (12–53)NCp7, we observed a gel shift of labeled tRNA but no band corresponding to the tRNA–RNA hybrid complex (Fig. 3, lanes 2–4) and aggregates unable to penetrate the gel at high concentrations of RB 2121 (Fig. 3, lane 4). We then analyzed the effect of RB 2121 on (12–53)NCp7-promoted hybrid formation. As expected, by competing with (12–53)NCp7 for the recognition of tRNA and RNA, RB 2121 inhibits the formation of the ternary complex leading only to the formation of aggregates (Fig. 3, lanes 7 and 8).
Figure 3.
RB 2121 inhibits NCp7-dependent annealing of tRNA3Lys to HIV-1 (77–257)RNA. Fixed concentrations of [33P]tRNA3Lys and RNA(77–257) (5 × 10−12 mol of each denatured RNA per assay) were incubated in 25 mM Tris⋅HCl, pH 7.5/0.1 mM MgCl2/80 mM NaCl buffer at 37°C in presence of 25 μM, 125 μM, and 250 μM RB 2121 (lanes 2–4) or with 13 μM (12–53)NCp7 in the presence of 0 μM, 100 μM, and 500 μM RB 2121 (lanes 6–8). Lanes 1 and 5 represent tRNA as a control (C). Samples were analyzed by native PAGE at 4°C, and complexes were visualized by autoradiography. h, high molecular weight complex; t, ternary complex made of tRNA3Lys, RNA(77–257), and (12–53)NCp7.
Using competition experiments, we studied the effect of RB 2121 on RT–NCp7 direct recognition (30). RT immobilized on a nitrocellulose membrane was preincubated with increasing concentrations of RB 2121 before addition of a constant amount of NCp7. As shown in Fig. 4, RB 2121 inhibited this interaction in a dose-dependent manner (Fig. 4a, lanes 3–5). Moreover, RB 2121 was also able to compete with prebound NCp7, but this competition required concentrations that were about four times higher (data not shown). In contrast, analogues of RB 2121, which are unable to overlap the corresponding determinants in NCp7 perfectly, did not affect NCp7–RT interaction (Fig. 4a, lane 6).
Figure 4.
Inhibition of NCp7–RT complex formation by RB 2121. (a) Far Western blot analysis with an anti-NCp7 antibody of the interaction between NCp7 (35 μg) and RT (2 μg) in the presence of 0 μg (lane 2), 90 μg (lane 3), 180 μg (lane 4), or 900 μg (lane 5) of RB 2121 or 900 μg of an inactive analogue (lane 6). (b) Quantification of the dose-dependent inhibition by RB 2121 of RT–NCp7 complex formation. Data are expressed as the mean ± SD from four separate experiments.
RB 2121 Suppresses HIV-1 Expression in CEM-4 Cells at an Early Step of Lai Virus Replication.
When incubated with infected cells, RB 2121 showed dose-dependent antiviral properties by inhibiting HIV-1 Lai replication in a CEM-4 assay (ref. 35; Fig. 5a). The anti-HIV-1 activity was assessed by evaluating RT activity in cell supernatants 6 days after infection. No difference was observed when RB 2121 was added to the cells 3 h before or after infection, because the mean IC50 values were 30 ± 7 μM and 43 ± 7 μM, respectively (Fig. 5a). Protection from virus-induced cytopathic effects was evaluated by comparing cell viability in treated or nontreated infected cells. The potency of RB 2121 to inhibit cytopathic effects was found to be 23 μM (data not shown). No cellular toxicity was observed up to 150 μM when noninfected CEM-4 cells were treated by RB 2121 for the same period of time.
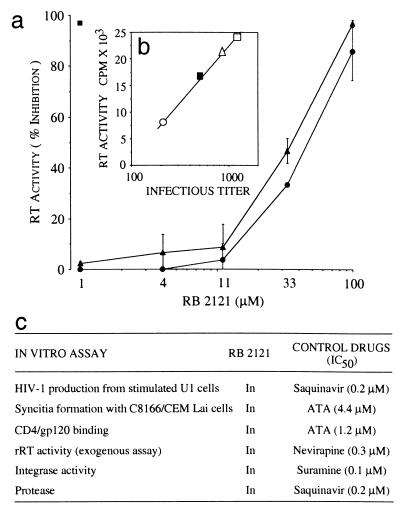
Figure 5.
Anti-HIV-1 activity of RB 2121. (a) Inhibition of RT activity in 6-day culture supernatants of infected CEM-4 cells; results represent the mean ± SD of three experiments. RB 2121 was added to CEM-4 cells before (▴) or after (●) HIV-1 infection. Percentage of inhibition obtained with 1 μM zidovudine (■). (b) Infectivity of virions produced in the presence of RB 2121. Regression analysis of RT activity (expressed in cpm) vs. the infectious titer in each supernatant tested: untreated culture (□), cultures treated with 1 μM (▵), 10 μM (■), and 100 μM (○) of RB 2121. (c) Lack of RB 2121 effects on HIV-1-specific activities and viral enzymes. RB 2121 (up to 100 μM) was unable to block virus entry or viral enzymes. All assays were carried out by described techniques (see Materials and Methods). Control drugs gave previously reported values.
To determine whether the antiviral activity of RB 2121 was caused by a defect in viral maturation, we have measured the effects of RB 2121 on the infectivity of virions produced during acute HIV-1 infection of CEM-4 cells. The results (Fig. 5b) show a linear relationship (r2 = 0.99) between the infectious titer and the p24 content and RT level in the cell supernatants. This correlation shows that even in the presence of high concentrations of RB 2121, the residual virus particles were infectious and that the inhibition of the spread of HIV in CEM-4 cultures was not caused by a defect in virus maturation.
The addition of phorbol ester or cytokines (tumor necrosis factor-α) was shown to stimulate transcription and viral production in chronically infected promonocytic U1 cells that harbor two inducible copies of a latent integrated HIV-1 provirus (36). The procedure was designed as a model for late events in HIV-1 replication. Pretreatment with RB 2121 for 5 h before stimulation did not inhibit phorbol 12-myristate 13-acetate-induced or tumor necrosis factor-α-induced production of virions in U1 cells, as measured by RT activity in culture supernatants (Fig. 5c). This result indicates that, in contrast to previously described zinc-finger ejectors (31, 32), the cyclic peptide did not inhibit posttranscriptional events of HIV replication. Because RB 2121 has no affinity for NCp7, it is therefore unable to interfere with the NCp7-promoted virion assembly (10). Moreover, this result shows that RB 2121 does not impair protease activity, leading us to conclude that the antiviral effect of RB 2121 could take place at an early step of the viral life cycle.
RB 2121 Inhibits HIV-1 Replication by Impairing the Proviral DNA Synthesis.
Different assays were used to clarify the mechanism of action of RB 2121. The data presented in Fig. 5a suggested that RB 2121 activity was not caused by an impairment of HIV-1 entry into the cells, because the compound displayed similar potencies when administrated before and after infection. This result was confirmed in a syncytia-formation assay in which RB 2121 was found to be ineffective up to 100 μM. RB 2121 did not inhibit the CD4–gp120 interaction, as determined by using an in vitro assay (ref. 37; Fig. 5c), eliminating a possible effect at the level of virion binding to the target cell. On the other hand, the antiviral activity of RB 2121 was not caused by a direct inhibition of HIV-1 enzymes acting early in the viral life cycle. Indeed, the cyclic peptide was unable to block RT, integrase, and viral protease activities, as determined by using previously described in vitro assays (ref. 35; Fig. 5c).
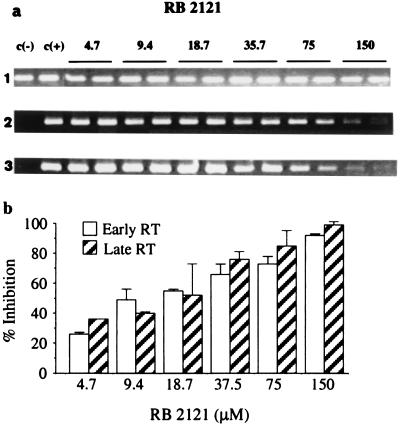
To account for these results, we hypothesized that the antiviral activity of RB 2121 occurred at the level of proviral DNA synthesis. Indeed, mutations in the HIV-1 genome of NCp7 determinants, used to generate RB 2121, were shown to result in virus particles with slight modifications in the core structure and defects in RNA packaging (21, 23–25). However, these modifications were not sufficient to explain the observed complete loss of infectivity, suggesting that NCp7 could be involved in RT-dependent steps. The effect of RB 2121 therefore was investigated in infected CEM-4 cells by monitoring PCR-amplified early and late reverse transcription sequences. Cells preincubated with RB 2121 were infected with HIV-1 Lai for 2 h, and, 24 h after infection, total DNA was extracted. Early and late RT-produced DNA sequences were amplified by PCR by using two different probes, enabling us to amplify 140-bp R/U5 and 293-bp R/Gag fragments that correspond, respectively, to events occurring between the first and the second strand transfer and to the late events of reverse transcription. RB 2121 induced a dose-dependent reduction in DNA products formed at both steps of retrotranscription, as compared with untreated cells (Fig. 6 a and b). When the experiment was analyzed after 16 h, RB 2121 seemed to inhibit the late reverse-transcript synthesis more efficiently rather than the early (data not shown), indicating that the NCp7 mimic impaired not only initiation of reverse transcription but also strand transfers or polymerization. All these results show that RB 2121 inhibits HIV-1 replication through disruption of NCp7-dependent steps, such as NCp7–nucleic acids (Fig. 3) or NCp7–RT recognition (Fig. 4), thus impairing the formation of a functional complex comprising RT, NCp7, and nucleic acids (11, 14, 18). This hypothesis highlights the importance of the structure of the NCp7 zinc-finger domain in promoting RT-induced provirus synthesis, supported by the drastic reduction in transcribed DNA levels and the complete loss of virus infectivity observed with the mutant His23Cys, characterized by a change in the conformation of the mutated zinc array (23, 25).
Figure 6.
Selective inhibition by RB 2121 of early and late HIV-1 DNA synthesis. (a) Gel electrophoresis of PCR products. Infected CEM-4 cells were treated by increasing concentrations of RB 2121 for 24 h. DNA was amplified for β-globin-generated (lane 1; 105 bp; as a control), early RT-generated (lane 2; 140 bp), and late RT-generated (lane 3; 293 bp) HIV-1 sequences. The specific dose-dependent inhibition of early and late HIV-1 transcripts by RB 2121 suggests an impairment of RT processivity. (b) Quantitative analysis (±SD) of the PCR results. Inhibition is expressed as percentage of controls (untreated infected cells).
All the Determinants of RB 2121 Are Crucial for Its Antiretroviral Activity.
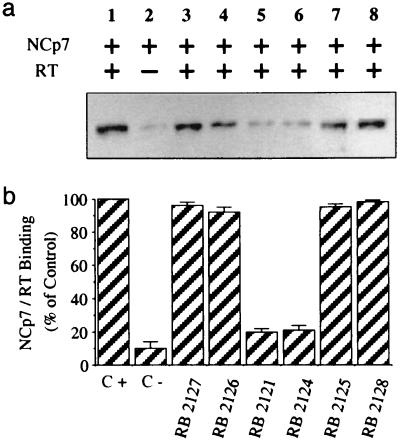
RB 2121 variants (Fig. 1c) were studied to try to correlate their ability to inhibit RT–NCp7 complex formation and their antiviral activity. For this purpose, RT immobilized on Sepharose beads was preincubated with the same concentrations of RB 2121 derivatives before the addition of NCp7. After incubation and washes, the collected samples were analyzed by Western blotting to quantify the amount of NCp7 bound to RT (Fig. 7). RB 2127, the linear analogue of RB 2121 that is unable to impair RT–NCp7 complex formation (Fig. 7, lane 3), confirms that the cyclic structure is required to present the main determinants in a spatial disposition, allowing the peptide to compete with NCp7 for its target recognition. The two cysteine residues seemed to be critical for the activity of RB 2121, because changing cysteines to serines in RB 2125 produced an inactive derivative (Fig. 7, lane 7). Trp-mutated RB 2126 and RB 2128 also were found to be inactive (Fig. 7, lanes 4 and 8), confirming the importance of Trp residue in the mechanism of action of RB 2121, in agreement with the critical role found for Trp-37 in NCp7 (27–29). Only the RB 2121 and its S-methylated cysteine-derivative RB 2124 were able to compete with NCp7 for RT recognition (Fig. 7, lanes 5 and 6). Accordingly, only RB 2121 (Fig. 5) and to a lesser extent RB 2124 (data not shown) showed antiretroviral activities in the cellular assay.
Figure 7.
Importance of structural determinants for the retroviral activity of RB 2121. (a) Immobilized RT was preincubated or not with 43 μM RB 2121 or its analogues (lanes 3–8) for 1.5 h before the addition of 0.85 μM NCp7 in TBS-T buffer. Bound NCp7 was recovered and measured by Western blot analysis by using antibody selective of NCp7. Lane C+ corresponds to the total binding of NCp7 to immobilized RT, and lane C− corresponds to the same experiment with beads without RT. (b) Quantification of the inhibition of RT–NCp7 interaction by RB 2121 and its analogues. Data are expressed as the mean ± SD from three separate experiments.
CONCLUSION
Emergence of drug-resistant viruses to currently used therapies against HIV-1 (4) indicates that a successful treatment of the disease could come from a combination of drugs with different antiviral mechanisms of action. Compounds such as RB 2121 are interesting in that they mimic the structural determinants of NCp7, which could not be mutated without complete loss of viral infectivity. Moreover, their mode of action is completely different from that of RT or protease inhibitors and from that of zinc ejectors, which have been shown to possess antiretroviral activity (31, 40) through disruption of Gag processing and subsequent defects in formation of infectious virus (32). One difficulty of the approach could be the high concentration of NCp7 as compared with enzymes in the virus. But, the affinity of NCp7 for RT is relatively weak (≈10−7 M; ref. 30), and most of these nucleocapsid molecules are bound to different nucleic acids present during the synthesis of the provirus.
RB 2121 can be considered as the lead of a new class of antiviral agents able to inhibit the NCp7-dependent formation of HIV-1 provirus. The affinity and bioavailability of these compounds could be improved by combinatorial chemistry to provide a drug for clinical investigation. This process of improvement should be facilitated by high-throughput screening assays based on inhibition of the RT–NCp7 complex formation.
Acknowledgments
We thank E. Remy for annealing experiments, H. Meudal and P. Petitjean for NMR studies and peptide synthesis, J. F. Ferron and M. F. Bachelot for their contribution in biochemical studies, and H. de Rocquigny for helpful discussions. This work was supported by the French program against AIDS (Agence Nationale de Recherche sur le SIDA and SIDACTION).
ABBREVIATIONS
- RT
reverse transcriptase
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
References
- 1.Erickson J W. Nat Struct Biol. 1995;2:523–529. doi: 10.1038/nsb0795-523. [DOI] [PubMed] [Google Scholar]
- 2.Cohen J. Science. 1997;227:32–33. doi: 10.1126/science.277.5322.32. [DOI] [PubMed] [Google Scholar]
- 3.Wong J K, Hezareh M, Gunthard H F, Havlir D V, Ignacio C C, Spina C M, Richman D D. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 4.Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, et al. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 5.Darlix J L, Gabus C, Nugeyre M T, Clavel F, Barré-Sinoussi F. J Mol Biol. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 6.Berg J M. Science. 1986;232:485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- 7.Darlix J L, Lapadat-Tapolsky M, de Rocquigny H, Roques B P. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 8.Aldovini A, Young R A. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berkowitz R D, Ohagen A, Höglund S, Goff S P. J Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poon D T K, Wu J, Aldovini A. J Virol. 1996;70:6607–6616. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barat C, Lullien V, Keith G, Nugeyre M T, Gruninger-Leitch F, Barré-Sinoussi F, LeGrice S F J, Darlix J L. EMBO J. 1989;8:3279–3285. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Rocquigny H, Gabus C, Vincent A, Fournié-Zaluski M C, Roques B P, Darlix J L. Proc Natl Acad Sci USA. 1992;89:6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darlix J L, Vincent A, Gabus C, de Rocquigny H, Roques B P. C R Acad Sci. 1993;316:763–771. [PubMed] [Google Scholar]
- 14.Peliska J A, Balasubramanian S, Giedroc D P, Benkovic S J. Biochemistry. 1994;33:13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Rodriguez L, Tsuchihasi Z, Fuentes G M, Bambara R A, Fay P J. J Biol Chem. 1995;270:15005–15011. doi: 10.1074/jbc.270.25.15005. [DOI] [PubMed] [Google Scholar]
- 16.Wu W, Henderson L E, Copeland T D, Gorelick R J, Bosche W J, Rein A, Levin J G. J Virol. 1996;70:7132–7142. doi: 10.1128/jvi.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Quan Y, Arts E J, Li Z, Preston B D, de Rocquigny H, Roques B P, Darlix J L, Kleiman L, Parniak M A, et al. J Virol. 1996;70:4996–5004. doi: 10.1128/jvi.70.8.4996-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cameron C E, Ghosh M, Le Grice S F J, Benkovic S J. Proc Natl Acad Sci USA. 1997;94:6700–6705. doi: 10.1073/pnas.94.13.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo J, Henderson L E, Bess J, Kane B, Levin J G. J Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morellet N, Jullian N, de Rocquigny H, Maigret B, Darlix J L, Roques B P. EMBO J. 1992;11:3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morellet N, de Rocquigny H, Mely Y, Jullian N, Demene H, Ottmann M, Gerard D, Darlix J L, Fournie-Zaluski M C, Roques B P. J Mol Biol. 1994;235:287–301. doi: 10.1016/s0022-2836(05)80033-6. [DOI] [PubMed] [Google Scholar]
- 22.Lee B M, De Guzman R N, Turner B G, Tjandra N, Summers M F. J Mol Biol. 1998;279:633–649. doi: 10.1006/jmbi.1998.1766. [DOI] [PubMed] [Google Scholar]
- 23.Déméné H, Dong C Z, Ottmann M, Rouyez M C, Jullian N, Morellet N, Mely Y, Darlix J L, Fournie-Zaluski M C, Saragosti S, et al. Biochemistry. 1994;33:11707–11716. doi: 10.1021/bi00205a006. [DOI] [PubMed] [Google Scholar]
- 24.Dorfman T, Luban J, Goff S P, Haseltine W A, Göttlinger H G. J Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorelick R J, Chabot D J, Ott D E, Gagliardi T D, Rein A, Henderson L E, Arthur L O. J Virol. 1996;70:2593–2597. doi: 10.1128/jvi.70.4.2593-2597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu Q, Darlix J L. J Virol. 1996;70:5791–5789. doi: 10.1128/jvi.70.9.5791-5798.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morellet N, Déméné H, Theilleux V, Huynh-Dinh T, de Rocquigny H, Fournié-Zaluski M C, Roques B P. J Mol Biol. 1998;283:419–434. doi: 10.1006/jmbi.1998.2098. [DOI] [PubMed] [Google Scholar]
- 28.De Guzman R N, Wu Z R, Stalling C C, Pappalardo L, Borer P N, Summers M F. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 29.Remy E, de Rocquigny H, Petitjean P, Muriaux D, Theilleux V, Paoletti J, Roques B P. J Biol Chem. 1998;273:4819–4822. doi: 10.1074/jbc.273.9.4819. [DOI] [PubMed] [Google Scholar]
- 30.Druillennec S, Caneparo A, de Rocquigny H, Roques B P. J Biol Chem. 1999;274:11283–11288. doi: 10.1074/jbc.274.16.11283. [DOI] [PubMed] [Google Scholar]
- 31.Rice W G, Supko J G, Malspeis L, Buckheit R W, Clanton D, Bu M, Graham L, Schaeffer C A, Turpin J A, Domagala J, et al. Science. 1995;270:1194–1197. doi: 10.1126/science.270.5239.1194. [DOI] [PubMed] [Google Scholar]
- 32.Turpin J A, Terpening S J, Schaeffer C A, Yu G, Glover C J, Felsted R L, Sausville E A, Rice W G. J Virol. 1996;70:6180–6189. doi: 10.1128/jvi.70.9.6180-6189.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laoui A, Luttmann C, Morize I, Pantel G, Morgat A, Rubin-Carrez C, Laroche V, Guitton J D, Gigonzac O, James-Surcouf E, et al. In: New Perspectives in Drug Design. Dean P M, Jolles G, Newton C G, editors. New York: Academic; 1995. pp. 255–284. [Google Scholar]
- 34.Tanchou V, Delaunay T, de Rocquigny H, Bodeus M, Darlix J L, Roques B P, Benarous R. AIDS Res Hum Retroviruses. 1994;10:983–993. doi: 10.1089/aid.1994.10.983. [DOI] [PubMed] [Google Scholar]
- 35.Mayaux J F, Bousseau A, Pauwels R, Huet T, Henin Y, Dereu N, Evers M, Soler F, Poujade C, De Clercq E, et al. Proc Natl Acad Sci USA. 1994;91:3564–3568. doi: 10.1073/pnas.91.9.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weissman D, Poli G, Bousseau A, Fauci A S. Proc Natl Acad Sci USA. 1993;90:2537–2541. doi: 10.1073/pnas.90.6.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schols D, Baba M, Pauwels R, de Clercq E. J Acquir Immune Defic Syndr. 1989;2:10–15. [PubMed] [Google Scholar]
- 38.Escaich S, Wallon M, Baginski I, Ritter J, Philippe N, Bertrand Y, Claris O, Raudrant D, Sepetjan M, Trepo C. J Acquir Immune Defic Syndr. 1991;4:130–135. [PubMed] [Google Scholar]
- 39.Dong C Z, de Rocquigny H, Remy E, Mellac S, Fournié-Zaluski M C, Roques B P. J Peptide Res. 1997;50:269–278. doi: 10.1111/j.1399-3011.1997.tb01468.x. [DOI] [PubMed] [Google Scholar]
- 40.Huang H, Chopra R, Verdine G L, Harrison S C. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]