Abstract
Photoactivation of the retinal photoreceptor rhodopsin proceeds through a cascade of intermediates, resulting in protein–protein interactions catalyzing the activation of the G-protein transducin (Gt). Using stabilization and photoregeneration of the receptor’s signaling state and Gt activation assays, we provide evidence for a two-site sequential fit mechanism of Gt activation. We show that the C-terminal peptide from the Gt γ-subunit, Gtγ(50–71)farnesyl, can replace the holoprotein in stabilizing rhodopsin’s active intermediate metarhodopsin II (MII). However, the peptide cannot replace the Gtβγ complex in direct activation assays. Competition by Gtγ(50–71)farnesyl with Gt for the active receptor suggests a pivotal role for Gtβγ in signal transfer from MII to Gt. MII stabilization and competition is also found for the C-terminal peptide from the Gt α-subunit, Gtα(340–350), but the capacity of this peptide to interfere in MII-Gt interactions is paradoxically low compared with its activity to stabilize MII. Besides this disparity, the pH profiles of competition with Gt are characteristically different for the two peptides. We propose a two-site sequential fit model for signal transfer from the activated receptor, R*, to the G-protein. In the center of the model is specific recognition of conformationally distinct sites of R* by Gtα(340–350) and Gtγ(50–71)farnesyl. One matching pair of domains on the proteins would, on binding, lead to a conformational change in the G-protein and/or receptor, with subsequent binding of the second pair of domains. This process could be the structural basis for GDP release and the formation of a stable empty site complex that is ready to receive the activating cofactor, GTP.
Rhodopsin is a prototypical G-protein-coupled receptor in retinal rods (1, 2). Available information supports a mechanism in which the initial isomerization of the chromophore 11-cis-retinal, and thus the formation of the agonistic all-trans-retinal, leads to crucial contacts between the ligand and the apoprotein opsin. These steric constraints result in a defined arrangement of donor and acceptor groups for proton translocations leading to subsequent tautomeric conformations of the receptor, identified as “metarhodopsin” photointermediates, each with a characteristic absorption spectrum. Metarhodopsin I (MI, λmax = 478 nm) is in a pH- and temperature-dependent equilibrium with metarhodopsin II (MII, λmax = 380 nm), distinguished by its deprotonated Schiff base linkage [and broken salt bridge (3, 4)] between the retinal and Lys296. MII has been shown to catalyze retina rod cell-specific G-protein (Gt) activation through nucleotide exchange (5, 6).
Despite recent progress in structure determination of both Gt and rhodopsin, the molecular mechanism of signal transfer between the two proteins is poorly understood. Interacting surfaces of rhodopsin and Gt include intracellular loops of the receptor and domains on both Gt α- and Gt γ-subunits (1, 7–9). C-terminal domains of Gt α- and Gt γ-subunits, Gtα(340–350) and Gtγ(60–71)farnesyl, have been studied most extensively by site-directed mutagenesis and peptide competition experiments (10–17). Synthetic peptides representing Gtα(340–350) and Gtγ(60–71)farnesyl were shown to compete for Gt binding sites on rhodopsin by mimicking the ability of the heterotrimeric Gt to stabilize MII (10, 15). Gtγ(60–71)farnesyl (DKNPFKELKGGC-farnesyl) and Gtα(340–350) (IKENLKDCGLF) have unique individual amino acid requirements for rhodopsin binding (10, 13, 15, 16). The major difference between the two domains is that Gtγ(60–71)farnesyl has the absolute requirement for a farnesyl lipid group to establish rhodopsin binding (15). These apparent differences in the structural requirements of Gtα(340–350) and Gtγ(60–71)farnesyl for binding to MII and their location on the opposite sides of the rhodopsin interacting surface of Gt suggest that determinants of MII⋅Gtα(340–350) and MII⋅Gtγ(60–71)farnesyl recognition are different. However, it has remained unclear when and how these two domains are involved in the overall mechanism of signal transfer from rhodopsin to Gt. Here, the interactions of the synthetic peptides Gtα(340–350) and Gtγ(50–71)farnesyl with rhodopsin have been studied in (i) flash photolysis experiments monitoring extra MII formation, (ii) flash photolysis of MII monitoring the peptide-dependent reformation of the protonated retinal Schiff base and salt bridge with Glu113, and (iii) fluorescence and near-infrared light-scattering assays measuring Gt activation.
MATERIALS AND METHODS
Peptides. Peptide synthesis and purification were as described before (15, 18). After purification, all peptides were lyophilized, dissolved in water, and pH was adjusted to 7.5 with NaOH, followed by desalting on Sephadex G-25 in water and lyophilization and storage at −20°C under nitrogen. Immediately before the experiments, the peptides were dissolved in appropriate buffers or deionized water to obtain stock solutions of 1–10 mM. Amino acid sequences for the peptides were as follows. Gtα(340–350) (IKENLKDCGLF) (10); high-affinity analog of Gtα(340–350) [Gtα(340–350)HAA; VLEDLKSCGLF] (14); Gtγ(60–71)farnesyl (DKNPFKELKGGC-farnesyl); Gtγ(50–71)farnesyl (EDPLVKGIPEDKNPFKELKGGC-farnesyl) (15). Gtγ(50–71)farnesyl and Gtγ(60–71)farnesyl have similar biochemical profiles and can be used interchangeably.
UV/Vis Spectroscopy.
The amount of extra MII was monitored by time-resolved UV/Vis spectroscopy by using a dual wavelength spectrophotometer (Shimadzu UV300). Recorded traces represent readings of a 380 nm/417 nm absorbance difference from the samples containing 5–10 μM of washed rod outer-segment membranes prepared as before (19). All measurements were done in 100 mM Na–Hepes pH 7.9/50 mM NaCl/1 mM DTT/1 mM MgCl2/1 mM EDTA at 1.5°C. Cuvette path length was 2 mm. Twelve percent of rhodopsin was flash-activated by 500 ± 20 nm light.
Photoregeneration from the Signaling State.
Photoregeneration of rhodopsin from the signaling state was performed essentially as described (20). Briefly, 2 μM [for measurements with Gtα(340–350)HAA] or 4 μM [for measurements with Gtγ(50–71)farnesyl] affinity-purified rhodopsin [prepared essentially as described (20, 21)] in 100 mM Mes (pH 6) or 100 mM Hepes (pH 8), 130 mM NaCl, 0.03% (wt/vol) n-dodecyl β-d-maltopyranoside (DM) was fully bleached in the presence of the peptides by using 543 nm light. Samples with Gtγ(50–71)farnesyl contained 0.003% (wt/vol) DM. Photoregeneration was started with a flash of blue light (412 ± 7 nm filter). The contribution of the 470-nm side product to the measurement signal (20) was minimized by choosing the measuring wavelength of 543 nm. The absorption changes reflect the reprotonation of the retinal Schiff base bond.
Fluorescence Spectroscopy.
Traces of Gt activation are a percent change of fluorescence emission at 340 nm recorded after exciting the sample at 300 nm essentially as described before (22, 23). Rhodopsin (50 nM) in washed rod outer segment membranes (WM), or 50 nM affinity purified rhodopsin in 0.02% (wt/vol) DM (20, 21) were reconstituted with 330 nM Gt [prepared essentially as described (24)] and desired concentrations of peptides in a final buffer: 20 mM 1,3-bis[tris(hydroxymethyl)methylamino]propane (BTP), pH 7.5/130 mM NaCl/1 mM MgCl2. The samples were exposed to light, and Gt activation was initiated by injection of GTPγS to a final concentration of 10 μM. All recordings were under continuous exposure to 543.5 nm laser light and constant stirring at 20°C for WM and vesicle samples or 10°C for the samples in DM. For experiments requiring phospholipid vesicles with a low rhodopsin content, purified bovine rhodopsin (3 μM) was reconstituted with phosphatidylcholine from egg yolk in a 1:1,500 molar ratio by microdialysis as described (23). Data are evaluated as fluorescence change relative to the initial fluorescence intensity. For comparison, traces from samples containing peptides are normalized to the amplitude obtained with the peptide-free sample. Purified Gt was separated into α- and βγ-subunits by chromatography on a cibacron blue column (HiTrap Blue, Amersham Pharmacia: 10 mM BTP, pH 7.5/1 mM MgCl2/1 mM DTT/0–0.3 M NaCl gradient over three column volumes to elute Gtβγ, and after additional four column volumes step to 1 M NaCl to elute Gtα).
Light Scattering Measurements.
The changes in a dissociation signal were recorded as before (25, 26). Measuring conditions were 3 μM rhodopsin (WM), 0.4 μM Gt, 500 μM GTP, 20°C; 0.9% of rhodopsin was flash-activated. Buffer was 20 mM BTP adjusted to pH, 130 mM NaCl, 1 mM MgCl2.
Molecular Modeling.
Surface calculations of Gt have been performed on IBM RS6000 by using molmol (27). The rhodopsin-bound conformation of Gtα(340–350) determined by nuclear magnetic resonance spectroscopy (Protein Databank ID no. 1AQG) was docked to Gt [Protein Databank ID no. 1GOT (28)] as before (18), and final molecular coordinates were used to calculate a solvent accessible area of Gt. Surface mapping of the rhodopsin interacting domains of Gt other than Gtα(340–350) (10) and Gtγ(50–71)farnesyl (15) were as follows: Gtα(8–23) (10); Gtα(305–315) (10, 29); solvent-accessible parts of Gtβ(280–340) (30).
EXPERIMENTAL RESULTS
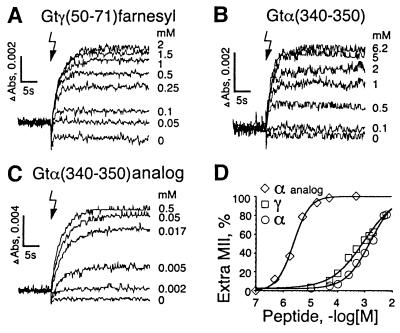
Both Peptides Interact with the MII State. Both Gtγ(50–71)farnesyl and Gtα(340–350) favor MII formation (Fig. 1 A and B). Half-maximum concentrations for the formation of extra MII are in the same range for both peptides but differ drastically from the high-affinity analog of Gtα(340–350), identified in an earlier study (ref. 14; Fig. 1 C and D). For the native peptides (Fig. 1 A and B), observed initial rates of MII formation are identical over a range of peptide concentrations. This shows that conformational changes of rhodopsin are rate-limiting in these experiments and argues for the specific binding of both peptides to MII once it is formed from its precursor MI. It also argues that the peptides do not influence the rate of formation of the MII photoproduct, which is a very sensitive parameter of detergent-like effects on membrane structure.
Figure 1.
Gt-derived peptides increase the formation of metarhodopsin II (MII) photointermediate. At pH 7.9, 1.5°C flash photolysis of rhodopsin (indicated by the flash symbol) predominantly produces the MI photoproduct (λmax = 478 nm), which is in a dynamic equilibrium with a small proportion of MII (λmax = 380 nm, isosbestic point for MI/MII at λ = 417 nm). Heterotrimeric Gt or Gt mimetic peptides shift the MI–MII equilibrium toward MII in a concentration-dependent manner to produce extra MII. The amount of extra MII was monitored by UV/Vis spectroscopy. Traces represent readings of a 380–417 nm absorbance difference (ΔAbs) from the samples containing 10 μM (A and C) or 5 μM (B) of washed-rod outer-segment membranes as described (19). Extra MII produced by the indicated concentrations (mM) of Gtγ(50–71)farnesyl (A), Gtα(340–350) (B), and the Gtα(340–350) high-affinity analog (VLEDLKSCGLF) (C). Amplitudes of the extra MII signals from A–C are plotted vs. peptide concentration (D, ΔAbs at saturation taken from C).
With the high-affinity analog (Fig. 1C), a variation of the initial rate is seen, which can be consistently explained with the reduced bimolecular collision rate, caused by the much lower concentrations of this peptide.
Both Peptides Block Photoregeneration from the Interactive State.
In the rhodopsin system, interactions of the activated receptor with a binding partner can be investigated by flash photolysis of the receptor–ligand complex (20). A flash of blue light probes the situation in the very moment of the flash, yielding information on the fraction of rhodopsin bound and stability of the complex. The assay is complementary to extra-MII formation but is independent of membrane or micelle properties. For the G-protein, it could be shown (20) that Gt blocks photoregeneration to the ground state in a GTP-dependent manner at the step where the Schiff base bond of the retinal is reprotonated [presumably restoring the broken salt bridge (3)].
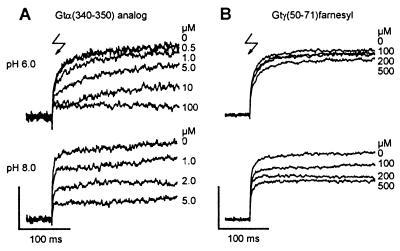
In Fig. 2, it is first seen that in all records, the control reaction (without peptide added) is somewhat slower at pH 6 than at pH 8. The reaction is, however, complete at any pH. These observations were already made in ref. 20. Both Gtγ(50–71)farnesyl and Gtα(340–350)HAA reduce the amount of photoregenerated rhodopsin in a dose-dependent manner (Fig. 2), and in the same range of concentrations where they stabilize the formation of MII.
Figure 2.
Effect of Gt-derived peptides on photoregeneration from metarhodopsin II. Gt-derived peptides, Gtα(340–350) high-affinity analog (A) and Gtγ(50–71)farnesyl (B) block photoregeneration from the bound state in a concentration-dependent manner. Blockage is more effective at pH 8 (Lower) than at pH 6 (Upper). At pH 6, Gtα(340–350) high-affinity analog shows a concentration-dependent complex dissociation and reversion to the dark form. Under these measuring conditions, ca. 15% of the signal amplitude is caused by the fast formation of an additional photoproduct, which is not affected by ligand binding (20) (vertical scale bars are 8·10−5 (A) and 2·10−4 (B) absorption units).
At pH 8, blocking of photoregeneration by Gtα(340–350)HAA is stable over 250 ms, showing that virtually no dissociation of the peptide from its binding site occurs. The rhodopsin molecules not occupied by Gtα(340–350)HAA react with their normal kinetics. At pH 6, the initial suppression of the absorption change (reflecting the fraction of rhodopsin bound to and blocked by peptide immediately before the flash) is hard to extract from the data, because a slow Schiff base reprotonation reaction is seen, which indicates a relatively high dissociation rate of the R*-peptide complex.
These results show that the Gtα(340–350)HAA peptide interacts with MII in a pH-dependent manner. The R*-peptide complex appears stable at pH 8 on the time scale of the experiment, but is weakened at pH 6, leading to complex dissociation and a subsequent reprotonation of the Schiff base.
In contrast, the Gtγ(50–71)farnesyl does not show any measurable R*-peptide complex dissociation at either pH. However, the blockade is more expressed at pH 8 than at pH 6.
Peptides Interfere Differently with Gt Activation.
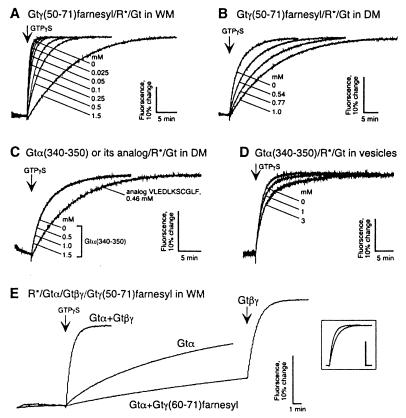
MII is the catalytically active intermediate of photoexcited rhodopsin (R*) (1, 5, 6). We went on to examine whether binding to MII by the two peptides would inhibit Gt activation as expected. Gtα(340–350) has been shown to effectively compete with Gt for MII stabilization (10). Gtγ(60–71)farnesyl has been reported to inhibit Gt binding to the photoexcited rhodopsin in urea washed rod outer segment membranes (15). We first used fluorescence spectroscopy to study the effects of the peptides on rhodopsin-catalyzed Gt activation (31). In WM, Gtγ(50–71)farnesyl slowed down the MII-catalyzed activation of Gt in a concentration-dependent manner (Fig. 3A). This effect can be attributed to interference of Gtγ(50–71)farnesyl in specific rhodopsin–Gt interactions as suggested before (15), or to the effect on the rhodopsin lipid microenvironment (32) because of the hydrophobic farnesyl group on the peptide. To discriminate between these two possibilities, rhodopsin was affinity-purified and the Gt activation assay was performed in the detergent DM. In the absence of a lipid bilayer, Gtγ(50–71)farnesyl was able to inhibit Gt activation with an efficacy similar to that observed in the presence of the lipid bilayer (Fig. 3B). This result argues for the specific interference of Gtγ(50–71)farnesyl in functionally crucial protein–protein contacts between rhodopsin and Gt. It is also consistent with the lack of any detergent-like effects of Gtγ(50–71)farnesyl on extra MII formation noted above (Fig. 1).
Figure 3.
Rate of rhodopsin (MII)-catalyzed Gt activation determined from changes of intrinsic Gtα fluorescence. Active rhodopsin-catalyzed formation of a Gt–GTPγS complex triggered by GTPγS addition results in an intrinsic fluorescence change of Gt. Inhibition of rhodopsin-catalyzed Gt activation by Gtγ(50–71)farnesyl in WM (A) and DM (B). Lack of effect of Gtα(340–350) on Gt activation in DM (C); similar results were obtained in WM with Gtα(340–350) up to 2 mM (data not shown). Competition of the native Gtα(340–350) peptide demonstrated for phosphatidylcholine vesicles reconstituted with low amounts of rhodopsin (rhodopsin content is approximately 15–20 times lower than in WM) (D). Gtγ(50–71)farnesyl does not replace Gtβγ in rhodopsin catalyzed activation of Gt (E). Activation traces from three independent experiments are superimposed: 1, control activation of Gt reconstituted from purified Gtα (450 nM) and Gtβγ (1 μM) subunits; 2, activation of the Gtα alone; 3, activation of Gtα in the presence of 200 μM of Gtγ(50–71)farnesyl, followed by the rescue addition of the Gtβγ. (Inset) Superimposed activation traces from experiment 1 (Gtα+Gtβγ) and experiment 3 after final addition of Gtβγ (Gtα+Gtγ(50–71)farnesyl+Gtβγ). The traces are corrected for the gross fluorescence change because of the addition of Gtβγ. Note that under the conditions of these experiment (see Materials and Methods) most of the G-protein is present in soluble form (26).
In contrast to Gtγ(50–71)farnesyl, Gtα(340–350) failed to inhibit Gt activation in an identical fluorescence assay, both in washed rod outer segment membranes (data not shown) and detergent solution (Fig. 3C). Only with a concentration 20–30 times higher than the IC50 of Gtα(340–350)HAA in the MII stabilization assay is the onset of competition by Gtα(340–350)HAA seen (Fig. 3C). Thus, despite its higher affinity for MII, this analog exhibited behavior similar to Gtα(340–350), showing a major discrepancy between the abilities to stabilize MII and to compete for Gt activation.
With the fluorescence assay, the Gt activation rate is saturated (33) when the content of active rhodopsin in the disc membrane exceeds 20% of the total native amount (22). Because of this effect, which arises from rate limitation by the membrane association of Gt, the assay tends to underestimate weak competition under conditions of full photoexcitation. This explains why the effect of native Gtα(340–350) becomes unmeasurably small. In special vesicle preparations with high lipid/rhodopsin ratio, this limitation can be overcome, and an effect of the native peptide can be observed (Fig. 3D).
Gtγ(50–71)farnesyl Peptide Cannot Replace the Gtβγ Complex in the Catalytic Interaction with the Receptor.
Because Gtγ(50–71)farnesyl can fully replace the holoprotein in the stabilization of the receptor in its MII state, the question arises whether the binding of this peptide to free receptor binding sites for the Gt γ-subunit suffices to induce nucleotide exchange in the Gt α-subunit. Addition of the two purified subunits in 1:2 stoichiometry gives a normal fluorescence change (Fig. 3E). A much lower activity is seen when the Gt α-subunit is present alone (plus a small residual amount of Gtβγ complex). Addition of Gtγ(50–71)farnesyl peptide in a subsaturating amount does not enhance this activity but suppresses it further. The final addition of Gtβγ complex restores the original activity to the degree expected from competition of Gtγ(50–71)farnesyl with the reconstituted Gtαβγ complex (compare Figs. 3 A and E Inset). The experiment shows that Gtγ(50–71)farnesyl cannot replace the Gtβγ complex as a helper in nucleotide exchange catalysis.
Quantitative Analysis by Real-Time Light Scattering Measurements.
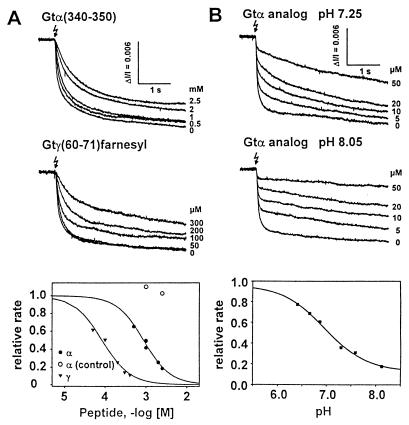
To quantitate the competition of the peptides in the native system, we used the real-time light scattering assay of Gt activation [dissociation signal (25, 26)]. With this technique, the competition of the Gtα(340–350) peptide is quantitatively measurable, showing that the IC50 of Gtα(340–350) in competing with Gt is higher than that of Gtγ(60–71) by a factor of 18 (Fig. 4A).
Figure 4.
Rate of receptor-catalyzed Gt activation from light scattering changes. After flash-induced (flash symbol) activation, rhodopsin catalyzes nucleotide exchange in Gt. GTP-bound Gt dissociates rapidly from the rod outer-segment membrane. All recordings are light scattering dissociation signals (25, 26), real-time measures of the rate at which membrane-bound Gt is rapidly activated. Binding of Gtα(340–350) or Gtγ(50–71)farnesyl inhibits the rate of Gt activation in a dose-dependent manner with an IC50 of 850 μM or 46 μM, respectively (see the dose-rate curves in Fig. 4A Bottom). Sequence of Gtα(control), IRENLKDCGAF. Competition by the Gtα(340–350) high-affinity analog (VLEDLKSCGLF) (Fig. 4B) is pH-dependent; the relative rate in the slow phase (from fits with two rate constants to records as shown above) shows an expressed pH dependence (Fig. 4B Bottom). Data are the relative rates, i.e., the measured rates in the presence of peptide (indicated concentrations in the left panel, 20 μM in the right panel) normalized to the rate without peptide.
Binding of the Gtα(340–350)HAA effectively traps active rhodopsin after a delay that is explained by the lower bimolecular reaction rate with this peptide, resulting from its lower concentration. The onset of trapping occurs earlier at higher peptide concentration. In the subsequent slow phase, active Gt is produced through the bottleneck of free receptor, arising from the off-rate of the peptide. The competition by Gtα(340–350)HAA shows a pH dependency (Fig. 4B) with a higher effectiveness at basic pH, in remarkable similarity to the photoregeneration results. These data show the additional pH dependence caused by the peptide on top of the intrinsic pH dependency of Gt activation in this assay (Fig. 5). Much less, if any, pH dependency is seen with the Gtγ(50–71)farnesyl peptide (not shown).
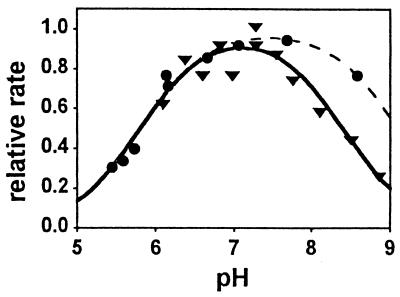
Figure 5.
pH-rate profile for rhodopsin-catalyzed Gt activation. pH dependence of the rate of the fast phase of the dissociation signal yields a “bell-shaped” pH-rate profile (different symbols identify two independent experiments). The data were fit to a product of two Henderson–Hasselbalch-type titration curves, yielding pKa1 = 5.8 (both curves), pKa2 = 8.4 (solid line) and 9.1 (dashed line; independent measurement with a second sample), respectively. For better comparison, data are normalized to the maximum rate obtained by the respective fit.
Proton Uptake Measurements.
The observed pH dependence of R*–peptide interactions could be caused by protonation changes of the receptor, peptides, or both. We tried to resolve the origin of the pH dependence by measuring the protonation state of the receptor on binding to the peptides. Unfortunately, the spectroscopic technique used for identification of the MIIa and MIIb subforms (21), which monitors flash-induced protonation changes by means of pH-indicator dyes, failed to give clear answers. This technique shows net protonation effects. Because the receptor and the peptides may both change their protonation state on binding, the source or target of protons released or taken up is hard to identify unambiguously.
DISCUSSION
Two-Site Interaction Between Receptor and G-Protein.
The present investigation confirms that synthetic peptides from the C termini of the Gt α- and γ-subunits, i.e., Gtα(340–350) (10, 14) and Gtγ(50–71)farnesyl (15) recognize the active form of rhodopsin with similar affinity and can replace the holoprotein in stabilizing the active Meta II photointermediate (Fig. 1). Although of similar affinity to the MII state, the peptides are different in their specific signaling interactions, as concluded from their different competition for active rhodopsin in the Gt activation process, both in membranes and in detergent solution (Fig. 3) and their distinct pH/rate profile [both in competition and photoregeneration (Figs. 2 and 4)]. Consistent correlation between the ability of the Gtγ(50–71)farnesyl to stabilize MII and compete for the rhodopsin catalyzed Gt activation and the inability of Gtγ(50–71)farnesyl to substitute for Gtβγ in Gtα activation are strong support for the direct involvement of the Gtβγ complex in a nucleotide exchange reaction on Gtα. Such direct participation (opposite to the current view of membrane/receptor targeting function of Gtβγ) was suggested recently (34, 35). It was based on virtual molecular displacement analysis, showing that Gβγ occupies the space of a nucleotide exchange factor for Gα, by analogy to the role of a nucleotide exchange factor EF-Ts recently crystallized with its partner, the elongation factor Tu (EF-Tu) (35). If, in fact, receptors use both Gα and Gβγ to transmit activating structural changes toward the nucleotide binding site, the receptor may reserve distinct signaling mechanisms for each of these interactions.
Our results argue that this indeed is the case: Because the peptides compete differently against the activation of Gt while they stabilize MII equally well, the peptide data are best explained by a “partial-agonist” behavior, with affinities for two different domains on the receptor (both present in the Meta II states, i.e., with deprotonated Schiff base). Gt may present binding sites (Fig. 6; see the discussion below) for binding to corresponding domains of photoexcited rhodopsin during different stages of the catalytic process. This should be reflected in the receptor surface, presumably in the loop structures. And indeed, loop mutations that affect defined steps in the catalytic sequence have been described (1, 23, 36), as well as a decoupled loop mobility indicative of a localized active conformation that is observed with the E134Q mutant in the dark state (37).
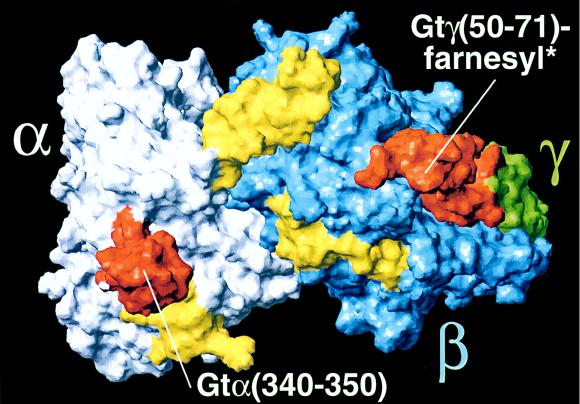
Figure 6.
Putative rhodopsin interacting surface of Gt. Gtα(340–350) and Gtγ(50–71)farnesyl are shown in orange at the opposite sides of the Gt surface. Other domains interacting with active rhodopsin are in yellow (9); also see Methods. Gtα(340–350) is shown in its conformation bound to the active receptor as obtained from NMR studies (18). *Only part of the Gtγ(50–71)farnesyl sequence is shown, namely Gtγ(50–66), as resolved by x-ray crystallography (28).
Sequential Fit of G-Protein Domains to the Active Receptor.
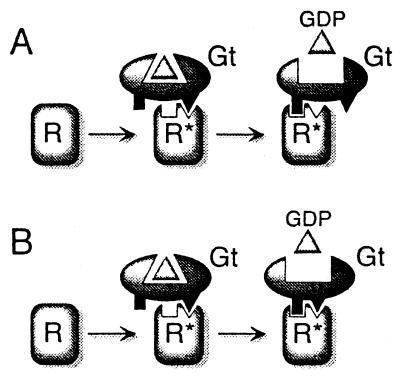
Two principal possibilities exist for the binding of two matching pairs of sites between the G-protein and the receptor, namely, (i) the two receptor binding sites operate sequentially, one site at a time (Fig. 7A), or (ii) the interaction with one site induces the interaction of the second site, so that both sites eventually contribute simultaneously to the catalytic reaction (Fig. 7B).
Figure 7.
Two-site sequential fit scheme of rhodopsin–transducin interaction. Scheme showing microscopic and gross-structural determinants of receptor-catalyzed G-protein activation. (A) Exclusive sequential fit, one site at a time, alternatively; (B) interaction with one site induces conformational change in Gt and simultaneous interaction at two sites; a concurrent conformational change in R*, not shown in the figure, is possible.
Though both C-terminal regions of Gtα and Gtγ have been localized to a common surface of Gt by x-ray studies (Fig. 6), the distance between these two sites in a GDP-bound state of Gt appears too large to interact with rhodopsin at the same time [estimated minimum of 40 Å between Gtα- and Gtγ- C termini, as compared with a maximum distance between loop sites at rhodopsin′s cytoplasmic surface of 35 Å (38)]. Thus, Gt holoprotein in its inactive conformation (Fig. 6) is not likely to present the binding sites on the Gt α- and γ-subunit at the right distance, though the accurate distances and positions of the binding sites on both the receptor and the G-protein require further experimental data.
We have shown in Fig. 3E that the Gtγ(50–71)farnesyl, at a concentration at which it successfully stabilizes MII, cannot replace the Gt βγ-complex in nucleotide exchange catalysis. This shows that nucleotide exchange catalysis eventually relies on the geometry of interaction in matching pairs of at least two sites between receptor and Gt holoprotein. If this means that, for catalysis, both sites have to bind simultaneously (Fig. 7B), a major conformational change in the Gt holoprotein or in the active receptor or both would be required. It would overcome the discrepancy between the relatively small size of the receptor and its large footprint on Gt.
Microscopic Recognition of Domains and Gross-Structural Interlocking in Receptor G-Protein Interaction.
The pH dependence of the R*-peptide interaction, as seen in the photoregeneration experiments and the light scattering assay of Gt activation, shows a discrepancy with the known behavior of the Gt holoprotein (Fig. 5).
In the photoregeneration experiments, the reversion of metarhodopsin II to its dark form was clearly blocked more efficiently at basic pH by both peptides. Because these measurements were performed with purified rhodopsin in dodecyl-maltoside, where it was shown that two isospectral subforms of MII, MIIa, and MIIb, exist (6), a first conclusion could be that the peptides prefer binding to the subform enriched at basic pH. This is MIIa, which is in equilibrium with the surface protonated isospectral MIIb conformation (21). The existence of such spectrally identical subforms in native membranes was supported by recent studies of UV/Vis absorption changes (39) and the electrical effects accompanying MII formation (40). However, peptide binding to MIIa would be in striking contrast with the behavior of the Gt holoprotein, which is known to bind to the MIIb form, at least in detergent solution (20).
The peptide efficiency in the light scattering activation assay is also pH-dependent, although to a varying degree. Competition by the Gtα(340–350)HAA peptide decreases the relative rate of activation to one-fourth at pH 8 compared with pH 6, in qualitative accordance with the higher efficiency of the peptides seen in the photoregeneration data. Gtγ(60–71)farnesyl peptide exhibited only a slight pH dependency but with the same general tendency, i.e., better inhibition at basic pH.
Interaction of R* with the holo-Gt (and also with rhodopsin kinase) has the well known “bell-shaped” pH-rate profile [Fig. 5 (3, 4)], dropping to low efficiencies at high and low pH with a maximum around pH 7. We can now propose that the increasing rate of Gt activation with increasing pH in the acidic branch of the profile reflects the pH dependency of microscopic recognition of domains, which occurs independently of the specific geometry in which they are arranged in conformations MIIa and MIIb, respectively. The decreasing rate at high pH can consistently be attributed (4) to the pH-dependent formation (6, 21) of the receptor conformation termed MIIb, which satisfies the gross-structural constraints of catalytic receptor/G-protein interaction. Although we believe that MIIa is a necessary step in reaching rhodopsin’s active conformation (39), its role in nucleotide exchange catalysis (Fig. 7) is not clear yet. Binding of holo-transducin in the MIIa conformation may occur, but it is unknown whether a signal is thereby transmitted. With the kinetic parameters under cellular conditions, Gt will not frequently encounter MII in the MIIa form.
Presence of the Two-Step Mechanism in Other Systems.
Our results demonstrate that individual domains of Gt can discriminate between their respective binding domains on active states of rhodopsin. A scheme of receptor activation with analogies to the sequence of rhodopsin’s intermediate states (41) applies to ligand-activated receptors (1, 42, 43). It will be of interest to explore whether the mechanism elucidated here applies to G-protein-coupled receptors in general.
Acknowledgments
We are indebted to Peter Henklein (Institut für Biochemie, Charité) for peptide synthesis. This work was supported by Deutsche Forschungsgemeinschaft (DFG) and Fonds de Chemischen Industrie (FCI) grants to K.P.H.
ABBREVIATIONS
- G-protein
heterotrimeric guanine nucleotide binding protein
- Gt
retina rod cell-specific G-protein
- Gtα
Gtβ, and Gtγ, alpha-, beta-, and gamma-subunits of Gt
- Gtα(340–350)HAA
high-affinity analog of Gtα(340–350)
- MII
metarhodopsin II
- MI
metarhodopsin I
- WM
washed rod outer segment membranes
- DM
n-dodecyl β-d-maltopyranoside
- BTP
1,3-bis[tris(Hydroxymethyl)methylamino]propane
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Helmreich E J, Hofmann K P. Biochim Biophys Acta. 1996;1286:285–322. doi: 10.1016/s0304-4157(96)00013-5. [DOI] [PubMed] [Google Scholar]
- 2.Sakmar T P. Prog Nucleic Acid Res Mol Biol. 1998;59:1–34. doi: 10.1016/s0079-6603(08)61027-2. [DOI] [PubMed] [Google Scholar]
- 3.Cohen G B, Oprian D D, Robinson P R. Biochemistry. 1992;31:12592–12601. doi: 10.1021/bi00165a008. [DOI] [PubMed] [Google Scholar]
- 4.Hofmann, K. P., in Proceedings of the 224th Symposium of the Novartis Foundation, ed. Goode, J., Wiley, Chichester, U.K., in press.
- 5.Kibelbek J, Mitchell D C, Beach J M, Litman B J. Biochemistry. 1991;30:6761–6768. doi: 10.1021/bi00241a019. [DOI] [PubMed] [Google Scholar]
- 6.Hofmann K P, Jäger S, Ernst O P. Isr J Chem. 1995;35:339–355. [Google Scholar]
- 7.Bohm A, Gaudet R, Sigler P B. Curr Opin Biotechnol. 1997;8:480–487. doi: 10.1016/s0958-1669(97)80072-9. [DOI] [PubMed] [Google Scholar]
- 8.Sprang S R. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 9.Hamm H E. J Biol Chem. 1998;273:669–672. doi: 10.1074/jbc.273.2.669. [DOI] [PubMed] [Google Scholar]
- 10.Hamm H E, Deretic D, Arendt A, Hargrave P A, König B, Hofmann K P. Science. 1988;241:832–835. doi: 10.1126/science.3136547. [DOI] [PubMed] [Google Scholar]
- 11.Hamm H E, Rarick H M. Methods Enzymol. 1994;237:423–436. doi: 10.1016/s0076-6879(94)37079-6. [DOI] [PubMed] [Google Scholar]
- 12.Acharya S, Saad Y, Karnik S S. J Biol Chem. 1997;272:6519–6524. doi: 10.1074/jbc.272.10.6519. [DOI] [PubMed] [Google Scholar]
- 13.Garcia P D, Onrust R, Bell S M, Sakmar T P, Bourne H R. EMBO J. 1995;14:4460–4469. doi: 10.1002/j.1460-2075.1995.tb00125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin E L, Rens Domiano S, Schatz P J, Hamm H E. J Biol Chem. 1996;271:361–366. doi: 10.1074/jbc.271.1.361. [DOI] [PubMed] [Google Scholar]
- 15.Kisselev O G, Ermolaeva M V, Gautam N. J Biol Chem. 1994;269:21399–21402. [PubMed] [Google Scholar]
- 16.Kisselev O G, Pronin A, Ermolaeva M V, Gautam N. Proc Natl Acad Sci USA. 1995;92:9102–9106. doi: 10.1073/pnas.92.20.9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kisselev O G, Ermolaeva M V, Gautam N. J Biol Chem. 1995;270:25356–25358. doi: 10.1074/jbc.270.43.25356. [DOI] [PubMed] [Google Scholar]
- 18.Kisselev O G, Kao J, Ponder J W, Fann Y C, Gautam N, Marshall G R. Proc Natl Acad Sci USA. 1998;95:4270–4275. doi: 10.1073/pnas.95.8.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pulvermüller A, Maretzki D, Rudnicka Nawrot M, Smith W C, Palczewski K, Hofmann K P. Biochemistry. 1997;36:9253–9260. doi: 10.1021/bi970772g. [DOI] [PubMed] [Google Scholar]
- 20.Arnis S, Hofmann K P. Biochemistry. 1995;34:9333–9340. doi: 10.1021/bi00029a008. [DOI] [PubMed] [Google Scholar]
- 21.Arnis S, Hofmann K P. Proc Natl Acad Sci USA. 1993;90:7849–7853. doi: 10.1073/pnas.90.16.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jäger S, Palczewski K, Hofmann K P. Biochemistry. 1996;35:2901–2908. doi: 10.1021/bi9524068. [DOI] [PubMed] [Google Scholar]
- 23.Ernst O P, Hofmann K P, Sakmar T P. J Biol Chem. 1995;270:10580–10586. doi: 10.1074/jbc.270.18.10580. [DOI] [PubMed] [Google Scholar]
- 24.Heck M, Hofmann K P. Biochemistry. 1993;32:8220–8227. doi: 10.1021/bi00083a024. [DOI] [PubMed] [Google Scholar]
- 25.Kühn H, Bennett N, Michel Villaz M, Chabre M. Proc Natl Acad Sci USA. 1981;78:6873–6877. doi: 10.1073/pnas.78.11.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schleicher A, Hofmann K P. J Membr Biol. 1987;95:271–281. doi: 10.1007/BF01869489. [DOI] [PubMed] [Google Scholar]
- 27.Koradi R, Billeter M, Wüthrich K. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 28.Lambright D G, Sondek J, Bohm A, Skiba N P, Hamm H E, Sigler P B. Nature (London) 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 29.Onrust R, Herzmark P, Chi P, Garcia P D, Lichtarge O, Kingsley C, Bourne H R. Science. 1997;275:381–384. doi: 10.1126/science.275.5298.381. [DOI] [PubMed] [Google Scholar]
- 30.Taylor J M, Jacob Mosier G G, Lawton R G, VanDort M, Neubig R R. J Biol Chem. 1996;271:3336–3339. doi: 10.1074/jbc.271.7.3336. [DOI] [PubMed] [Google Scholar]
- 31.Guy P M, Koland J G, Cerione R A. Biochemistry. 1990;29:6954–6964. doi: 10.1021/bi00482a003. [DOI] [PubMed] [Google Scholar]
- 32.O’Brien D F. Methods Enzymol. 1982;81:378–384. doi: 10.1016/s0076-6879(82)81055-0. [DOI] [PubMed] [Google Scholar]
- 33.Hofmann K P. In: GTPases in Biology—Handbook of Experimental Pharmacology, Vol. 108/II. Dickey B, Birnbaumer L, editors. Berlin: Springer; 1993. pp. 267–290. [Google Scholar]
- 34.Iiri T, Farfel Z, Bourne H R. Nature (London) 1998;394:35–38. doi: 10.1038/27831. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Jiang Y, Meyering Voss M, Sprinzl M, Sigler P B. Nat Struct Biol. 1997;4:650–656. doi: 10.1038/nsb0897-650. [DOI] [PubMed] [Google Scholar]
- 36.Franke R R, König B, Sakmar T P, Khorana H G, Hofmann K P. Science. 1990;250:123–125. doi: 10.1126/science.2218504. [DOI] [PubMed] [Google Scholar]
- 37.Kim J M, Altenbach C, Thurmond R L, Khorana H G, Hubbell W L. Proc Natl Acad Sci USA. 1997;94:14273–14278. doi: 10.1073/pnas.94.26.14273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unger V M, Hargrave P A, Baldwin J M, Schertler G F. Nature (London) 1997;389:203–206. doi: 10.1038/38316. [DOI] [PubMed] [Google Scholar]
- 39.Szundi I, Mah T L, Lewis J W, Jäger S, Ernst O P, Hofmann K P, Kliger D S. Biochemistry. 1998;37:14237–14244. doi: 10.1021/bi981249k. [DOI] [PubMed] [Google Scholar]
- 40.Dickopf S, Mielke T, Heyn M P. Biochemistry. 1998;37:16888–16897. doi: 10.1021/bi981879m. [DOI] [PubMed] [Google Scholar]
- 41.Fahmy K, Siebert F, Sakmar T P. Biophys Chem. 1995;56:171–181. doi: 10.1016/0301-4622(95)00030-2. [DOI] [PubMed] [Google Scholar]
- 42.Samama P, Cotecchia S, Costa T, Lefkowitz R J. J Biol Chem. 1993;268:4625–4636. [PubMed] [Google Scholar]
- 43.Gudermann T, Kalkbrenner F, Schultz G. Annu Rev Pharmacol Toxicol. 1996;36:429–459. doi: 10.1146/annurev.pa.36.040196.002241. [DOI] [PubMed] [Google Scholar]