Abstract
Tandem repeat amplification of resistance determinants in Haemophilus influenzae plasmids is associated with the occurrence of separate circular DNA molecules. They were demonstrated to represent mono- and multimeric forms of the amplifiable segments of the plasmids which comprise the respective resistance transposons and an additional region designated as an amplification sequence. The latter region mediates the recombinational events involved in amplification. The DNA circles apparently lack the ability to replicate autonomously but most probably provide an effective means for the translocation of resistance genes from one plasmid to another.
Full text
PDF
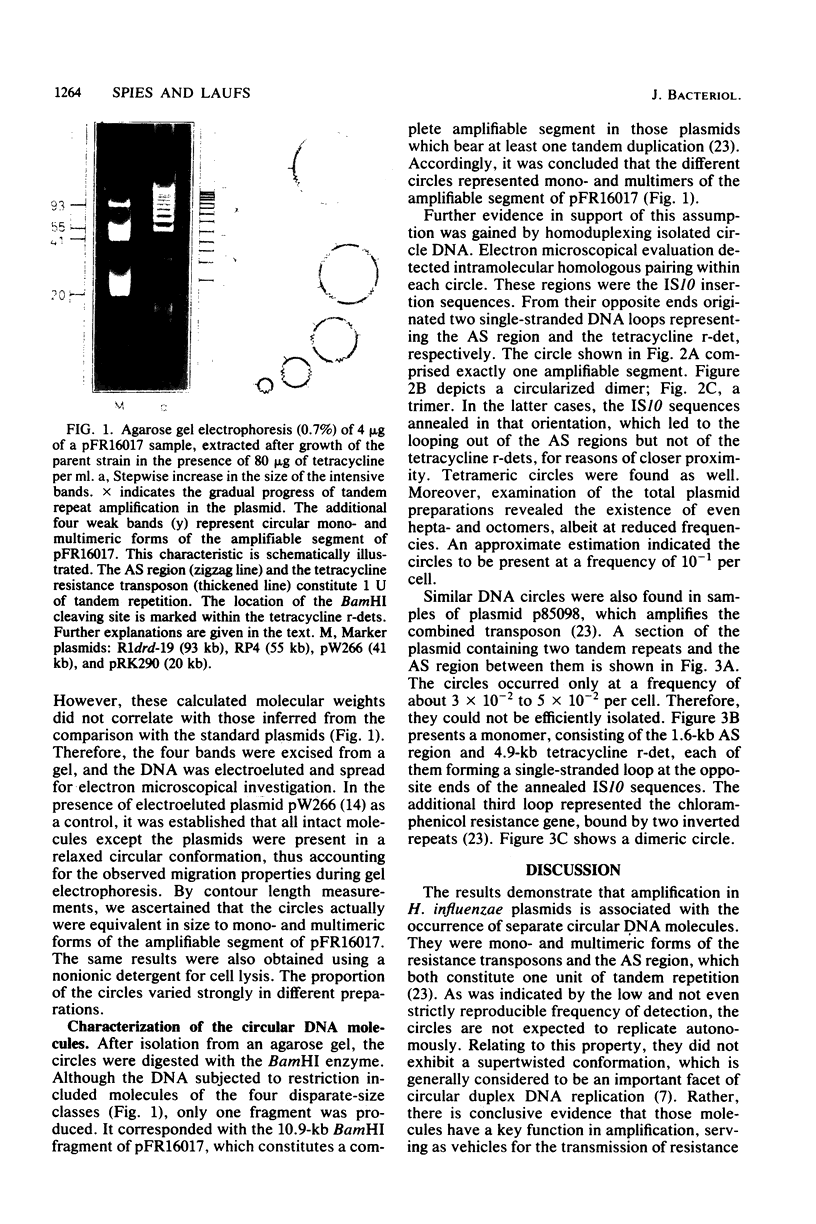
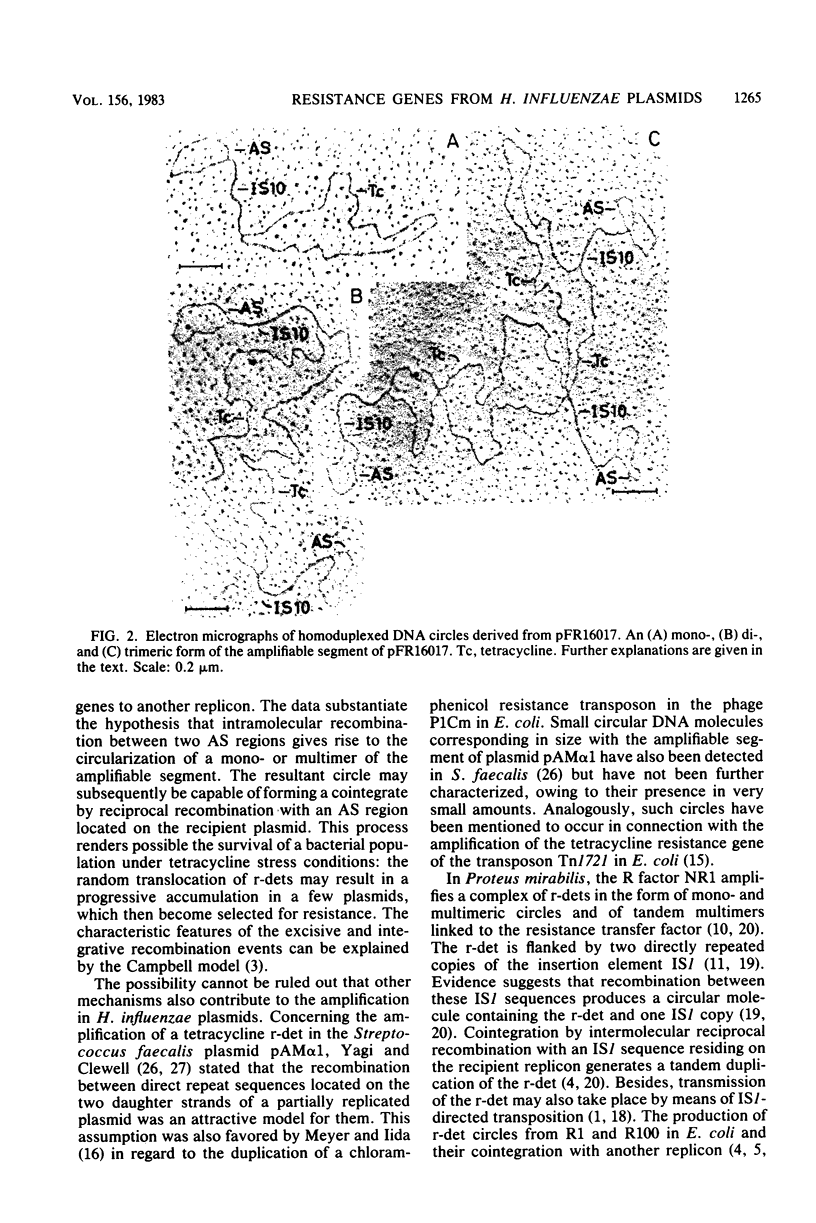


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arber W., Iida S., Jütte H., Caspers P., Meyer J., Hänni C. Rearrangements of genetic material in Escherichia coli as observed on the bacteriophage P1 plasmid. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1197–1208. doi: 10.1101/sqb.1979.043.01.136. [DOI] [PubMed] [Google Scholar]
- Barth P. T., Datta N., Hedges R. W., Grinter N. J. Transposition of a deoxyribonucleic acid sequence encoding trimethoprim and streptomycin resistances from R483 to other replicons. J Bacteriol. 1976 Mar;125(3):800–810. doi: 10.1128/jb.125.3.800-810.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M., Clerget M., Caro L. IS1-promoted events associated with drug-resistance plasmids. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):157–165. doi: 10.1101/sqb.1981.045.01.025. [DOI] [PubMed] [Google Scholar]
- Chandler M., Silver L., Lane D., Caro L. Properties of an autonomous r-determinant from R100.1. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1223–1231. doi: 10.1101/sqb.1979.043.01.139. [DOI] [PubMed] [Google Scholar]
- Coleman D. C., Foster T. J. Analysis of the reduction in expression of tetracycline resistance determined by transposon Tn10 in the multicopy state. Mol Gen Genet. 1981;182(1):171–177. doi: 10.1007/BF00422786. [DOI] [PubMed] [Google Scholar]
- Cozzarelli N. R. DNA gyrase and the supercoiling of DNA. Science. 1980 Feb 29;207(4434):953–960. doi: 10.1126/science.6243420. [DOI] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund T., Normark S. Recombination between short DNA homologies causes tandem duplication. Nature. 1981 Jul 16;292(5820):269–271. doi: 10.1038/292269a0. [DOI] [PubMed] [Google Scholar]
- Hashimoto H., Rownd R. H. Transition of the R factor NR1 and Proteus mirabilis: level of drug resistance of nontransitioned and transitioned cells. J Bacteriol. 1975 Jul;123(1):56–68. doi: 10.1128/jb.123.1.56-68.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Otsubo E., Davidson N., Saedler H. Electron microscope heteroduplex studies of sequence relations among bacterial plasmids: identification and mapping of the insertion sequences IS1 and IS2 in F and R plasmids. J Bacteriol. 1975 May;122(2):764–775. doi: 10.1128/jb.122.2.764-775.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn G., Laufs R., Kaulfers P. M., Kolenda H. Molecular nature of two Haemophilus influenzae R factors containing resistances and the multiple integration of drug resistance transposons. J Bacteriol. 1979 May;138(2):584–597. doi: 10.1128/jb.138.2.584-597.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R. A., Berg D. E., Allet B., Reznikoff W. S. Restriction enzyme cleavage map of Tn10, a transposon which encodes tetracycline resistance. J Bacteriol. 1979 Jan;137(1):681–685. doi: 10.1128/jb.137.1.681-685.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs R., Riess F. C., Jahn G., Fock R., Kaulfers P. M. Origin of Haemophilus influenzae R factors. J Bacteriol. 1981 Aug;147(2):563–568. doi: 10.1128/jb.147.2.563-568.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattes R., Burkardt H. J., Schmitt R. Repetition of tetracycline resistance determinant genes on R plasmid pRSD1 in Escherichia coli. Mol Gen Genet. 1979 Jan 10;168(2):173–184. doi: 10.1007/BF00431443. [DOI] [PubMed] [Google Scholar]
- Meyer J., Iida S. Amplification of chloramphenicol resistance transposons carried by phage P1Cm in Escherichia coli. Mol Gen Genet. 1979 Oct 3;176(2):209–219. doi: 10.1007/BF00273215. [DOI] [PubMed] [Google Scholar]
- Meyers J. A., Sanchez D., Elwell L. P., Falkow S. Simple agarose gel electrophoretic method for the identification and characterization of plasmid deoxyribonucleic acid. J Bacteriol. 1976 Sep;127(3):1529–1537. doi: 10.1128/jb.127.3.1529-1537.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo E., Zenilman M., Ohtsubo H. Plasmids containing insertion elements are potential transposons. Proc Natl Acad Sci U S A. 1980 Feb;77(2):750–754. doi: 10.1073/pnas.77.2.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne K., Cohen S. N. Occurrence of insertion sequence (IS) regions on plasmid deoxyribonucleic acid as direct and inverted nucleotide sequence duplications. J Bacteriol. 1975 May;122(2):776–781. doi: 10.1128/jb.122.2.776-781.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöffl F., Arnold W., Pühler A., Altenbuchner J., Schmitt R. The tetracycline resistance transposons Tn1721 and Tn1771 have three 38-base-pair repeats and generate five-base-pair direct repeats. Mol Gen Genet. 1981;181(1):87–94. doi: 10.1007/BF00339010. [DOI] [PubMed] [Google Scholar]
- Silver L., Chandler M., Lane H. E., Caro L. Production of extrachromosomal r-determinant circles from integrated R100.1: involvement of the E. coli recombination system. Mol Gen Genet. 1980;179(3):565–571. doi: 10.1007/BF00271746. [DOI] [PubMed] [Google Scholar]
- Spies T., Laufs R., Riess F. C. Amplification of resistance genes in Haemophilus influenzae plasmids. J Bacteriol. 1983 Aug;155(2):839–846. doi: 10.1128/jb.155.2.839-846.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. P., Greenberg J., Rownd R. H. Generation of miniplasmids from copy number mutants of the R plasmid NR1. J Bacteriol. 1977 Dec;132(3):986–995. doi: 10.1128/jb.132.3.986-995.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H., Hashimoto H., Mitsuhashi S. Salmonella typhimurium LT2 mutation affecting the deletion of resistance determinants on R plasmids. J Bacteriol. 1980 Apr;142(1):145–152. doi: 10.1128/jb.142.1.145-152.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y., Clewell D. B. Amplification of the tetracycline resistance determinant of plasmid pAM alpha 1 in Streptococcus faecalis: dependence on host recombination machinery. J Bacteriol. 1980 Aug;143(2):1070–1072. doi: 10.1128/jb.143.2.1070-1072.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y., Clewell D. B. Identification and characterization of a small sequence located at two sites on the amplifiable tetracycline resistance plasmid pAMalpha1 in Streptococcus faecalis. J Bacteriol. 1977 Jan;129(1):400–406. doi: 10.1128/jb.129.1.400-406.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]