Abstract
Accurate placement of the division septum at the midpoint of Escherichia coli cells requires the combined action of a general division inhibitor (MinC), a site-specific suppressor of division inhibition (MinE), and an ATPase (MinD) that is required for proper functioning of both MinC and MinE. We previously showed that a functional MinE-Gfp fusion accumulates in a ring structure at/near the middle of cells. Here we show that functional Gfp-MinD accumulates alternately in either one of the cell halves in what appears to be a rapidly oscillating membrane association-dissociation cycle imposed by MinE. The results indicate that MinD represents a novel type of dynamic cellular element in bacteria, with multiple roles in directing the division apparatus to the middle of the cell.
Prokaryotic cells prepare for cytokinesis by assembly of the tubulin-like GTPase FtsZ into a ring-shaped structure at the site of division. Other proteins then are recruited to the FtsZ ring to form the mature septal ring organelle that mediates cell envelope invagination (1–4). In Escherichia coli, the division septum is placed accurately at the midpoint of the cell. Potential division sites (PDSs) are also present near the cell poles, but usage of these sites is specifically repressed by the products of the min operon MinC, D, and E (5–8). The MinC protein can inhibit FtsZ ring assembly at all PDSs, but requires MinD for this function (7, 9, 10). The MinD protein is well conserved and belongs to a family of ill-understood prokaryotic ATPases (11, 12). Mutants lacking MinC or MinD produce numerous minicells, reflecting frequent aberrant septation near the cell poles (7, 9, 13). The MinE protein suppresses the MinCD-mediated division block specifically at midcell, ensuring proper placement of the septum (7, 14–16). We recently showed that a functional MinE-Gfp fusion accumulates in a ring structure at/near the middle of cells (16). Surprisingly, assembly of the MinE ring also required MinD, and evidence indicates a role for MinD in directing MinE to the cell membrane (16). Whether MinD also is involved in the subsequent accumulation of MinE at midcell is not known.
Here we report on the remarkably dynamic properties of functional Gfp-tagged MinD in vivo. The results have important implications for the mechanism of division site selection in E. coli and reveal unforeseen complexity in the spatial organization of bacterial cells.
MATERIALS AND METHODS
Strains, Plasmids, and Phages.
Strains PB103 (dadR trpE trpA tna) and PB104 (PB103, minD1) (17), PB114 (PB103, ΔminCDE∷aph) (7), DX1 (PB114, recA∷Tn10) (11), and DR102/pDB346 (PB114, ftsZ0 recA∷Tn10/PλR∷ftsZ cI857) (16) have been described. DR104 (PB104, recA∷Tn10) was obtained by P1-mediated transduction of recA∷Tn10 to PB104.
Plasmids pDR119 (bla+ lacIq+ Plac∷gfpmut2-minD) and pDR122 (bla+ lacIq+ Plac∷gfpmut2-minD minE) are derivatives of pMLB1113 (7) and were constructed in several steps. Plasmid pGfpmut2 (18) was used as a template for a PCR using primers 5′-CGGGGATCCTCTAGATTTAAGAAG-3′ and 5′-GATGCTAGCTTTGTATAGTTCATCCATG-3′, which were designed to introduce XbaI and NheI sites (underlined), respectively, upstream and downstream of the gfpmut2 ORF. Treatment with XbaI and NheI resulted in a 739-bp fragment, which was ligated to similarly treated pET-21c (Novagen), yielding pDR107c. In a second PCR, EcoRI and SalI sites were introduced on either side of the minD ORF by using primers 5′-CAAGGAATTCATATGGCACGCATTATTGTTG-3′ and 5′-GTACCGTCGACTTATCCTCCGAACAAGCG-3′. The 820-bp EcoRI–SalI fragment was ligated to EcoRI- and SalI-digested pDR107c, resulting in plasmid pDR117. Plasmid pDR119 was obtained by inserting the 1,669-bp BglII–HindIII fragment of pDR117 into the BamHI and HindIII sites of pMLB1113. Replacement of the 2,538-bp PacI–SacI fragment of pDR119 with the 2,858-bp PacI–SacI fragment of pDB175 (7) yielded pDR122.
Phages λDR119 (imm21 bla+ lacIq+ Plac∷gfpmut2-minD) and λDR122 (imm21 bla+ lacIq+ Plac∷gfpmut2-minD minE) were obtained by crossing pDR119 and pDR122, respectively, with λNT5 (7). Both plasmids and phages encode a 58-kDa fusion protein in which the complete Gfpmut2 peptide (18) is fused to the N terminus of the complete MinD peptide with the linker peptide ASMTGGQQMGRIRIH. In addition, pDR122 and λDR122 contain native minE, which is present in its natural context, immediately downstream of the minD coding sequence (7).
Growth Conditions.
Cells were grown at 30°C in M9 minimal salts medium supplemented with 50 μg/ml of tryptophan, 0.2% casamino acids, 0.2% maltose, and isopropyl β-d-thiogalactoside (IPTG) (as indicated), to an OD (600 nm) of 0.2–0.4. Doubling times ranged from 175 to 200 min.
Cells grown in LB medium (≈50-min doubling time) showed very similar behavior of Gfp-MinD (not shown). However, oscillation parameters were difficult to determine accurately under these conditions because of a phenomenon that is likely related to our previous finding that the MinE-Gfp ring disassembled relatively rapidly on adherence of LB-grown cells to microscope slides (16). Thus, whereas virtually all cells that were in suspension showed the typical segregated localization pattern of Gfp-MinD, adherence of these cells to the surface of microscope slides appeared to cause a dispersal of Gfp-MinD throughout the cell body after only one or two oscillation cycles. In contrast, cells grown in minimal medium displayed sustained segregation/oscillation of Gfp-minD until the fluorescence signal became too weak to detect. Similarly, we have found that the apparent instability of the MinE ring during microscopy is significantly suppressed by growth of cells in minimal medium.
Microscopy and Other Methods.
For fluorescence and differential interference contrast (DIC) imaging, cells were immediately applied to a microscope slide and viewed with a Zeiss Axioplan-2 microscope outfitted with a Hamamatsu C4742–95 progressive scan cooled charge-coupled device camera and a plan-NEOFLUAR (100×, numerical aperture = 1.3) oil immersion objective by using either a 495-nm dichroic mirror, a 450- to 490-nm excitation filter and a 500- to 550-nm barrier filter (fluorescent images), or Nomarski optics (DIC images). For phase micrographs, cells were treated as described (7) and viewed with a plan-APO (100×, numerical aperture = 1.4) objective. Images were captured by using qed software and further manipulated by using Adobe photoshop. Exposure times for fluorescent images ranged from 0.5 to 1.0 sec. The positions of at least 113 division septa in randomly chosen cells were determined to calculate the percentage of polar septa. Western analyses were performed essentially as described (11).
RESULTS
Gfp-MinD Is Functional.
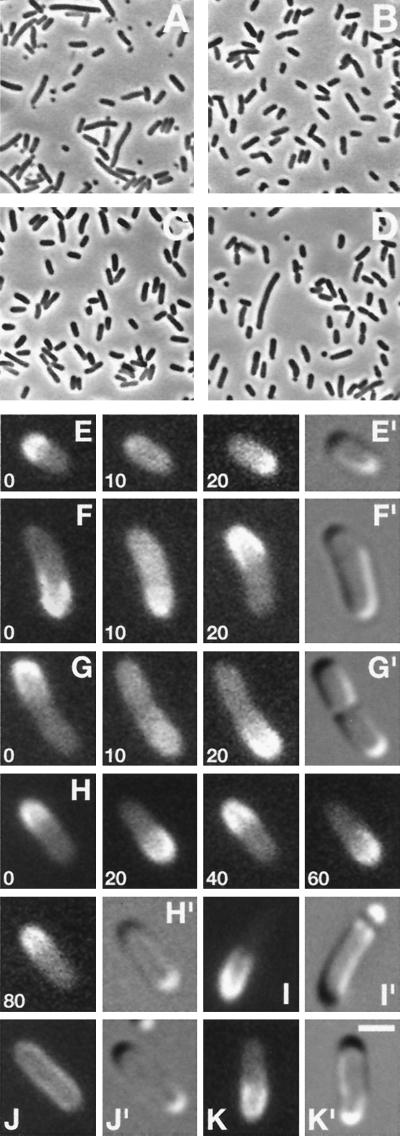
To monitor the location of MinD in living cells, we constructed lysogenic phages λDR119 (Plac∷gfp-minD) and λDR122 (Plac∷gfp-minD minE). Both phages express a bright variant of Gfp (Gfpmut2, ref. 18) fused to the N terminus of the complete MinD peptide (Gfp-MinD) under control of the IPTG-inducible lac promotor. In addition, λDR122 coexpresses native MinE with Gfp-MinD. The functionality of Gfp-MinD was tested by determining whether the phages could complement the classical minD1 mutation (5, 19) in strain DR104 (minD1 recA∷Tn10). As controls, the phages also were introduced into the wild-type (WT) strain PB103 and into strain PB114 (ΔminCDE), which lacks the complete min operon. As expected, both DR104(λDR119) and DR104(λDR122) lysogens showed the minicell phenotype (Min−) in the absence of inducer (Fig. 1A). In contrast, both lysogens reverted to a WT division phenotype in the presence of low concentrations of IPTG (37 μM), demonstrating that attachment of the Gfp moiety did not substantially interfere with MinD function (Fig. 1B). Under either condition, both PB103 lysogens were WT, whereas both PB114 lysogens were Min− (Table 1), because of the absence of MinC.
Figure 1.
Dynamic properties of functional Gfp-MinD in live cells. Phase (A–D), fluorescence (E–K), and differential interference contrast (E′–K′) micrographs showing properties of Gfp-MinD. Cells were grown either in the absence (A and C) or presence of 25 μM (D and I), or 37 μM (B, E–H, J, and K) IPTG. (A and B) Correction of minD1 by gfp-minD in strain DR104(λDR119) [minD1 recA∷Tn10 (Plac∷gfp-minD)]. (C and D) The induction of minicell formation by moderate overexpression of Gfp-MinD in strain PB103/pDR119 (wt/Plac∷gfp-minD). (E–H) Time-lapse images showing Gfp-MinD oscillation in normally dividing cells of strains DR104(λDR122) [minD1 recA∷Tn10 (Plac∷gfp-minDE)] (F and G), and PB103(λDR122) [wt(Plac∷gfp-minDE)] (E and H). Times are indicated in sec. (I) Gfp-MinD segregation and minicell formation in a PB103/pDR119 cell. (J and K) Gfp-MinD localization in strain PB114 (ΔminCDE) lysogenic for either λDR119 (Plac∷gfp-minD) (J) or λDR122 (Plac∷gfp-minDE) (K). [Bar represents 1 μm (E–K) or 4 μm (A–D).]
Table 1.
Biological activity, cellular distribution, and oscillation parameters of Gfp-MinD
| Exp. | Strain | Genotype | Phenotype
|
Dist. | Dwell (range) | Shift (range) | Cycle | |
|---|---|---|---|---|---|---|---|---|
| −IPTG | +IPTG | |||||||
| 1 | PB103(λDR119) | wt(Plac∷gfp-minD) | WT | WT | O | 33 (19–69) | 15 (7–21) | 96 |
| 2 | PB103(λDR122) | wt(Plac∷gfp-minDE) | WT | WT | O | 9 (5–14) | 8 (6–12) | 34 |
| 3 | PB103/pDR119 | wt/Plac∷gfp-minD | WT | Min− | O | 93 (27–290) | 22 (13–45) | 230 |
| 4 | PB103/pDR122 | wt/Plac∷gfp-minDE | WT | WT | O | 9 (5–16) | 10 (6–20) | 38 |
| 5 | PB114(λDR119) | [ΔminCDE(Plac∷gfp-minD)] | Min− | Min− | M | NA | NA | NA |
| 6 | PB114(λDR122) | [ΔminCDE(Plac∷gfp-minDE)] | Min− | Min− | O | 10 (5–17) | 10 (6–14) | 40 |
| 7 | DR104(λDR119) | minD1 recA∷Tn10(Plac∷gfp-minD) | Min− | WT | O | 35 (17–68) | 27 (13–49) | 124 |
| 8 | DR104(λDR122) | minD1 recA∷Tn10(Plac∷gfp-minDE) | Min− | WT | O | 10 (6–16) | 10 (5–17) | 40 |
| 9 | DR104(λDR122) | As exp. 8, but treated with CAM | Min− | WT | O | 10 (7–15) | 10 (6–13) | 40 |
Cells were grown in the absence or presence of 25 μM (experiments 3 and 4), 37 μM (experiments 1, 2, and 5–7) or 50 μM (experiments 8 and 9) IPTG. To determine the division phenotype, cells were chemically fixed and observed by phase microscopy. To determine the location and oscillation parameters of Gfp-MinD, cells grown in the presence of IPTG were immediately observed by fluorescence microscopy. The dwell period was defined as the period in which the location of Gfp-MinD appeared static, and the shift period as the period between two consecutive dwell periods. One complete oscillation cycle equals two dwell plus two shift periods. Values are given in s and represent the average of at least 26 events occurring in 20 individual cells. Experiment 9 was identical to 8, except that 45 min before examination chloramphenicol (CAM; 25 μg/ml) was added to the culture, resulting in complete cessation of growth within 5 min. Ranges are given in parentheses. WT, less than 4% of total septa were polar; Min−, minicell producing, more than 35% of septa were polar; O, segregating and oscillating; M, present along entire membrane, no net movement obvious; NA, not applicable.
Segregation and Oscillation of Gfp-MinD.
Microscopic inspection of the normally dividing DR104 and PB103 lysogens revealed a remarkable distribution of fluorescence (Fig. 1 E–H). Based on immuno-electron microscopy and cell fractionation studies, we previously concluded that MinD associates peripherally with the inner membrane (11). Accordingly, Gfp-MinD predominantly localized to the cell periphery. At any one time, however, the protein appeared to be present in only one of the cell halves in the majority of cells. Thus, fluorescence was present along one polar cap and a variable portion of the adjacent cylindrical part, up to approximately the middle of the cell. Even more striking was the observation that the protein dwelled for only 10–30 sec (depending on the phage used, see below) in this position, and then moved to the opposite cell half, where it dwelled again for 10–30 sec before shifting back to its original position, and so on (Fig. 1 E–H). This repeated relocation of the protein at regular intervals is illustrated in Fig. 1H, showing a typical cell of PB103(λDR122) in which Gfp-MinD relocated four times within a span of 80 sec. By using low excitation intensities to minimize signal degradation, we recorded up to 20 relocation events per cell within 350 sec before the signal became too weak to detect. During the shift period, the peripheral staining pattern became ill defined whereas fluorescence in the cytoplasm increased noticeably (Fig. 1 E–G, 10 sec), suggesting that the protein dissociated from the membrane before reassociating in the opposite cell half. This remarkable oscillatory behavior was observed in virtually all cells, including very small cells (Fig. 1E) and constricting cells (1G). Light intensities could be varied widely without affecting average dwell and shift times (not shown), indicating that oscillation was not induced by microscopy itself. Furthermore, identical behavior was observed in cells in which protein synthesis had been blocked with chloramphenicol (Table 1), excluding the formal possibility that what appeared as bulk movement reflected site-specific synthesis or degradation of Gfp-MinD.
Segregation/Oscillation of Gfp-MinD Requires MinE But Not MinC.
The behavior of Gfp-MinD in PB114(λDR122) cells (MinC−) was identical to that observed in normally dividing cells of strains PB103(λDR122) and DR104(λDR122), demonstrating that segregation and oscillation of the protein did not require MinC (Fig. 1K, Table 1). This finding was in stark contrast to the behavior of Gfp-MinD in PB114(λDR119) cells (MinC−, MinE−). The protein clearly still accumulated at the periphery of these cells, implying that neither MinE nor MinC is required for the apparent affinity of MinD for the cell membrane. However, Gfp-MinD failed to segregate as observed above, but was evenly distributed along the entire membrane (Fig. 1J). These results show that the cellular location of Gfp-MinD depends on MinE and strongly suggest that the MinE ring is instrumental in sequestering Gfp-MinD in either one cell half.
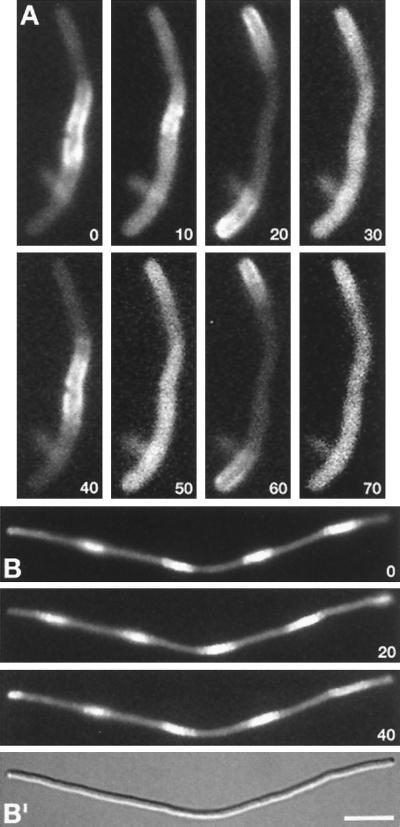
Segregation/Oscillation of Gfp-MinD in FtsZ− Filaments.
The MinE ring and septal ring are separate structures and FtsZ− filaments contain multiple MinE rings (16). To determine the distribution of Gfp-MinD in such filaments, we examined cells of strain DR102(λDR122)/pDB346 [ΔminCDE∷aph ftsZ0 recA∷Tn10 (Plac∷gfp-minD minE)/PλR∷ftsZ cI857] in which ftsZ expression was shut off by growth at 30°C (16). Time-lapse images of a short filament are shown in Fig. 2A. The filament appears to consist of one central and two polar segments, suggesting the presence of two MinE rings in this particular cell (Fig. 3C). At time zero (t = 0), Gfp-MinD is present at the cell’s periphery within the central segment, but is absent from both polar segments. Ten seconds later, the fusion is present throughout the filament, suggesting release from the membrane. At t = 20 sec, Gfp-MinD is located exclusively in the polar segments and has clearly accumulated at the cell’s periphery, indicating reassociation with the membrane at this stage. Next (t = 30 sec), fluorescence again is seen throughout the filament body, after which (t = 40 sec) the protein returns to the membrane in the central segment, completing one cycle. Longer filaments showed increased segmentation, but similar behavior of Gfp-MinD. Thus, the signal always seemed to move from fluorescent segments to neighboring nonfluorescent segment(s) and back again (Fig. 2B). Moreover, movement of Gfp-MinD between segments occurred in a coordinated manner, suggesting that relocation is triggered by a single signal that is sensed throughout the filaments, or that movement in one part of the cell quickly induces movement in adjacent parts. As in FtsZ+ MinE− cells (Fig. 1J), Gfp-MinD failed to accumulate in segments, but was evenly distributed along the periphery of FtsZ− MinE− filaments of strain DR102(λDR119)/pDB346 (not shown). These results show that FtsZ/septal rings are not required for Gfp-MinD segregation/oscillation and further support a determining role for the MinE ring in this phenomenon.
Figure 2.
Gfp-MinD localization in FtsZ− filaments. Time-lapse fluorescence images of a short (A) and long (B) filament of strain DR102(λDR122)/pDB346 [ΔminCDE∷aph ftsZ0 recA∷Tn10 (Plac∷gfp-minDE)/PλR∷ftsZ cI857]. Times are indicated in sec. (B′) A differential interference contrast image of the filament in B. Cells were grown at 30°C (resulting in repression of ftsZ expression) in the presence of 37 μM IPTG. [Bar represents 2.5 μm (A) or 5.0 μm (B).]
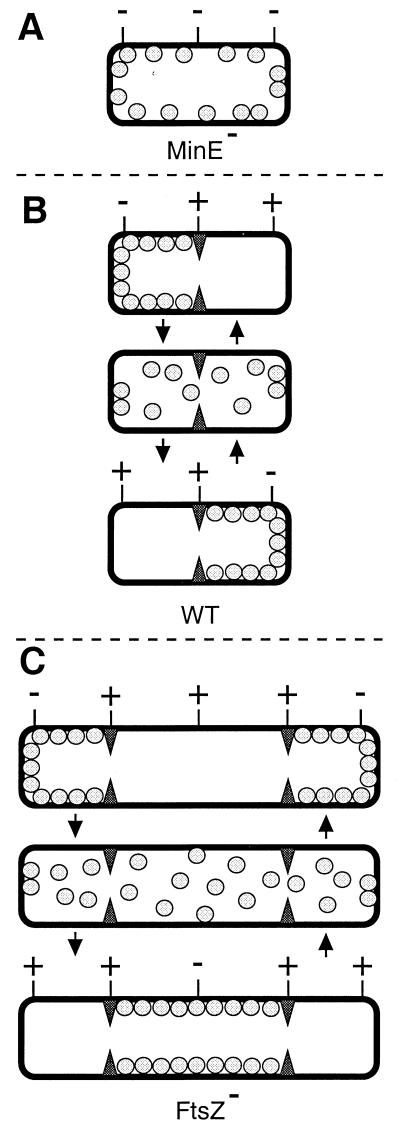
Figure 3.
Model for MinD and MinE action in preventing aberrant septation events. MinD is represented by gray spheres, the MinE ring by filled triangles, and PDSs by either a − (blocked by MinC/MinD action) or + (not blocked by MinC/MinD, available for assembly of FtsZ ring) sign. (A) In the absence of MinE, MinD is distributed evenly over the membrane. Provided MinC is present, this prevents septal ring formation at all PDSs, resulting in the formation of nonseptate filaments (7). (B) In WT cells, MinD oscillates from one side of the MinE ring to the other, alternately blocking division at each of the polar PDSs. For simplicity, it is assumed that the MinC/MinD division block is relieved as soon as MinD leaves a PDS, although it may well remain refractive to FtsZ assembly for some period afterward. (C) In the absence of FtsZ, multiple MinE rings define three or more cell segments. As in WT cells, MinD oscillates between the segments flanking each MinE ring.
Oscillation Frequency and Division Site Selection.
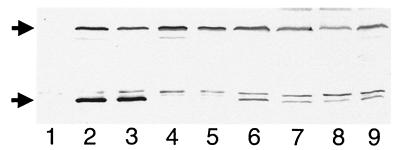
Additional observations suggested that MinE is not only required for the segregation of Gfp-MinD, but also affects its oscillation frequency. As shown in Table 1, λDR122 lysogens consistently displayed a ≈3-fold shorter oscillation cycle than λDR119 lysogens. Both types of lysogens expressed Gfp-MinD to a similar level (Fig. 4, lanes 2–5), suggesting that the increased dynamism of Gfp-MinD in λDR122 lysogens was caused by the coexpression of minE with gfp-minD. Also, because DR104 and PB103 carrying either phage were WT under these conditions, we conclude that oscillation cycles between 0.5 and 2.0 min are compatible with a normal division pattern (Table 1).
Figure 4.
Identification of Gfp-MinD with MinD-specific antiserum. Immunoblot showing Gfp-MinD (58.1 kDa, upper arrow) and native MinD (29.6 kDa, lower arrow) as detected with MinD-specific antiserum. Cells were grown in the presence of 37 μM (lanes 1–5) or 25 μM (lanes 6–9) IPTG to an OD (600 nm) of 0.3, and whole-cell extracts were prepared. Lanes 1–5 contained 20 μg total protein of strains PB114 [ΔminCDE] (lane 1); PB103(λDR119) [wt(Plac∷gfp-minD)] (lane 2); PB103(λDR122) [wt(Plac∷gfp-minDE)] (lane 3); PB114(λDR119) [ΔminCDE (Plac∷gfp-minD)] (lane 4); and PB114(λDR122) [ΔminCDE (Plac∷gfp-minDE)] (lane 5). Lanes 6–8 contained, respectively, 4, 2, and 1 μg of PB103/pDR119 (wt/Plac∷gfp-minD), and lane 9 contained 2 μg of PB103/pDR122 (wt/Plac∷gfp-minDE). Samples in lanes 6–9 were mixed with appropriate amounts of PB114 extract such that each lane contained 20 μg of total protein.
Evidence that longer oscillation cycles of MinD affect its proper function came from the study of cells in which Gfp-MinD was expressed at a higher level from multicopy plasmids. As with native MinD (7), expression of Gfp-MinD at low levels had no effect on the division pattern of WT cells (see above), whereas expression at sufficiently high levels induced a MinC-dependent division block (not shown). Interestingly, expression of either native MinD (not shown) or Gfp-MinD at intermediate levels resulted in incorrect placement of the septum, rather than in a division block. Thus, strain PB103/pDR119 (wt/Plac∷gfp-minD) was WT in the absence of inducer (Fig. 1C) but produced a significant number of minicells (45% of septa were polar) in the presence of 25 μM IPTG (Fig. 1D). In contrast, PB103/pDR122 (wt/Plac∷gfp-minD minE) produced very few minicells at this low concentration of inducer (4% polar septa). As judged from Western analyses, Gfp-MinD levels in cells carrying either plasmid were almost identical and 5- to 10-fold higher than in the corresponding lysogens (Fig. 4). Notwithstanding the fact that high levels of MinE can by itself cause a Min− phenotype (7, 14, 15), these results indicate that the Min− phenotype induced by moderate overexpression of MinD actually is suppressed by coexpression of MinE. At this level of expression, Gfp-MinD still clearly accumulated in either one of the cell halves in both PB103/pDR119 and PB103/pDR122 (Fig. 1I, Table 1). However, whereas the average oscillation cycle in PB103/pDR122 cells was equivalent to that in the PB103(λDR122) lysogen, this value was almost 6-fold higher in PB103/pDR119, and we observed the protein dwell for up to 5 min in individual cells of this strain (Table 1). These results indicate that Gfp-MinD and MinE affect the oscillation frequency of the former in opposite manners, with the frequency inversely related to the MinD/MinE ratio in the cell. Furthermore, the extended oscillation cycle of Gfp-MinD in PB103/pDR119 provides a credible explanation for asymmetric septation in these cells because prolonged dwelling of MinD in one cell half might reasonably be expected to increase the chance of uninhibited FtsZ ring assembly at the polar PDS in the opposite, unoccupied half. Therefore, these results support the notion that oscillation of MinD is not merely compatible with its function, but that a certain minimum oscillation frequency is, in fact, required to efficiently prevent septation at both polar PDSs.
DISCUSSION
Previously, we found that functional MinE-Gfp accumulates in an FtsZ-independent ring structure at/near the middle of cells (16). Here, we showed that functional Gfp-MinD displays a surprisingly asymmetric and highly dynamic localization pattern, which also does not require FtsZ, but depends on MinE. In aggregate, our results support a model (Fig. 3) in which (i) the MinE ring prevents MinC/MinD action at midcell, (ii) MinD accumulates alternately at the membrane on either side of the MinE ring, (iii) the MinD-dependent division block is quickly reversible, and (iv) frequent relocation of MinD ensures that aberrant FtsZ ring assembly is prevented in both cell halves. Both genetic studies in E. coli and two-hybrid assays have indicated that MinE is able to suppress MinCD-mediated division inhibition by interfering with the interaction between MinC and MinD (9, 13, 20). The present observations suggest an additional mechanism of suppression, i.e., by being sequestered on either side of the MinE ring, MinD simply may be prevented from occupying membrane at the proper division site at midcell. Possibly, both mechanisms play complementary roles in suppressing MinCD action in the middle of normally dividing cells.
The unique in vivo properties of Gfp-MinD deepen our understanding of division site selection in E. coli, but also present many new challenges. As shown previously (16), MinD is required for assembly of the MinE ring. Conversely, as shown here, MinE is required for MinD segregation/oscillation. How the two proteins establish and maintain such interdependent, but distinct, localization patterns is far from clear. The marked asymmetric accumulation of Gfp-MinD at the membrane in either one of the cell halves, as well as the finding that increased levels of Gfp-MinD reduces its relocation frequency, are suggestive of cooperative assembly of the protein into some higher-order, semistable structure. In addition, our results implicate MinE not only in the segregation of MinD, but also in stimulating relocation of the protein to the opposite cell half. Possibly, the MinE ring both acts as a gasket to sequester MinD in one of the cell halves, as well as induces disassembly of MinD from the membrane, which then is followed by reassembly of MinD on the other side of the ring. In any event, how MinD assembles at the membrane, and how MinE might promote its relocation are important questions requiring further experimentation.
The observation that Gfp-MinD continues to oscillate in deeply constricting cells (Fig. 1G) also raises a number of interesting questions. Does oscillation continue until septal closure and, if so, is MinD unequally distributed over the two daughter cells, or is septal closure coordinated with the oscillation cycle such that it coincides with a MinD shift phase? We previously failed to detect clear MinE-Gfp ring structures in deeply constricted cells (16). This could mean that MinD continues to oscillate even after the MinE ring is largely disassembled. Alternatively, the exact timing of MinE disassembly may be determined by specific growth conditions, or native MinE may behave differently in this respect than the tagged version. Further careful observations on the behavior of both MinD and MinE during the late stages of constriction should shed more light on these issues.
Perhaps the most pertinent question raised by this study is why should MinD oscillate between cell halves, rather than being simply distributed over both? We are intrigued by the notion that such an oscillator could, in principle, serve as a cellular measuring device. By continuously probing spatial boundaries, MinD might provide the cell with up-to-date information on the location of its middle. For example, if MinE, on forming a ring, became actively excluded from membrane occupied by MinD, then oscillation of the latter might drive the ring toward the cell center where, at equilibrium, the time-integrated concentration of MinD would be lowest. Such a mechanism is attractive because it might explain how the MinE ring is positioned and provide a compelling reason for MinD oscillation. Further work will be needed to explore this and other possibilities.
Finally, our findings highlight the fundamental differences between the mechanisms used by E. coli and Bacillus subtilis to impart topological specificity to MinCD-mediated division inhibition. B. subtilis lacks a MinE homologue (21–23), and site specificity is provided by an unrelated protein, DivIVA, which is absent in E. coli (24, 25). Recently, the localization of B. subtilis MinD was shown to depend on DivIVA, and both proteins were found to associate with the septal ring relatively late in the division cycle, but before the onset of constriction. In addition, both proteins were present at both polar caps, indicating they remain associated, seemingly indefinitely, with the newly formed cell poles after division is completed (25, 26). Furthermore, this localization pattern of MinD and DivIVA was shown to strictly depend on FtsZ and other septal ring components, and no movement of MinD was reported (26). Thus, whereas MinE allows FtsZ assembly at midcell by specifically protecting this site from MinCD action, DivIVA does so by attracting and/or tethering the bulk of MinD to its proper site of action, away from the desired future site of septal ring assembly. As such, division site selection provides a fascinating example of how different organisms reach a similar goal (medial division) by regulating the subcellular activity of a similar tool (MinC/MinD) in distinctly different manners.
Acknowledgments
We thank Cynthia Hale, Robert Hogg, and Timothy Nilsen for comments on the manuscript. This work was supported by National Science Foundation Young Investigator Award MCB94-58197 and generous donations from The Elizabeth M. and William C. Treuhaft Fund, The Frank K. Griesinger Trust, Arline H. Garvin, James S. Blank, Charles E. Spahr, Alfred M. Taylor, and Theodore J. Castele (to P.d.B.). D.M.R. was supported by a National Research Service Award Institutional Training Grant (T32GM08056) from the National Institutes of Health.
ABBREVIATIONS
- PDS
potential division site
- IPTG
isopropyl β-d-thiogalactoside
- WT
wild type
References
- 1.Bramhill D. Annu Rev Cell Dev Biol. 1997;13:395–424. doi: 10.1146/annurev.cellbio.13.1.395. [DOI] [PubMed] [Google Scholar]
- 2.Lutkenhaus J, Addinall S G. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 3.Margolin W. Trends Microbiol. 1998;6:233–238. doi: 10.1016/s0966-842x(98)01280-3. [DOI] [PubMed] [Google Scholar]
- 4.Rothfield L I, Justice S S. Cell. 1997;88:581–584. doi: 10.1016/s0092-8674(00)81899-1. [DOI] [PubMed] [Google Scholar]
- 5.Adler H I, Fisher W D, Cohen A, Hardigree A A. Proc Natl Acad Sci USA. 1967;57:321–326. doi: 10.1073/pnas.57.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teather R M, Collins J F, Donachie W D. J Bacteriol. 1974;118:407–413. doi: 10.1128/jb.118.2.407-413.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Boer P A J, Crossley R E, Rothfield L I. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 8.Donachie W D, Begg K J. J Bacteriol. 1996;178:5971–5976. doi: 10.1128/jb.178.20.5971-5976.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Boer P A J, Crossley R E, Rothfield L I. Proc Natl Acad Sci USA. 1990;87:1129–1133. doi: 10.1073/pnas.87.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bi E, Lutkenhaus J. J Bacteriol. 1993;175:1118–1125. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Boer P A J, Crossley R E, Hand A R, Rothfield L I. EMBO J. 1991;10:4371–4380. doi: 10.1002/j.1460-2075.1991.tb05015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koonin E V. J Mol Biol. 1993;229:1165–1174. doi: 10.1006/jmbi.1993.1115. [DOI] [PubMed] [Google Scholar]
- 13.de Boer P A J, Crossley R E, Rothfield L I. J Bacteriol. 1992;174:63–70. doi: 10.1128/jb.174.1.63-70.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao C-R, de Boer P A J, Rothfield L I. Proc Natl Acad Sci USA. 1995;92:4314–4317. doi: 10.1073/pnas.92.10.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pichoff S, Vollrath B, Touriol C, Bouché J-P. Mol Microbiol. 1995;18:321–329. doi: 10.1111/j.1365-2958.1995.mmi_18020321.x. [DOI] [PubMed] [Google Scholar]
- 16.Raskin D M, de Boer P A J. Cell. 1997;91:685–694. doi: 10.1016/s0092-8674(00)80455-9. [DOI] [PubMed] [Google Scholar]
- 17.de Boer P A J, Crossley R E, Rothfield L I. J Bacteriol. 1988;170:2106–2112. doi: 10.1128/jb.170.5.2106-2112.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cormack B P, Valdivia R H, Falkow S. Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 19.Labie C, Bouché F, Bouché J-P. J Bacteriol. 1990;172:5852–5855. doi: 10.1128/jb.172.10.5852-5855.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang J, Cao C, Lutkenhaus J. J Bacteriol. 1996;178:5080–5085. doi: 10.1128/jb.178.17.5080-5085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levin P, Margolis P S, Setlow P, Losick R, Sun D. J Bacteriol. 1992;174:6717–6728. doi: 10.1128/jb.174.21.6717-6728.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varley A W, Stewart G C. J Bacteriol. 1992;174:6729–6742. doi: 10.1128/jb.174.21.6729-6742.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S, Price C W. Mol Microbiol. 1993;7:601–610. doi: 10.1111/j.1365-2958.1993.tb01151.x. [DOI] [PubMed] [Google Scholar]
- 24.Cha J-H, Stewart G C. J Bacteriol. 1997;179:1671–1683. doi: 10.1128/jb.179.5.1671-1683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edwards D H, Errington J. Mol Microbiol. 1997;24:905–915. doi: 10.1046/j.1365-2958.1997.3811764.x. [DOI] [PubMed] [Google Scholar]
- 26.Marston A L, Thomaides H B, Edwards D H, Sharpe M E, Errington J. Genes Dev. 1998;12:3419–3430. doi: 10.1101/gad.12.21.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]