Abstract
Background
Current efforts within the biomedical ontology community focus on achieving interoperability between various biomedical ontologies that cover a range of diverse domains. Achieving this interoperability will contribute to the creation of a rich knowledge base that can be used for querying, as well as generating and testing novel hypotheses. The OBO Foundry principles, as applied to a number of biomedical ontologies, are designed to facilitate this interoperability. However, semantic extensions are required to meet the OBO Foundry interoperability goals. Inconsistencies may arise when ontologies of properties – mostly phenotype ontologies – are combined with ontologies taking a canonical view of a domain – such as many anatomical ontologies. Currently, there is no support for a correct and consistent integration of such ontologies.
Results
We have developed a methodology for accurately representing canonical domain ontologies within the OBO Foundry. This is achieved by adding an extension to the semantics for relationships in the biomedical ontologies that allows for treating canonical information as default. Conclusions drawn from default knowledge may be revoked when additional information becomes available. We show how this extension can be used to achieve interoperability between ontologies, and further allows for the inclusion of more knowledge within them. We apply the formalism to ontologies of mouse anatomy and mammalian phenotypes in order to demonstrate the approach.
Conclusion
Biomedical ontologies require a new class of relations that can be used in conjunction with default knowledge, thereby extending those currently in use. The inclusion of default knowledge is necessary in order to ensure interoperability between ontologies.
Background
As the volume of biomedical data and knowledge presented in scientific papers increases, there is an increasing need to support formal analyses of these data and to pre-process knowledge for further use in solving problems and developing and testing hypotheses. The precise capture of biological data and knowledge and their correct and consistent representation in computational form is a basic pre-requisite for achieving these goals. Ontologies may provide a basis for integrating, processing and applying biomedical data. Their integration into a common ontological framework is an indispensible step towards the development of expressive knowledge bases. Interoperability between these ontologies would facilitate the consistent use of biomedical data in the form of annotations, allow for queries over multiple ontologies and form a rich knowledge resource for biomedicine that could be further used in solving problems and stating hypotheses. Different ontologies have been developed by different groups with different intentions. As a result, translating a statement or transferring an annotation from one ontology to another may not always yield the correct results. The absence of clear principles for achieving interoperability between different ontologies hinders the development of advanced applications and analysis tools based on these ontologies. A number of biomedical ontologies exist, which cover domains such as anatomy [1], cell structure, biological processes, functions [2], diseases [3], development [4], experimental conditions, phenotypes, qualities [5] and relationships [6]. A subset of these are unified under the umbrella of the Open Biomedical Ontologies (OBO) Foundry [7]. The OBO Relationship Ontology [6], together with the principles set forth in the OBO Foundry [8] have contributed to better interoperability between a large number of these ontologies. We address here several remaining problems.
One particular difficulty in making these ontologies interoperable results from the existence of two particular types of biomedical ontologies. The first group describes a canonical or idealized view on a domain, such as an ontology of canonical anatomy. The other group describes phenotypes, properties or phenomena, that – when exemplified by individuals – may contradict knowledge represented in the first group. We call the former group canonical ontologies and the latter phenotype ontologies. An example of a canonical ontology is the Foundational Model of Anatomy [9] (FMA), which describes an idealized domain, i.e., it describes a prototypical, idealized human anatomy. Many ontologies describing structure, such as cell structure, histology or anatomy, are canonical in this sense. On the other hand, a phenotype ontology describes phenomena whose exemplification by individuals may lead to deviations from this idealized structure. For example, the Mammalian Phenotype Ontology [10] contains the term "absent tail" as a specific type of "abnormal tail morphology". When a researcher would like to refer to an individual mouse with an "absent tail", this mouse does not comply with the canonical, idealized mouse anatomy that excludes such abnormalities.
The integration of ontologies of these different types cannot be achieved using methods developed hitherto, and a new set of methods transcending the framework of classical logic must be introduced to avoid inconsistencies while preserving the specificity of both types of knowledge. We present an approach that uses nonmonotonic reasoning to integrate canonical and phenotype ontologies.
Methods and Results
GFO-Bio
Integrating ontologies is a powerful means for achieving interoperability. We adopt John Sowa's definition of ontology integration [[11], p. 494], which he characterized as the process of finding commonalities between different ontologies A and B and deriving a new, integrated ontology C that facilitates interoperability between information systems based on ontologies A and B. There are several approaches to achieving such an integration [12], but there is no generally accepted solution.
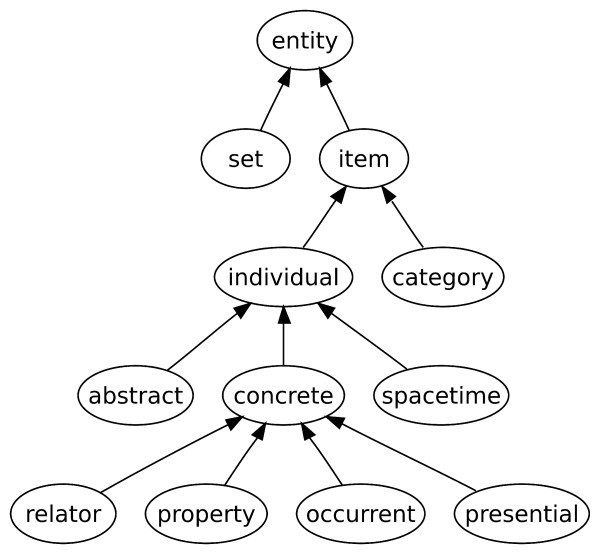
Our approach to integration is based on top-level ontologies [13]. For our study, we use the top-level ontology General Formal Ontology (GFO) [14]. GFO has several features that distinguish it from other top-level ontologies such as BFO [15,16] and DOLCE [17]. Among the relevant features are the inclusion of a theory of levels of reality [18], and the explicit incorporation of an ontological theory of higher-order categories (see figure 1 for an overview of selected categories and an explanation of higher-order categories). We have developed GFO-Bio [19], a core ontology [20] for biology. It is formalized in the Web Ontology Language [21] (OWL) and includes aspects of faceted classification [22] combined with GFO's theory of ontological levels of reality [18].
Figure 1.
The main categories of the General Formal Ontology (GFO) as outlined in [14, p. 70]. The basic distinction of GFO relevant for this work relates to items and is between individuals and categories. Individuals are items that cannot be instantiated. Categories are items that can have instances and may be predicated of other entities. The instances of categories of first order are individuals, while higher-order categories have categories as instances. For all items, the instance-of relationship is a relation of major importance, linking items (including categories) to the categories of which they are an instance.
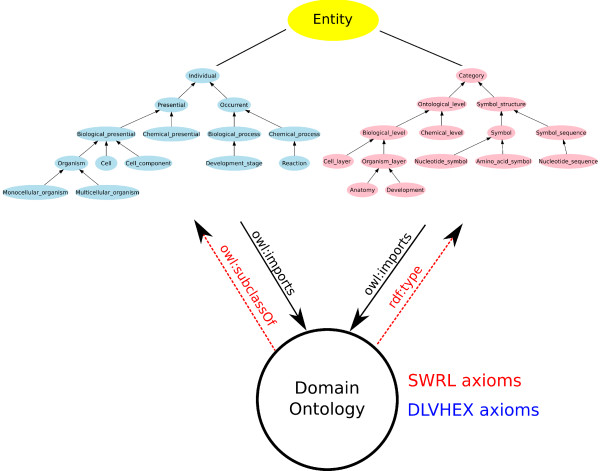
GFO-Bio comprises an ontology of individuals, similar to other established upper biomedical ontologies. "Biological individual" is introduced as a subclass of GFO's "Individual" category. The classes are defined or restricted using description logic statements. For example, a "Molecule" is a subclass of "Material object", which has as part at least two atoms. In addition, GFO-Bio contains another branch, in which categories themselves are further described and defined. This is an ontology of categories within the biomedical domain. It is this part of GFO-Bio that can directly represent directed acyclic graphs, which are commonly used for many biomedical ontologies. For an overview of the modules of GFO-Bio, see figure 2. In the remainder of this section, the basic ontology used can be considered to contain only two categories, "Individual" and "Category". We prefix relationships between categories with CC and relationships between individuals with II. Relationships between categories and individuals are prefixed with CI or IC respectively. For example, the relationship IC-instance-of is the instantiation relation, and the relation CC-isa is the is-a relation.
Figure 2.
The modules of the biological core ontology GFO-Bio. GFO-Bio comes in two modules. The first focuses on individuals and defines categories of individuals like "Cell" or "Organism" in the Web Ontology Language (OWL). It contains 168 categories and 73 relations. The second treats categories as instances in OWL and describes interrelations between domains or whole domain ontologies. It is based on a theory of levels of reality and contains 43 categories and two additional relations. Both modules are related using the Semantic Web Rule Language [44] and DLVHEX rules.
Default rules and default logic
Using GFO-Bio as a framework for integrating biomedical ontologies, we address the problem of accurately representing canonical and phenotype ontologies. A canonical anatomy ontology such as the Foundational Model of Anatomy [9] (FMA) establishes rules such as:
| Every instance of a human body has as part an appendix. |
This does not necessarily apply to every real human body: an individual human body may lack an appendix as part. However, the rule describes an idealized or canonical human. Phenotype ontologies describe phenomena, whose exemplification by individuals can be deviations from these idealizations. For example, an individual may be both an instance of a prototypical human body as described in the FMA (which implies an appendix as part) and an instance of the category "human body with absent appendix". In a classical logical framework, such as those commonly used in biomedical knowledge representation, e.g. in the form of OWL [21], a formalization of the conjunction of these two statements would lead to an inconsistency. A human body in the former case has an appendix as a part, while in the latter case it does not. Instantiating both categories creates the inconsistency. A logical inconsistency in the formal sense can only arise when the logical functor of negation is used. This functor is hidden in concepts such as "absent X", as used in the Mammalian Phenotype Ontology [10]. The formal detection of logical inconsistencies by inferences needs an explication of negation.
In order to avoid terms such as "absent X" and make the negation explicit, we adopt a modified form of the lacks relation [23], which we explicitly define as:
Individual p lacks category C with respect to relation R, if and only if there does not exist an x such that: pRx and x is an instance of C.
We use binary relations of the kind x lacks-R C instead of x lacks C with respect to R. For example, the fact that some individual x lacks a category C with respect to the relation has-part will be denoted as x lacks-part C.
Using the lacks relation may cause an inconsistency when a canonical ontology and a corresponding phenotype ontology are used in a classical logic formalism, such as first order logic [24] or description logic [25]. The reason is that classical formalisms enforce very strict interpretations, e.g. of quantifications like "every human", which results in monotonicity of these formalisms: the inferences drawn from a classical logical theory T remain true in every extension of T with additional facts.
In order to prevent inconsistencies, while at the same time preserving the intuition behind statements such as "a human has an appendix as part", the interpretation of such statements in the canonical ontology must be modified. We propose to use a nonmonotonic logic that treats the statements provided in a canonical ontology as true by default. Adding further knowledge, e.g. by referring to a phenotype ontology or using a statement involving the lacks relation (and therefore negation), may invalidate previously drawn conclusions.
Several ways of treating default rules and exceptions in logics have been proposed. The most popular among these proposals are default logic [26], circumscription [27,28] and autoepistemic logic [29,30]. We use default logic for our application, because it admits a transparent representation, and allows a semantically correct translation to a form of nonmonotonic, declarative logic programs called answer set programs [31].
In default logic, a default rule has the following form:
This means that if A() is true (prerequisite), and it is consistent to assume that B(), then C() can be derived. Intuitively, A() is a prerequisite, and assuming B() adds justification for deriving C() from A(). Thus, as long as B() can be assumed, default logic concludes C(). In order to formalize our example of humans normally having an appendix as part, we would use the following default rule:
Here, the precondition is Human(x), the fact that x is a human. Then, if it is consistent to assume that x has as part an instance of Appendix, it is concluded that x has as part an instance of Appendix. The definition of the relation IC-has-part follows the schema in table 1.
Table 1.
Schema of introduced relations
| Relation | Domain:Range | Definition |
| x II-R y | Individual:Individual | The individuals x and y stand in the relationship II-R. |
| x IC-R y | Individual:Category | There exists an individual z, such that: z IC-instance-of y and x II-R z. |
| x CC-R y | Category:Category | For all individuals a such that: a IC-instance-of x, a IC-R y. |
| x CC-canonical-R y | Category:Category | For all individuals a such that: a IC-instance-of x, by default, a IC-R y. |
| x II-lacks-R y | Individual:Individual | The individuals x and y do not stand in the relationship II-R. |
| x IC-lacks-R y | Individual:Category | The individual x does not stand in the relationship IC-R to y. |
| x CC-lacks-R y | Category:Category | For all individuals a such that: a IC-instance-of x, a IC-lacks-R y. |
For each relation used in a biomedical ontology, a number of relations between categories, individuals and between individuals and categories can be created. The CC-canonical-R relationship is a default relation that is accompanied by axioms in an answer set program to describe its semantics as a default.
Nonmonotonicity arises from "it is consistent to assume that x IC-has-part Appendix", which means that if x IC-has-part Appendix cannot be proven false from the given facts, its addition to the knowledge base does not lead to a contradiction. Adding the statement that x does not have an appendix as part (x IC-lacks-part Appendix) would lead to an inconsistency with x IC-has-part Appendix; therefore, this rule could no longer be used to derive that x has an appendix as part.
Answer-set programming, the formalism we use for our implementation, can mimic default rules. It uses two kinds of negation, called strong and weak negation. Strong negation is the classical (monotonic) negation, as used in the definition of the lacks relation. Weak negation, often denoted as not A, corresponds to the above statements "it cannot be proven that A is true", or "it is consistent to assume that A is false".
Formalizing defaults in biomedical ontologies
In a canonical ontology, relationships between its categories can be interpreted as default relations. By default, a human has some appendix as part. However, an instance of a human, such as John, may lack an appendix as a part; therefore, John is an instance of both "human" and "human without an appendix" (or "absent appendix"). In order to include canonical relationships between two categories, new relations must be introduced, such as CC-canonical-has-part. Then, the relationship between "human" and "appendix" becomes "human CC-canonical-has-part appendix". Further, this relationship corresponds to a default rule:
Using a class of lacks relationships as introduced by [23], we formalize the default operator in the rule above as:
In general, for each relation R between the categories in an ontology, we create several new relations: CC-R for the monotonic relationship between the categories, CC-canonical-R for the nonmonotonic default relationship between categories, IC-R for the monotonic relationship between an individual and a category, such as "John IC-has-part Appendix", meaning that John has some appendix as part, and II-R for the monotonic relationship between individuals. In addition, we introduce a class of lacks relationships. A schematic view of the new relationships introduced is shown in table 1. The schema is somewhat incomplete, because the introduction of canonical relations can be extended to the class of lacks relation, in the sense that some category may canonically lack some other category with respect to a relation R. In this case, the relation R must be replaced by lacks-R. This allows the treatment of exceptions between categories. For example, the category "Mouse with absent tail" can be defined as a mouse which lacks a tail as part.
Implementation
We have used a technique known as DL-programs [32] to implement rules together with the OWL version of GFO-Bio. The system DLVHEX allows for a bidirectional flow of information between an answer-set program and a description logic knowledge base or ontology; thus, it is well suited for our purposes. DLVHEX is based on the well-established datalog system DLV [33].
Relationships that are used in GFO-Bio are made available in the DLVHEX system. It then becomes possible to express the necessary axioms for relations of the kind CC-canonical-R. For example, for the relationship CC-canonical-has-part, the following axiom is added, corresponding to formula (5) in DLVHEX:
IC-has-part(X,Y) :- ind(X),class(Y),class(Z),inst(X,Z),
CC-canonical-has-part(Z,Y),
not IC-lacks-part(X,Y).
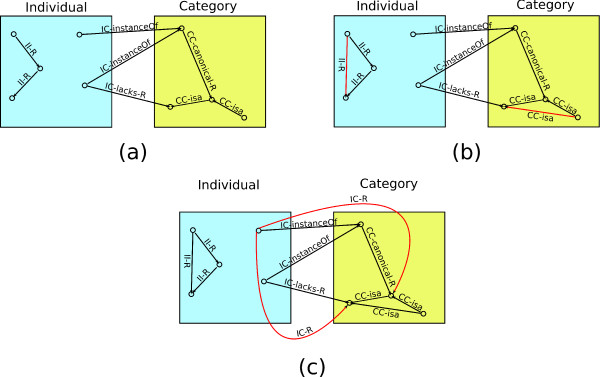
This means that if two categories Z and Y stand in the relation CC-canonical-has-part, and it cannot be proven that X IC-lacks-part Y (not IC-lacksPart(X,Y)), then it is concluded that an individual X, which is an instance of Z, stands in the relation IC-has-part to the category Y. A simple example illustrating this reasoning is shown in figure 3.
Figure 3.
In figure (a), the left side shows five individuals (instances of GFO-Bio's "Individual" category) and the right side contains four categories (instances of GFO-Bio's "Category" category). In addition, a number of relations are illustrated between the individuals, between the categories, and between individuals and categories. The relation R, denoted as II-R, is transitive. Figure (a) and the transitivity of II-R should be seen as the input ontology. In figure (b), the result of a classification using a description logic reasoner is illustrated. Here, the transitivity of the CC-isa relation and the relation II-R is resolved, reflected by the additional links. Figure (c) shows the result from applying the answer set rules formulated in DLVHEX. In this step, the default relationship between two categories, denoted by CC-canonical-R, is resolved. Two additional IC-R links are created for one individual. For the other individual, which instantiates the same category, these links are not created, because the IC-lacks-R relation blocks them.
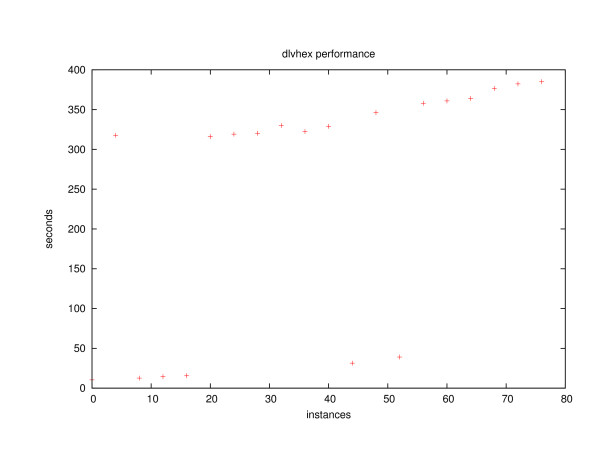
A plot showing the performance of our implementation for a common type of query on a mid-sized ontology is shown in figure 4. The sample test indicates that queries can be answered, but require several minutes. While this may be insufficient in practice for some applications, we believe that it shows that our implementation works, but needs further improvement. An extensive performance evaluation of the proposed method after some improvements on the implementation is subject to future work.
Figure 4.
This test uses the Adult Mouse Anatomy Ontology with 2740 classes. For testing, a fixed number of instances of Adult mouse is generated for a single run. Moreover, each of the mouse instances is assigned a single exception by means of a lacks-part tail statement. Using this dataset, a query for all parts of each mouse is run, which yields 2729 parts per mouse (derived from applicable canonical relationships). As intended, tails are not contained, and neither are any parts of tail like caudal vertebra. The graph shows the number of seconds for test runs with 0 to 80 mouse instances. For some numbers of instances, the time consumption is considerably less than for others. The difference of over 300 seconds is caused by the answer set solver DLV. We have been unable to examine the source code in order to determine the reason for this behaviour. The dataset used for this test can be accessed from the project webpage [45].
Ontology integration with GFO-Bio
Integrating biological ontologies using GFO-Bio involves several steps. First, an OWL-DL version of each ontology must be aquired or produced. OWL-DL is a sufficiently expressive language because negation is available and logical inconsistencies can be formally detected in the OWL-DL framework. For the purpose of this conversion, we provide a tool [19] that converts OBO format files [34] into OWL-DL. This conversion yields correct results for ontologies that are built according to the OBO Foundry principles, but may provide incorrect conversions for other ontologies available in the OBO format. The generated OWL-DL file must then be imported by GFO-Bio. Each top-level class of the imported ontology is then defined, at least partially, using categories from GFO-Bio's individual tree. For example, the "Cell" category of the Celltype Ontology [35] must be declared a subclass of (or an equivalent of) GFO-Bio's "Cell" category. Further, a second OWL-DL file can be produced for each integrated ontology containing the ontology's categories as instances of GFO-Bio's category branch. We also provide a tool for performing this conversion for OBO files. This file must be imported by GFO-Bio as well. In this file, relationships between categories, as directly expressed in the OBO-style directed acyclic graphs (DAGs), are modelled as relationships between OWL instances.
For example, the relationship expressed in the DAG of the Gene Ontology's cellular component ontology, "Membrane part-of Cell", is represented twice in GFO-Bio: First, "Membrane" and "Cell" are created as classes in OWL, and the following restriction created (in line with [36]):
SubClassOf(Membrane restriction(II-part-of someValuesFrom(Cell)))
In addition, the Gene Ontology's "Cell" category is declared equivalent to GFO-Bio's "Cell" category. Second, "Membrane" and "Cell" are treated as instances of GFO-Bio's "Category" class, and a relation CC-part-of ('CC' indicating the category-category reading of the relation) between "Membrane" and "Cell" is asserted:
Individual(Membrane value(CC-part-of Cell))
While neither the first nor the second step alone require more than the description logic fragment of OWL, in conjunction they result in an OWL-Full [21] ontology.
For an adequate integration of canonical and phenotype ontologies, nonmonotonically treated formulas must be added. This requires the addition of an answer set program for each relation CC-canonical-R and the corresponding relations IC-R and IC-lacks-R:
IC-R(X,Y) :- ind(X),class(Y),class(Z),inst(X,Z),
CC-canonical-R(Z,Y),
not IC-lacks-R(X,Y).
Additions to the OBO Relationship Ontology
The OBO Relationship Ontology [6] requires several additions for our proposal to succeed. First, the classes of lacks relations, as described in table 1, must be added. This will allow lacking body parts to be defined in ontologies such as the Mammalian Phenotype Ontology [10].
In the description logic variant of the Web Ontology Language [21,25] (OWL-DL), lacks relations can be expressed using negated statements. However, lacks relations are reduced to relations between individuals in a different way compared to what is done for most other relations in the OBO Relationship Ontology (cf. table 1). Ontologies developed directly in OWL-DL could use negation to avoid reference to lacks relations at all.
Second, canonical-R relations must be included as relations between categories, using the semantics introduced here. In particular, the canonical-R relations require a nonmonotonic knowledge representation formalism, and cannot be formalized using any form of classical logic. We presented one possible implementation using answer set semantics, but there are other alternatives. At its core, however, the definition of the canonical-R relations remains the same in all possible formalisms dealing with defaults: if it is consistent to assume that some relation holds, this relation holds.
Use case: Integration of Mouse Anatomy and Mammalian Phenotype Ontology
The method we propose can be used in conjunction with existing tools and ontologies. Little effort is required to modify current ontologies to fit within our proposed methodology. Below, we demonstrate how to re-interpret the Adult Mouse Anatomy Ontology [1] (MA) and the Mammalian Phenotype Ontology [10] (MP) to fit within our proposed framework, and discuss problems with the current formalization in the MP.
Mouse Anatomy
The Adult Mouse Anatomy Ontology (MA) uses two relationships, is-a and part-of. We introduce one new relationship to the MA, which we call canonical-has-part, and automatically add for each statement of the type
X part-of Y (6)
the new statement
Y canonical-has-part X. (7)
We believe that this will result in most cases in correctly interpreted default rules, but this method will generate some inadequate statements. Therefore, manual verification will be necessary. In addition, some of the generated statements may not contain default rules, but are universally true, while some of the currently present statements involving part-of may not be universally true, but represent default rules. Therefore, automatically generating default rules from existing statements can only be the first step, and in the continued development of the MA, a distinction must be made between default rules and universally true statements. This may make it necessary to include additional relationships between categories in the MA, e.g. canonical-part-of and has-part.
Mammalian Phenotype Ontology
The Mammalian Phenotype Ontology (MP) defines, among others, categories labelled by terms such as absent-X. In these terms, the hidden negation must be made explicit. The MP is available in two versions, one containing only is-a relations, and another experimental version that attempts to define terms using relationships such as inheres-in [17] and categories from PATO [37] (an ontology of phenotypic properties), MA and others. The MP provides property concepts such as absent_tail, although we believe that it would be more adequate to term this property absence of a tail, because absent_tail suggests a reading as an object concept, namely as a tail which is absent. These properties can be composed with object concepts, e.g. adult mouse, in order to refer to more specific object concepts like an adult mouse without a tail (without explicating all of these in advance). Formally, the category absent_tail is defined as the intersection of PATO:lacking physical parts, inheres-in MA:adult mouse, and towards MA:tail. The translation to OWL [36] yields
EquivalentClasses(absent_tail
intersectionOf(
PATO:lacking_physical_parts
restriction(inheres-in someValuesFrom(MA:adult_mouse))
restriction(towards someValuesFrom(MA:tail))))
Such formalization has the problem that a reduction of absent_tail to relationships between individuals is inappropriate. It becomes manifest in restriction(towards someValuesFrom(MA:tail)), which enforces the existence of an instance of MA:tail in the OWL model [38] – yet, ontologically, there is no instance of tail if the mouse does not have a tail. If "absent tail" is taken literally, i.e., a "virtual" tail with a property of "absence" is accepted in the OWL model, the immediate objection is weakened. However, this would either imply that the towards link points to an arbitrary tail of some other entity, or that the mouse with the absent tail does have a tail (which may have the propery of being "absent"). This causes at the very least some inconveniences, e.g. when querying such a model for tails, one must pay attention to exclude all virtual tails. We expect that a general application of this approach to many object classes cannot be controlled with reasonable effort. The underlying problem here is a different kind of concept composition compared to that which is commonly employed in description logics and the semantics of the OBO file format [36]. For composing the concept absent_tail with recourse to tail, a direct link from absent_tail to the OWL class MA:tail would be required instead of a link to instances of the class MA:tail. However, links on the class level are not available if the formalization is supposed to adhere to the decidable description logic variant of OWL.
Other problems occur if the lack of a certain part is considered with respect to (sub-)parts of it. For example, a caudal vertebra is a part of a tail. The logical definition for absent caudal vertebra can be formulated similarly to the one for the absent_tail. Then the question arises to which entity the PATO property inheres-in should link. If tail is chosen, this definition cannot be applied to mice lacking a tail, because then there is nothing to inhere in. Another option is to link to mouse instead. In both cases, whenever a mouse lacks a tail as part, it also lacks all parts of the tail as part. Since each instance of caudal vertebra which is a part of a mouse is part of its tail, a mouse without tail lacks a caudal vertebra. This conclusion cannot be drawn in the approach currently taken by the MP. Moreover, we do not want to conclude that a mouse has a caudal vertebra as part when it lacks a tail.
Therefore, our suggestion is to introduce the relationship lacks-part and to define terms of the type absent_X as standing in the lacks-part relationship to X. Then, absent_X terms refer to categories of objects instead of properties, and those categories can be viewed as a reification of the binary lacks-part relation. For example, absent_tail would be defined as
[Term]
id: MP:0003456
name: absent tail
relationship: lacks_part MA:0000008 ! tail
By design, this use of lacks-part does not have the problem of non-existent instances. It also does not permit the conclusion that a mouse has a caudal vertebra as part when a mouse lacks a tail.
Discussion
Meaningful integration of the numerous biomedical ontologies is a major task with many challenges. Currently, the infrastructure for such integration is developed in the form of top-level ontologies, biomedical core ontologies and logic-based inference systems.
Concept conversion
The formalism we introduced requires reformulating the definitions for the categories expressed in phenotype ontologies. Categories in the form "absent-X" should be defined by, e.g., CC-lacks-part X, where X is a category in some canonical ontology. In some cases, this conversion can be done automatically using simple pattern matches. The Mammalian Phenotype Ontology [10] contains 395 categories of the type "absent-X", which indicate a CC-lacks-part relationship. However, it is likely that an amount of manual curation will be required to convert relevant concepts into the required form. We believe that the advantages gained by having a common framework for integrating a large number of biomedical ontologies justifies this effort, in particular since it also allows for a semantically richer definition of terms.
Defaults and canonical knowledge
We introduce the notion of "default knowledge" as a technical term within the formalism we propose. We do not discuss what a "default" is, or when a piece of knowledge becomes a default, in contrast to merely contingent knowledge. Developers of domain ontologies must decide this. Widespread acceptance of some fact, its sanction by scientific discourse, or its implicit use in scientific writing may provide starting points for finding defaults. These principles have been used to construct the Foundational Model of Anatomy [9] (FMA). The Mammalian Phenotype Ontology classifies categories almost exclusively under categories named "abnormal-X". The ontology of phenotypic properties (PATO) contains the property of being "abnormal". Each of the corresponding categories and annotations can be investigated and the corresponding default rule identified. Not all pieces of information contained in ontologies such as the FMA will be default knowledge, but we expect that a significant number of facts can be translated to the formalisms we propose, thereby making the nature of the fact as a default explicit.
There is a difference between canonical and default knowledge, in particular in the context of anatomy. Canonical human anatomy, for example, describes an idealized, prototypical human being. This does not necessarily coincide with a normal human being, i.e. in the sense of statistically averaged values. Defaults, on the other hand, tend to capture in their commonsense usage the normal cases of a category. We believe that the framework of default logic, compared with other systems, provides the most adequate interpretation for canonical knowledge. However, while certainly needed, a precise distinction between normal, default and canonical knowledge is out of the scope of this study.
Comparison with other approaches
The important role of accommodating exceptions and defaults in biomedical knowledge representation has been recognized previously [39], where patterns to deal with a variety of cases were introduced and discussed. These cases are based on the description logic fragment of OWL [21], and therefore monotonic logic. In [39], three types of exceptions that occur in biomedical knowledge bases are distinguished:
1. Single exceptions: "Arteries carry oxygenated blood" except for the pulmonary artery. In [39], it is proposed to reformulate this statement to "Arteries except the pulmonary artery carry oxygenated blood".
2. Exceptions due to context: "The normal human manus has five digits", with "human" and "normal" being treated as explicit contexts.
3. Unpredictable number of exceptions, exceptions from exceptions, etc., such as drug uses, contraindications and interactions.
We offer a method for representing these types of exceptions using a nonmonotonic knowledge representation formalism. We use answer set programs to provide the semantics for treating knowledge in OWL as default knowledge with additional exceptions. This does not exclude the possibility to treat these types of exceptions exclusively in a monotonic logic such as OWL where appropriate, for which [39] provides a solution. The solution in [39] to the example of arteries carrying oxygenated blood, except the pulmonary artery, has the problem that it must be explicitly known that some artery is not the pulmonary artery, in order to conclude that this artery carries oxygenated blood. There may be cases where this is not wanted, especially if the exception occurs very rarely. In particular, if there is only one rare exception to a rule and some statement influencing the property which changes with this exception is asserted, then the knowledge engineer will usually make this exception explicit, and ignore it otherwise. Then, a question whether an artery carries oxygenated blood evaluates to true, except when it is proven that this artery is the pulmonary artery. On the other hand, the solution proposed by [39] is guaranteed to provide the correct inference in every case. Depending on the users and uses of a knowledge base or ontology, different representations for this case may be selected, and in many cases the treatment in [39] is adequate.
Case two is solved by explicitly introducing a context argument, in the form of additional properties, e.g., by introducing some relation hasAnatomicalStatus which maps to "normal". Then, a Mouse that has an anatomical status "normal" could have, e.g., a tail and a head as part. If a mouse had no tail, it can be concluded that it is an anatomically abnormal mouse. However, then it would be impossible to conclude that it still has a head. An extension to the solution in [39] would be to make the context more fine-grained, by specifying mouse with anatomically normal tails, heads, and so on. This comes down to specifying an enormous number of exceptions in a monotonic logic, and in order to obtain a correct answer to a query for all the parts of some individual mouse, all these exceptions must be explicitly excluded. It would not be possible to simply state that some entity is a mouse in order to obtain its parts. Instead it is required to specify explicitly which parts are normal and abnormal, which means in essence to add the answers to the query asked.
The third case in [39] is closest in spirit to our work, as one of the proposals is to use a hybrid reasoning system in order to deal with it. We have extended this idea by giving a formal account of our treatment of exceptions, which is based on a well-studied nonmonotonic logic, and is implemented in a computationally tractable framework. It can also be used in conjunction with appropriate upper ontologies. Further, we have shown how to use this formalism to achieve interoperability between canonical and phenotype ontologies in biology. And finally, we give an implementation of our ontology and support for reasoning over exceptions. This could be achieved because recent years have seen an increasing effort in developing reasoners for the Semantic Web and extending them in various ways, among them the implementation we are using, DLVHEX.
We believe that our solution to the problem of exceptions and deviations from a canonical ontology is more general than the proposal in [39]. In our opinion, the knowledge contained in a canonical ontology is inherently default knowledge. There is no adequate solution for representing this type of knowledge in a monotonic knowledge representation formalism. Representation in monotonic logic requires exceptions to be encoded in the ontology either as a list of exceptions to an axiom, or using a general "abnormality" predicate. For example, the fact that mice usually have some tail as part can be represented as "Mouse has-part Tail except when ..." followed by a complete list of exceptions. Alternatively, "Mouse" can be replaced by "normal mouse" in the rule, and a mouse without a tail is not normal. The first solution requires complete knowledge of all known exceptions. These must additionally be explicitly excluded in every query for parts of the mouse. The second way does not require this knowledge of exceptions, but allows for no further inferences once a mouse is known to be not normal. Defaults and exceptions cannot be dealt with in a monotonic logic without substantially modifying the canonical ontology, and limiting the ability to query the ontology.
Limitations
A major drawback of the system we are using, DLVHEX, is its use of RACER [40] as a description logic reasoner and of DLV [33] as a datalog system. RACER and DLV are proprietary software. In order to be of general use and high quality, an implementation entirely based on free software is beneficial, if not necessary [41,42].
A number of formalisms have been proposed as a solution to handling defaults in Semantic Web representation languages or other knowledge representation formalisms. Many require modifying the language, and therefore changing tools that are used to develop ontologies. Many biomedical ontologies are developed using tools such as OBO-Edit [43] by biology experts, but not necessarily experts in logic or formal ontology. The solution we propose requires no changes to existing tools, since we are using a hybrid reasoning mechanism. Tools that are currently in use can be used further by the ontology developers. The additional semantic features that allow for the treatment of canonical relations as defaults are maintained separately from the ontologies in which they are used.
Conclusion
In this paper we tackle the problem of integrating biomedical ontologies to facilitate interoperability among them and thus among information systems based on them. We particularly focus on adequately treating two kinds of ontologies, namely canonical and phenotype ontologies, e.g., the Mouse Anatomy ontology and the Mammalian Phenotype ontology. Given this distinction, we have argued that canonical ontologies represent default knowledge. Their integration with ontologies covering phenotypes may thus lead to inconsistencies if used within a classical logic framework, because some phenotypic descriptions are exceptions to defaults. We have shown how existing techniques from knowledge representation can be used to resolve these problems. Moreover, our solution uses the biological core ontology GFO-Bio as an ontological foundation, which provides support for our solution through higher order categories and relations. Integrating canonical and phenotype ontologies, however, requires both an appropriate ontological basis as well as a nonmonotonic representation formalism.
Our work primarily extends the OBO Relationship Ontology [6], and requires few changes to domain ontologies. In particular, our proposal does not require modifications in the tools that domain ontology developers use for curating ontologies, or changes in the way these ontologies are developed and stored. Our solution remains fully compatible with the OBO representation format, and addresses all logical, formal and computational requirements in our proposed extension to the OBO Relationship Ontology. It is there that nonmonotonic semantics must be made available to users. In its current form, based on a classical, monotonic logic, the OBO Relationship Ontology cannot support interoperability between all ontologies that will become part of the OBO Foundry, in particular between anatomy and phenotype ontologies. Our proposal aims to bring about effective interoperability and integration between the ontologies in the OBO Foundry without the need to modify the representation formalism or the tools used in ontology curation and analysis.
Availability and Requirements
Project name: GFO-Bio/NMR
Project homepage: http://bioonto.de/pmwiki.php/Main/NonmonotonicReasoning
Operating systems: GFO-Bio and axiomatization are platform independent, reasoning using DLVHEX requires GNU/Linux or Mac OS X.
Programming languages: OWL, Semantic Web Rule Langue (SWRL), datalog, Java Other requirements: Parts of our implementation require DLVHEX, DLV and RACER.
License: Modified BSD License. DLVHEX requires RACER and DLV. Both are proprietary software.
Competing interests
The authors declare that there are no competing interests.
Authors' contributions
HH conceived the initial idea on using nonmonotonic reasoning for integrating ontologies, RH the relevance for anatomy and phenotype ontologies. HH, FL, JK and RH designed the framework for consistent integration of anatomy and phenotype ontologies. RH implemented the framework and performed the tests. All authors contributed to writing the paper, have read and approved the final version of this manuscript.
Acknowledgments
Acknowledgements
We are grateful to Roberto Poli for enlightening discussions and his initial ideas on facet analysis and the theory of levels of reality. We thank Kay Prüfer for his comments on an early draft of this paper, and Roman Schindlauer and Thomas Krennwallner for their patch for and help with DLVHEX. We thank two anonymous reviewers for their valuable comments.
We thank the Max Planck Society and the Institute of Medical Informatics, Statistics and Epidemiology at the University of Leipzig for funding.
Thanks to Christine Green for her help in preparing the English manuscript.
Contributor Information
Robert Hoehndorf, Email: hoehndorf@eva.mpg.de.
Frank Loebe, Email: loebe@informatik.uni-leipzig.de.
Janet Kelso, Email: kelso@eva.mpg.de.
Heinrich Herre, Email: heinrich.herre@imise.uni-leipzig.de.
References
- Hayamizu TF, Mangan M, Corradi JP, Kadin JA, Ringwald M. The Adult Mouse Anatomical Dictionary: a tool for annotating and integrating data. Genome Biology. 2005;6:R29. doi: 10.1186/gb-2005-6-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrisholm R, Kibbe W, Osborne J, Wolf W, Doyle M, Doyle AO, Smith M, Zhu J. Disease Ontology http://diseaseontology.sourceforge.net
- The Plant Ontology Consortium Plant Ontology (PO) http://www.plantontology.org
- Gkoutos GV, Green ECJ, Mallon AM, Hancock JM, Davidson D. Building Mouse Phenotype Ontologies. In: Altman RB, Dunker AK, Hunter L, Jung TA, Klein TE, editor. Proceedings of the 9th Pacific Symposium on Biocomputing (PSB 2004), Hawaii, USA, Jan 6–10. London: World Scientific; 2004. pp. 178–189. [DOI] [PubMed] [Google Scholar]
- Smith B, Ceusters W, Klagges B, Köhler J, Kumar A, Lomax J, Mungall C, Neuhaus F, Rector AL, Rosse C. Relations in biomedical ontologies. Genome Biology. 2005;6:R46. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Open Biomedical Ontologies (OBO) Foundry http://obofoundry.org
- OBO Foundry Principles http://obofoundry.org/crit.shtml
- Rosse C, Mejino JLV., Jr A Reference Ontology for Biomedical Informatics: the Foundational Model of Anatomy. Journal of Biomedical Informatics. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Smith CL, Goldsmith CAW, Eppig JT. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biology. 2004;6:R7. doi: 10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa JF. Knowledge Representation: Logical, Philosophical and Computational Foundations. Pacific Grove: Brooks/Cole; 2000. [Google Scholar]
- Kalfoglou Y, Schorlemmer M. Ontology mapping: the state of the art. The Knowledge Engineering Review. 2003;18:1–31. doi: 10.1017/S0269888903000651. [DOI] [Google Scholar]
- Wache H, Vögele T, Visser U, Stuckenschmidt H, Schuster G, Neumann H, Hübner S. Ontology-based Integration of Information – A Survey of Existing Approaches. In: Stuckenschmidt H, Gómez-Pérez A, Gruninger M, Uschold M, editor. Proceedings of the IJCAI'01 Workshop on Ontologies and Information Sharing, Seattle, Washington, USA, Aug 4–5. 2001. pp. 108–117. [Google Scholar]
- Herre H, Heller B, Burek P, Hoehndorf R, Loebe F, Michalek H. General Formal Ontology (GFO) – A Foundational Ontology Integrating Objects and Processes [Version 1.0] Onto-Med Report 8, Research Group Ontologies in Medicine, Institute of Medical Informatics, Statistics and Epidemiology, University of Leipzig, Leipzig, Germany. 2006.
- Grenon P. BFO in a Nutshell: A Bi-categorial Axiomatization of BFO and Comparison with DOLCE. IFOMIS Report 06/2003, Institute for Formal Ontology and Medical Information Science (IFOMIS), University of Leipzig, Leipzig, Germany. 2003.
- Grenon P. Spatio-temporality in Basic Formal Ontology: SNAP and SPAN, Upper-Level Ontology, and Framework for Formalization: PART I. IFOMIS Report 05/2003, Institute for Formal Ontology and Medical Information Science (IFOMIS), University of Leipzig, Leipzig, Germany. 2003.
- Masolo C, Borgo S, Gangemi A, Guarino N, Oltramari A. WonderWeb Deliverable D18: Ontology Library (final) Tech rep, Laboratory for Applied Ontology – ISTC-CNR, Trento, Italy. 2003.
- Poli R. The Basic Problem of the Theory of Levels of Reality. Axiomathes. 2001;12:261–283. doi: 10.1023/A:1015845217681. [DOI] [Google Scholar]
- Hoehndorf R, Loebe F, Poli R, Kelso J, Herre H. GFO-Bio: A biomedical core ontology http://onto.eva.mpg.de/gfo-bio.html
- Valente A, Breuker J. Towards principled core ontologies. In: Gaines BR, Musen MA, editor. Proceedings of the 10th Knowledge Acquisition Workshop (KAW'96), Banff, Alberta, Canada, Nov 9–14. 1996. pp. 301–320. [Google Scholar]
- McGuinness DL, van Harmelen F. OWL Web Ontology Language Overview. W3C recommendation, World Wide Web Consortium (W3C) 2004.
- Bowker L. Multidimensional Classification of Concepts and Terms. In: Wright SE, Budin G, editor. Handbook of Terminology Management, Basic Aspects of Terminology Management. Vol. 1. Amsterdam: John Benjamins; 1997. [Google Scholar]
- Ceusters W, Elkin P, Smith B. Referent Tracking: The Problem of Negative Findings. In: Hasman A, Haux R, van der Lei J, De Clercq E, Roger-France F, editor. Ubiquity: Technologies for Better Health in Aging Societies: Proceedings of MIE Volume 124 of Studies in Health Technology and Informatics. Amsterdam: IOS Press; 2006. pp. 741–746. [PubMed] [Google Scholar]
- Hilbert D, Ackermann W, Luce RE. Principles of Mathematical Logic. American Mathematical Society; 1999. [Google Scholar]
- Baader F, Calvanese D, Mcguinness DL, Nardi D, Patel-Schneider PF. The Description Logic Handbook: Theory, Implementation, and Applications. New York, NY, USA: Cambridge University Press; 2003. [Google Scholar]
- Reiter R. A logic for default reasoning. Artificial Intelligence. 1980;13:81–132. doi: 10.1016/0004-3702(80)90014-4. [DOI] [Google Scholar]
- McCarthy J. Circumscription – A form of non-monotonic reasoning. Artificial Intelligence. 1980;13:27–39. doi: 10.1016/0004-3702(80)90011-9. [DOI] [Google Scholar]
- McCarthy J. Applications of circumscription to formalizing common-sense knowledge. Artificial Intelligence. 1986;28:89–116. doi: 10.1016/0004-3702(86)90032-9. [DOI] [Google Scholar]
- Konolige K. On the relation between default and autoepistemic logic. Artificial Intelligence. 1988;35:343–382. doi: 10.1016/0004-3702(88)90021-5. [DOI] [Google Scholar]
- Gabbay DM, Hogger CJ, Robinson JA, (Eds) Handbook of Logic in Artificial Intelligence and Logic Programming, Nonmonotonic Reasoning and Uncertain Reasoning. Vol. 3. Oxford, UK: Oxford University Press; 1994. [Google Scholar]
- Lifschitz V. Answer set programming and plan generation. Artificial Intelligence. 2002;138:39–54. doi: 10.1016/S0004-3702(02)00186-8. [DOI] [Google Scholar]
- Eiter T, Ianni G, Schindlauer R, Tompits H. A Uniform Integration of Higher-Order Reasoning and External Evaluations in Answer Set Programming. In: Pack Kaelbling L, Saffiotti A, editor. Proceedings of the Nineteenth International Joint Conference on Artificial Intelligence (IJCAI'05), Edinburgh, Scotland, UK, Jul 30 -Aug 5. Denver, Colorado: Professional Book Center; 2005. pp. 90–96. [Google Scholar]
- Leone N, Pfeifer G, Faber W, Eiter T, Gottlob G, Perri S, Scarcello F. The DLV System for Knowledge Representation and Reasoning. ACM Transactions on Computational Logic. 2006;7:499–562. doi: 10.1145/1149114.1149117. [DOI] [Google Scholar]
- File Format Guide http://www.geneontology.org/GO.format.shtml
- Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biology. 2005;6:R21. doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golbreich C, Horrocks I. The OBO to OWL mapping, GO to OWL 1.1! In: Golbreich C, Kalyanpur A, Parsia B, editor. Proceedings of the OWLED 2007 Workshop on OWL: Experiences and Directions, Innsbruck, Austria, Jun 6–7, Volume 258 of CEUR Workshop Proceedings. Aachen, Germany: CEUR-WS.org; 2007. [Google Scholar]
- OBO Phenotype Group PATO – Phenotypic Quality Ontology http://www.bioontology.org/wiki/index.php/PATO:Main_Page
- Patel-Schneider PF, Hayes P, Horrocks I. OWL Web Ontology Language – Semantics and Abstract Syntax. 2004. http://www.w3.org/TR/owl-semantics/
- Rector AL. Defaults, Context, and Knowledge: Alternatives for OWL-Indexed Knowledge Bases. In: Altman RB, Dunker AK, Hunter L, Jung TA, Klein TE, editor. Proceedings of the 9th Pacific Symposium on Biocomputing (PSB 2004), Hawaii, USA, Jan 6–10. London: World Scientific; 2004. pp. 226–237. [DOI] [PubMed] [Google Scholar]
- Haarslev V, Möller R. Racer: A Core Inference Engine for the Semantic Web. In: Sure Y, Corcho O, editor. Proceedings of the 2nd International Workshop on Evaluation of Ontology-based Tools (EON2003), Sanibel Island, Florida, USA, Oct 20, Volume 87 of CEUR Workshop Proceedings. Aachen, Germany: CEUR-WS.org; 2003. pp. 27–36. [Google Scholar]
- Raymond ES. The Cathedral and the Bazaar. Knowledge, Technology, and Policy. 1999;12:23–49. doi: 10.1007/s12130-999-1026-0. [DOI] [Google Scholar]
- Stallman RM, Gay J, Lessig L. Free Software, Free Society: Selected Essays of Richard M Stallman. Boston: Free Software Foundation; 2002. [Google Scholar]
- Day-Richter J, Harris MA, Haendel M, The Gene Ontology OBO-Edit Working Group. Lewis S. OBO-Edit – An Ontology Editor for Biologists. Bioinformatics. 2007;23:2198–2200. doi: 10.1093/bioinformatics/btm112. [DOI] [PubMed] [Google Scholar]
- Horrocks I, Patel-Schneider PF, Boley H, Tabet S, Grosof B, Dean M. SWRL: A Semantic Web Rule Language Combining OWL and RuleML. 2003. http://www.daml.org/2003/11/swrl/
- The Bio-Ontology Research Group Leipzig Project Nonmonotonic Reasoning. 2007. http://bioonto.de/pmwiki.php/Main/NonmonotonicReasoning