Abstract
Drosophila Enabled (Ena) was first identified as a genetic suppressor of mutations in the Abelson tyrosine kinase and subsequently was shown to be a member of the Ena/vasodilator-stimulated phosphoprotein family of proteins. All members of this family have a conserved domain organization, bind the focal adhesion protein zyxin, and localize to focal adhesions and stress fibers. Members of this family are thought to be involved in the regulation of cytoskeleton dynamics. The Ena protein sequence has multiple poly-(l-proline) residues with similarity to both profilin and src homology 3 binding sites. Here, we show that Ena can bind directly to the Drosophila homolog of profilin, chickadee. Furthermore, Ena and profilin were colocalized in spreading cultured cells. We report that the proline-rich region of Ena is responsible for this interaction as well as for mediating binding to the src homology 3 domain of the Abelson tyrosine kinase. These data support the hypothesis that Ena provides a regulated link between signal transduction and cytoskeleton assembly in the developing Drosophila embryo.
Regulation of the architecture of the actin cytoskeleton in response to extracellular cues is an important mechanism by which cells control their morphology and motility (1–3). Thus, there is increasing interest in identifying proteins that are links between signal transduction and the actin cytoskeleton. Members of the Enabled (Ena)/vasodilator-stimulated phosphoprotein (VASP) family of proteins, including Ena, Murine Enabled (Mena), VASP, and Ena/VASP-Like (EVL), are candidates for such a link. Ena, VASP, and Mena are localized to the actin cytoskeleton, focal adhesions, and sites of cell–cell contact when expressed in cultured cells (4–6). VASP and Mena have been implicated directly in cytoskeleton assembly through their interactions with profilin and their involvement in directing actin-filament assembly at one pole of the intracellular pathogen Listeria monocytogenes (5, 7–9). The observation that VASP can substitute partially for a loss of Ena in vivo suggests that Ena may have a similar cellular function (4).
The Ena/VASP proteins share an overall structural domain organization consisting of conserved N and C termini separated by a central proline-rich region of variable length and similarity (4–6). Proline-rich sequences are known to mediate binding to the actin-binding protein, profilin, and src homology 3 (SH3) domains. Consistent with this fact, both Ena and Mena bind SH3 domains in vitro (5, 10, 11), whereas VASP and Mena bind profilin (5, 9). The functional similarity between Ena and VASP in vivo suggests that Ena also may bind profilin. That Ena/VASP proteins can bind profilin is of particular interest, because profilin is critical for normal development. Yeast carrying mutations in profilin grows more slowly and is often multinucleated, unlike wild-type yeast (12). Mice lacking profilin die early in development (13). Drosophila without profilin die late during embryonic development and have abnormalities in cell migration during axonogenesis and oogenesis and aberrations in actin structures (14, 15).
Profilins are small, evolutionarily conserved proteins that bind actin monomers and regulate their polymerization (13, 16, 17). In addition to actin and poly-(l-proline), profilin is also a ligand for phosphoinositides (18). Although profilin binds poly-(l-proline) regardless of whether it is in a complex with actin (19), phosphoinositides dissociate the profilin–actin complex (18). This observation suggests that profilin may mediate a link between signal transduction pathways and changes in the actin cytoskeleton. Consistent with this suggestion, profilin associates with proteins such as Arp2/3 in regions of the cell where there is active cytoskeletal rearrangement (20, 21). Profilin also has been linked to signal transduction pathways through its interaction with PIP2 and regulation of PIP2’s hydrolysis by unphosphorylated phospholipase C (18, 22).
The Ena/VASP family members Ena and Mena also interact with SH3 domains (5, 10, 11). Although the precise sequences involved in these interactions have not been well characterized, they are assumed to take place through their proline-rich sequences. The physiological relevance of these interactions is not clear. Studies have shown that interaction of Ena with the Abelson (Abl) SH3 domain is not important for substrate recognition by the Abl kinase. However, phosphorylation of Ena reduces its ability to interact with the Abl SH3 domain, suggesting that this interaction is regulated by the Abl kinase (10).
In this study, we characterized molecular interactions between the Ena proline-rich region and two potential ligands: chickadee, the Drosophila homolog of profilin, and the SH3 domain. We report evidence that chickadee is a ligand for the Ena protein. Furthermore, we have colocalized Ena and profilin in spreading cultured cells. The SH3 binding specificity of Ena was examined in more detail. We report that these proteins recognize proline-rich sequences in Ena and identify some of the amino acids important for ligand binding. Taken together, these data provide further evidence that Ena may serve as a regulated link between Abl signaling and cytoskeletal dynamics in the developing Drosophila embryo.
MATERIALS AND METHODS
Molecular Biology.
DNA was purified by standard techniques (23). Site-directed mutagenesis of Ena was performed by the method of Deng and Nickoloff (24). Mutagenic primers were 25–40 nt in length and contained four to eight mismatches. For Ena8 P → A, which carries mutations in the putative Abl SH3 binding site, oligonucleotides containing base changes that changed prolines 407, 412, 413, 414, 470, 474, 475, and 476 to alanine were incorporated into a full-length Ena cDNA. The entire mutagenized fragments of DNA were sequenced.
Transfections, Immunoprecipitations, and Western Blot Analysis.
Drosophila S2 cells were transfected transiently with the Ena cDNA in the pPac-PL expression vector. Cells were harvested after 60 h and lysed in immunoprecipitation buffer (0.5% Triton X-100/50 mM Tris, pH 8.0/150 mM NaCl/5 mM EDTA/1 mM Na3VO4/1 mM Pefabloc/1 μg/ml pepstatin/1 μg/ml leupeptin/1 μg/ml aprotinin) or His buffer (0.5% Triton X-100/20 mM sodium phosphate, pH 7.8/500 mM NaCl/1 mM Na3VO4/1 mM Pefabloc/1 μg/ml pepstatin/1 μg/ml leupeptin/1 μg/ml aprotinin). After lysis, cell debris was pelleted at 12,000 × g for 20 min. Immunoprecipitations were carried out by using anti-Ena antibody as described (11). Proteins were resolved on SDS/7.5% polyacrylamide gels, transferred to poly(vinylidene difluoride) membrane, and blotted with anti-Ena or anti-chickadee (15) antibodies.
Purification of Fusion Proteins.
The Drosophila Abl SH3 domain (amino acids 82–325) was subcloned into the BamHI site of the pGEX2TK vector (Amersham Pharmacia). T. Pawson (Samuel Lunenfield Research Institute, Toronto) kindly provided all other SH3 domain constructs. Glutathione S-transferase (GST)–EnaC (the C terminus of Ena) was constructed by subcloning Ena amino acids 440–684 into pGEX5X (Amersham Pharmacia). GST fusion proteins were expressed in Escherichia coli DH5-α, prepared as described (12), and quantitated by Coomassie blue staining.
Solution Binding Assays and ena Mutant-Rescue Crosses.
Transfected S2 cell lysates or whole-fly lysates were incubated with equivalent amounts of GST, GST–SH3, GST-EnaC, or GST–Ena immobilized on glutathione Sepharose (Amersham Pharmacia) for 1 h at 4°C. Beads were washed twice with immunoprecipitation buffer and then boiled in SDS sample buffer. Bound proteins were analyzed by SDS/PAGE, followed by Western blot analysis. For peptide-competition assays, fusion proteins were preincubated for 30 min with the following peptides: Abl-Pro-AEPPPYPPPPIPGGK, Src-Pro-AERSSRPLPPIPGGK, ena-A-1-PGGPPAPAPPPPPPS, Ena-A-2-PGGPGAPPPPPPPPG, ena-Src-1-PPQAPQPPLQNGGMY, ena-Src-2-PAPAPPPPPPSFGGA, and ena-drk-1-PGYGGPPVPPPQQQA (Biosynthesis, Lewisville, TX). ena mutant-rescue crosses were carried out as described, except that the ena mutations ena210 and ena87 were used (4, 10).
Yeast Two-Hybrid System Screen.
A cDNA encoding the C-terminal 243 amino acids of Ena was fused to the sequence encoding the GAL4 DNA binding domain in the pAS1-CYH2 vector to create pAS-EnaC. The Saccharomyces cerevisiae strain Y190, which contains the reporter genes HIS3 and LacZ, was cotransformed with pAS-EnaC and the Drosophila larval-library pAct (gift of Stephen J. Ellidge, Baylor College of Medicine, Houston), in which cDNAs are fused to the sequence encoding the GAL4 activation domain. Transformants (n = 2.05 × 107) were screened for activation of the reporter genes by spreading the cells on medium lacking histidine and supplemented with 30 mM 3-aminotriazole. Colonies that grew in the absence of histidine appeared within 7 days after plating and were assayed for β-galactosidase production by a membrane-transfer assay. Plasmid DNA from the positive clones was isolated and sequenced.
Immunofluorescence Microscopy of Transfected Cells.
Ptk2 cells (CCL56, American Type Culture Collection) were grown in MEM (Life Technologies, Gaithersburg, MD), supplemented with 10% (vol/vol) FCS. Cells were transfected by the calcium phosphate method with pCMV/Ena (4). After 48 h, cells were washed once with PBS and incubated briefly with trypsin/EDTA solution (Life Technologies) to remove the cells from the Petri dish. Cells were then plated on coverslips in MEM/10% (vol/vol) FCS and allowed to spread for 30 min. Cells were washed with PBS, fixed, extracted in −20°C methanol, and incubated with 10 μg/ml affinity-purified rabbit antiserum raised against amino acids 55–235 of Ena and, for the detection of profilin, with tissue-culture supernatant of the 2H11 Hybridoma (gift of B. M. Jockusch, Zoological Institute, Technical University of Brunswick, Brunswick, Germany; ref. 25), diluted 1:4. Primary antibodies were detected by tetramethylrhodamine B isothiocyanate-labeled donkey anti-rabbit antibodies (Ena staining) and dichlorotriazinyl aminofluorescein-labeled goat anti-mouse antibodies (Dianova, Hamburg, Germany). Cells were examined with a Leitz Aristoplan microscope equipped with epifluorescence.
RESULTS
Isolation of the Drosophila Profilin Homolog, Chickadee, in a Yeast Two-Hybrid Screen.
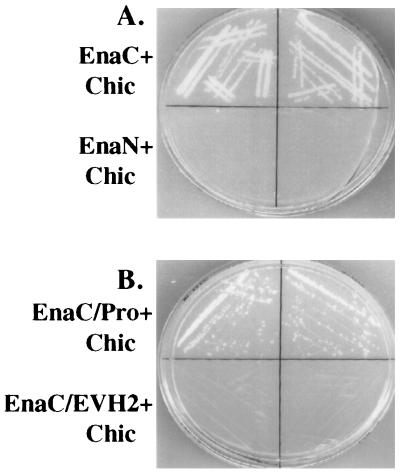
To identify target proteins for the C-terminal 243 amino acids of Ena, we performed a yeast two-hybrid screen. The C-terminal 243 amino acids of Ena, which include the consensus binding site for profilin (5, 9, 26, 27) and a proline-rich consensus site for binding the Abl SH3 domain (28), were fused to the DNA binding domain of the yeast transcription factor GAL4 and used to screen a Drosophila third-instar larval library whose inserts were fused to the activation domain of GAL4. The separately expressed domains are unable to activate transcription of the reporter genes HIS3 and LacZ unless a protein–protein interaction takes place (29). Of 20.5 million clones screened, 9 interacted with Ena as assessed by expression of both the HIS and LacZ reporter genes. One of these clones carried a cDNA encoding full-length chickadee, the Drosophila homolog of profilin. The interaction was specific, because a construct with the Ena N-terminal domain fused to the DNA binding domain of GAL4 did not interact with the same isolated chickadee clone (Fig. 1A). Of the seven remaining clones, two were partial Ena cDNAs (4) and the other five are unique sequences that are yet to be described.
Figure 1.
Identification of chickadee as a binding partner for Ena in a yeast two-hybrid screen. (A) The first 235 amino acids of Ena (EnaN) and last 243 amino acids of Ena (EnaC) were fused to the sequence encoding the GAL4 DNA binding domain in the pAS1-CYH2 vector. These two constructs were cotransformed with GAL4AD-chickadee (Chic), a clone in which the full-length chickadee coding sequences were fused to the sequence encoding the GAL4 activation domain and which was isolated in a yeast two-hybrid screen by using the C-terminal 243 amino acids of Ena as bait. From each of the cotransformations, two independent clones were spread on medium lacking histidine and analyzed for growth. EnaC, which contains two putative profilin binding sites, can interact with chickadee, whereas EnaN, which contains no putative binding sites for profilin, cannot. (B). Amino acids 440–490 from EnaC containing the putative profilin binding sites (EnaC/Pro) and amino acids 490–684 from EnaC lacking any putative profilin binding sites (EnaC/EVH2) were fused to the DNA binding domain of the yeast transcription factor GAL4 and cotransformed with GAL4AD chickadee. Cotransformed yeast was spread on medium lacking histidine and analyzed for growth. EnaC/Pro can interact with chickadee, whereas EnaC/EVH2 cannot.
The region of the Ena protein used as bait in the yeast two-hybrid screen contains several matches to a putative profilin binding site (4, 9, 26, 27). To test whether these sequences were important for mediating the interaction with chickadee, DNA encoding Ena amino acids 440–490, which contains these putative binding sites, and DNA encoding Ena amino acids 490–684 were fused to the DNA binding domain of the yeast transcription factor GAL4. Yeast were cotransformed with each of these constructs, and the chickadee cDNA was fused to the activation domain of GAL4 and tested for activation of transcription of the reporter genes HIS3 and LacZ. An interaction was detected when chickadee was cotransformed with Ena amino acids 440–490 and not Ena amino acids 490–684, suggesting that this interaction is mediated by proline-rich sequences in Ena (Fig. 1B).
Association of Drosophila Ena with Chickadee in Vitro.
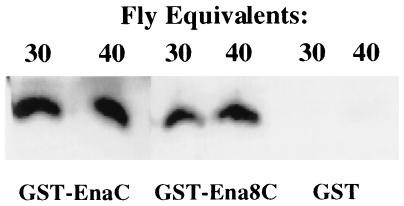
To test the specificity of the interaction between Ena and chickadee detected in the yeast two-hybrid assay, we tested the ability of GST–Ena fusion proteins to precipitate chickadee from a complex cell lysate. Ena amino acids 374–684, which contain the Ena sequences present in the yeast two-hybrid expression construct as well as additional Ena proline-rich sequences with consensus binding sites for profilin, were expressed as a GST fusion protein. This fusion protein was tested in solution binding assays for binding to chickadee in lysates prepared from adult Drosophila. The Ena fusion protein precipitated chickadee from the fly lysate, unlike GST alone (Fig. 2), and we conclude that Ena and chickadee interact in vitro.
Figure 2.
Binding of Ena protein to the Drosophila profilin homolog, chickadee. Serial dilutions of whole adult Drosophila lysates were split into two equal aliquots and bound in solution to an equal amount of either GST alone or GST fused to Ena amino acids 374–684. Retained proteins after solution binding were detected with anti-chickadee antibodies. The GST–Ena protein bound reproducibly to chickadee. The faint band seen in the GST alone bands is nonspecific Ena binding to the GST control.
Colocalization of Ena and Profilin in Cultured Cells.
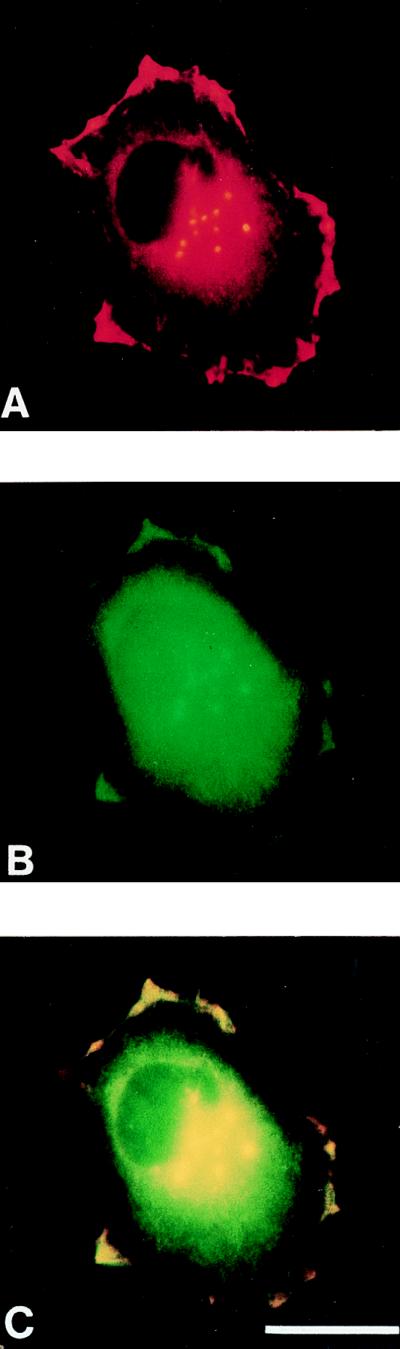
Profilin has been shown to be localized to cortical microfilament webs and leading lamellae of spreading or locomoting cells (30). In Drosophila, profilin is expressed ubiquitously throughout development, e.g., there are high levels of profilin in the ventral nerve cord of stage 16 embryos (15). The Ena protein is localized to actin stress fibers and focal adhesions in cultured cells (4) and is localized to the axonal tracts of the developing Drosophila embryonic central nervous system, although the small size of these cells makes higher-resolution localization difficult (11). Because Ena and chickadee interact in vitro and are expressed in the nervous system of Drosophila embryos, we speculated that these two proteins might interact in vivo in regions of dynamic actin remodeling. We therefore compared the subcellular distribution of transfected Drosophila Ena and endogenous profilin in spreading cultured Ptk2 cells. Ena and profilin were colocalized to the periphery of the spreading cells (Fig. 3). The colocalization, together with the biochemical interactions, suggests that Ena and profilin associate in vivo.
Figure 3.
Colocalization of Ena and profilin in cultured spreading PTK2 cells. PTK2 cells were transfected with an expression vector carrying the ena cDNA. On the second day after transfection, cells were treated with trypsin and allowed to spread on coverslips for 30 min. Cells were fixed and made permeable in methanol and double-labeled for Ena (A) and profilin (B). C merges A and B. Both Ena and profilin can be detected at the periphery of the spreading cells. (Bar = 20 μm.)
Solution Binding of Ena to SH3 Domains.
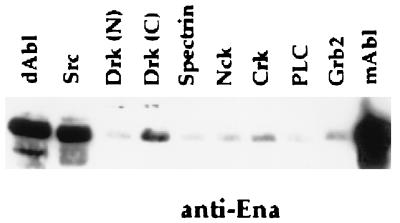
The proline-rich region of Ena contains multiple consensus binding sites for SH3 domain in addition to the profilin binding sequences (28). It has been shown with a filter binding assay that Ena binds the Abl and Src SH3 domains in vitro (10, 11). We decided to examine the SH3 binding specificity of Ena further by using a solution binding assay. Ena was expressed in Drosophila S2 cells, and the transfected cell lysates were incubated with a series of GST–SH3 fusions. Ena bound specifically to the Drosophila and murine Abl–SH3 domains and the murine src SH3 domain. Ena also bound to the C-terminal but not to the N-terminal SH3 domain of Drk (Fig. 4).
Figure 4.
SH3 domain binding of Ena. S2 cells were transfected with Ena under the control of an actin promoter. Proteins retained after solution binding to equal amounts of various GST–SH3 domains were blotted with anti-Ena N-terminal antibodies. None of the proteins bound to the GST negative control (data not shown). Ena binds to the murine and Drosophila Abl SH3 domains, the src SH3 domain, and the C-terminal SH3 domain of Drk.
Mapping of the Abl SH3 Binding Site in Ena.
Consensus sequences have been identified for binding to the Abl, Src, and Drk SH3 domains by using combinatorial peptides (31, 32). To identify those regions of the Ena proline-rich sequence that could be mediating SH3 domain binding, the proline motifs in Ena most similar to these published consensus sequences were identified, and peptides were synthesized to these regions. The two Ena peptides most closely matching the Abl consensus binding motif (Fig. 5A) partially and specifically blocked Ena binding to the Abl SH3 domain at 30 μM (ena-A-1) and 10 μM (ena-A-2), respectively, although they did not block Src SH3 domain binding (Fig. 5B). Peptides derived from Ena proline-rich sequences most closely matching the optimal sequences for Src or Drk SH3 binding did not compete for Ena binding with any of the SH3 domains tested (data not shown).
Figure 5.
Proline-rich motifs in Ena involved in SH3 binding. (A) A-1 represents Ena amino acids 401–415, and A-2 represents Ena amino acids 464–478. Asterisks indicate the residues chosen for mutagenesis from proline to alanine to generate Ena8. (B) S2 cells were transfected with Ena, and the lysates were incubated with glutathione-Sepharose-bound GST–AblSH3 or SrcSH3 preincubated with the indicated concentrations of peptides. Bound proteins were blotted with anti-Ena antibodies. The optimal consensus binding motifs for Abl (Abl–Pro) and Src (Src–Pro) specifically blocked binding of Ena to the Abl or to the Src SH3 domains, respectively. Ena A-1 at 30 μM and Ena A-2 at 10 μM specifically blocked binding of Ena to the AblSH3 domain. (C) Wild-type ena or ena8 was transfected into S2 cells. Serial 2-fold dilutions of transfected cell lysates were incubated with glutathione-Sepharose-bound GST–AblSH3 (lanes 1–3). Bound proteins were blotted with anti-Ena antibodies. Binding of the mutant Ena8 to the Abl SH3 domain was significantly reduced. The difference in binding between wild-type Ena and Ena8 was more evident at lower concentrations of protein. Expression levels of wild-type and mutant Ena were equivalent (lane 4).
To determine whether the proline motifs identified in the peptide binding experiment as Abl SH3 binding sites were sufficient to mediate Abl SH3 binding, site-directed mutagenesis was employed to change eight prolines to alanine, thereby eliminating many of the PXXP motifs present in the sites. Serial two-fold dilutions of transfected cell lysates containing either the mutant Ena protein (Ena8 P → A) or wild-type Ena were tested for solution binding to the Abl SH3 domain. At higher concentrations of protein, it was difficult to detect an effect of the proline-to-alanine mutations on binding. However, at lower concentrations of the Ena proteins, binding of the mutant protein was markedly reduced compared with the wild-type Ena protein (Fig. 5C).
To examine the in vivo effect of the proline-to-alanine mutations on Ena function, transgenes expressing wild-type Ena and the Ena8 P → A mutant proteins were tested for their ability to rescue ena mutant lethality. The ena8 P → A transgene rescued the embryonic lethality associated with loss-of-function mutations in ena as well as the wild-type ena transgene. The ena8 P → A-rescued flies were phenotypically normal and had viability and fertility comparable to wild-type ena-rescued flies (data not shown). Thus, the proline-to-alanine mutations present in Ena8 P → A are not sufficient to disrupt an essential function of the Ena protein.
Chickadee Binding to Ena8 P → A.
The data reported thus far have identified two ligands for the proline-rich region of Ena and what may be binding sites for these ligands. Interestingly, there is some overlap in the binding sites for the Abl SH3 domain and some of the putative binding sites for P → A profilin. Because mutation of the prolines in these overlapping regions reduces binding to the Abl SH3 domain, we wondered whether these mutations also would disrupt binding to chickadee. The mutant ena8 P → A cDNA was subcloned in the pGex expression vector, and the resulting mutant Ena fusion protein, GST–Ena8 P → A, was compared with wild-type GST–Ena in solution binding assays for its ability to pull down chickadee from serial dilutions of lysates prepared from adult Drosophila. The GST–Ena8 P → A fusion protein and wild-type GST–Ena pulled down approximately equivalent amounts of chickadee (Fig. 6), suggesting that distinct amino acids may be important for Ena binding to profilin and the Abl-SH3 domain, despite the overlap observed in some of their putative binding sites.
Figure 6.
Chickadee binding to the Ena mutant protein Ena8 P → A. Whole Drosophila lysates from 60 or 80 adult flies were split into three equal aliquots and bound in solution to equal amounts of GST alone, GST fused to Ena amino acids 374–684, or GST fused to Ena8 P → A amino acids 374–684. Proteins retained after solution binding were detected with anti-chickadee antibodies. Reproducibly, the GST–Ena protein, the GST–Ena8 P → A protein, and GST alone bound equally well to chickadee.
DISCUSSION
Previous studies of the Drosophila Ena protein have implicated it as both a downstream component of Abl-mediated signaling and a protein involved in cytoskeleton assembly (4, 10, 11). The Ena protein has a central proline-rich region that contains multiple sequences with similarity to previously identified profilin and Abl SH3 domain binding sites. We have examined these potential interactions and report here that Ena is a ligand for both the Drosophila homolog of profilin, chickadee, and SH3 domains. Our data support the developing hypothesis that Ena may serve as a bridge between Abl-mediated signaling and cytoskeletal dynamics.
The interaction between Ena and chickadee was observed initially in a yeast two-hybrid screen and was confirmed subsequently in a solution binding assay. Ena deletions leaving only proline-rich sequences retain their ability to interact with chickadee in the yeast two-hybrid assay, suggesting that this interaction occurs through these sequences. Although attempts were made to coimmunoprecipitate Ena and chickadee, these experiments were unsuccessful. Interestingly, coimmunoprecipitation of profilin with other poly-(l-proline)-containing ligands, such as VASP, Mena, and p140mDia, have not been reported (5, 9, 33). The reasons why these coimmunoprecipitations have not been reported are unclear, although these ligands’ interactions with profilin may be too transient or weak to survive the process of immunoprecipitation.
We have shown that Ena and profilin are colocalized at the periphery of spreading transfected mammalian cells, lending further support to the in vivo relationship between these two proteins. Colocalization to regions of dynamic cytoskeletal activity is consistent with a role for these two protein in regulating cytoskeletal dynamics. Recent data (D. Van Vactor and C. Goodman, personal communication) provide in vivo confirmation of the importance of the interaction between Ena and profilin. In a genetic screen to identify genes involved in outgrowth of motor neurons, two alleles of chickadee were identified. These mutant chickadee alleles have defects in embryonic motor-neuron outgrowth and a slight disorganization of the longitudinal pathways in the embryonic central nervous system. Other studies looking more closely at phenotypes in the nervous system of Abl mutations showed that Abl and profilin mutant animals have nearly identical phenotypes in the intersegmental nerve b. In both Abl and chickadee mutant animals, the intersegmental nerve b stops short of its distal target muscle, consistent with the Abl tyrosine kinase signaling pathway and chickadee playing a role in the same cellular processes. Given the similarity in these phenotypes, the researchers examined potential genetic interactions between alleles of chickadee and Ena. Interestingly, transheterozygous combinations of Ena and chickadee alleles result in an enhancement of a subset of the phenotypes seen in either mutant alone (D. Van Vactor, personal communication). Taken together with our biochemical data, this evidence suggests compellingly that chickadee and Ena—and, by analogy, the Abl signaling pathway—may interact in vivo during development of the embryonic nervous system.
That chickadee is linked to a signaling molecule is consistent with the increasing body of evidence suggesting that profilin is recruited to sites of actin polymerization by signaling molecules. Also consistent with this theory are the binding of profilin to Ena/VASP family members and the interactions between profilin, p140mDia, a downstream effector of rho (33), and N-WASP, an effector of Cdc42 (34). These interactions may serve to recruit profilin to specific sites in the cell where actin remodeling is taking place in response to external signals. The localization of profilin to the lamellipodia of fibroblasts (35) and PtK2 epithelial cells (25) as well as to areas of the cell where focal contacts are being formed and focal-adhesion proteins are localized (36) is strong support for a role for profilin in signaling and cytoskeletal changes.
In addition to binding profilin, the central proline-rich region of Ena is also a ligand for SH3 domains. SH3 domains mediate protein–protein interactions important for a large number of cellular activities, including regulation of catalytic activity, recruitment of substrates, subcellular localization of proteins, and forming signaling complexes (37–40). We have investigated the specificity of Ena’s interactions with SH3 domains and show that the Abl, Src, and Drk SH3 domains seem to be the preferred in vitro ligands for Ena, although it is not yet clear what SH3 domain-containing proteins may interact with Ena in vivo. Interestingly, VASP binding to Abl and Src SH3 domains also has been observed in vitro (S.M.A.-D. and F.M.H., unpublished data; S. Feller and U.W., unpublished data). SH3 domains are known to interact with proline-rich regions of their ligands (29). Consistent with this interaction, we have identified two poly-(l-proline) motifs in Ena that mediate binding to the Abl SH3 domain. Interestingly, the Abl SH3 binding sites that we have mapped in Ena also map closely to several of the putative chickadee binding sites in Ena, but the mutations that affect Ena binding to SH3 domains have no effect on binding to chickadee in the assays performed. These results suggest that there may be some specificity in sites recognized by these distinct proteins. It is worth noting that no SH3 domains were isolated in the yeast two-hybrid screen that identified profilin as a binding partner for Ena. Perhaps the proline-rich sequences present in the Ena bait are not the most important for binding to SH3 domains. Alternatively, the conditions in the yeast two-hybrid screen may not favor detection of an interaction between an SH3 domain and proline-rich sequences. Another possibility is that Ena’s interaction with the Abl SH3 domain may be less physiologically relevant than that with chickadee. Indeed, we have shown that mutations that disrupt binding to the Abl SH3 domain in vitro have no effect in vivo when they are expressed from a heterologous promoter. It will be important to identify critical amino acids for the interaction between chickadee and Ena and to examine whether these mutations have any in vivo effects.
Studies have shown that the Ena protein is linked both to Abl-mediated signaling and cytoskeletal assembly (4, 10, 11). Our findings that Ena binds both SH3 domains and the actin-binding protein profilin lend further support to these observations. Our previous report that Ena phosphorylation by Abl attenuates SH3 binding raises the possibility that this modulation in SH3 binding may regulate its interaction with other proteins during axon outgrowth in the central nervous system of the developing Drosophila embryo. One obvious candidate for such a protein is the Drosophila profilin homolog chickadee. Interaction of Ena/VASP family members with profilin is believed to be critical for their role in promoting actin polymerization. Thus, this interaction would provide a mechanism for regulating actin assembly by Ena and thus influence localized actin filament assembly that would alter growth-cone motility in response to extracellular signals. Interaction of Ena with both SH3 domains and profilin further supports the view that Ena provides a bridge between signal transduction and the cytoskeleton.
Acknowledgments
We thank Drs. David Van Vactor and Zach Wills for helpful discussions and sharing unpublished results. This work was supported by National Institutes of Health Grants CA 48582 (to F.M.H.) and GM50877 and a grant from the Deutsche Forschungsgemeinschaft (to U.W.). S.M.A. was supported by National Institutes of Health Postdoctoral Training Grant CA9681 and a fellowship from the National Cancer Institute.
ABBREVIATIONS
- VASP
vasodilator-stimulated phosphoprotein
- SH3
src homology 3
- GST
glutathione S-transferase
References
- 1.Carlier M F. Curr Opin Cell Biol. 1998;10:45–51. doi: 10.1016/s0955-0674(98)80085-9. [DOI] [PubMed] [Google Scholar]
- 2.Dramsi S, Cossart P. Annu Rev Cell Dev Biol. 1998;14:137–166. doi: 10.1146/annurev.cellbio.14.1.137. [DOI] [PubMed] [Google Scholar]
- 3.Elch M D, Mallavarapu A, Rosenblat J, Mitchison T J. Curr Opin Cell Biol. 1997;9:54–61. doi: 10.1016/s0955-0674(97)80152-4. [DOI] [PubMed] [Google Scholar]
- 4.Ahern-Djamali S M, Comer A R, Bachmann C, Kastenmeier A S, Reddy S K, Beckerle M C, Walter U, Hoffmann F M. Mol Biol Cell. 1998;9:2157–2171. doi: 10.1091/mbc.9.8.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gertler F, Niebuhr K, Reinhard M, Wehland J, Soriano P. Cell. 1996;87:227–239. doi: 10.1016/s0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- 6.Haffner C, Jarchau T, Reinhard M, Hoppe J, Lohmann S M, Walter U. EMBO J. 1995;14:19–27. doi: 10.1002/j.1460-2075.1995.tb06971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakraborty T, Ebel F, Domann E, Niebuhr K, Gerstel B, Pistor S, Temm-Grove C J, Jockusch B M, Reinhard M, Walter U, et al. EMBO J. 1995;4:1314–1321. doi: 10.1002/j.1460-2075.1995.tb07117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pistor S, Chakraborty T, Walter U, Wehland J. Curr Biol. 1995;5:1–10. doi: 10.1016/s0960-9822(95)00104-7. [DOI] [PubMed] [Google Scholar]
- 9.Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch B M, Walter U. EMBO J. 1995;14:1583–1585. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comer A R, Ahern-Djamali S M, Juang J-L, Jackson P D, Hoffmann F M. Mol Cell Biol. 1997;18:152–160. doi: 10.1128/mcb.18.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gertler F B, Comer A R, Juang J-L, Ahern S M, Clark M J, Liebl E C, Hoffmann F M. Genes Dev. 1995;9:521–533. doi: 10.1101/gad.9.5.521. [DOI] [PubMed] [Google Scholar]
- 12.Haarer B K, Lillie S H, Adams A E M, Magdolen V, Bandlow W, Brown S S. J Cell Biol. 1990;110:105–114. doi: 10.1083/jcb.110.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sohn R H, Goldschmidt-Clermont P S. BioEssays. 1994;16:465–472. doi: 10.1002/bies.950160705. [DOI] [PubMed] [Google Scholar]
- 14.Cooley L, Verheyen E, Ayers K. Cell. 1992;69:173–184. doi: 10.1016/0092-8674(92)90128-y. [DOI] [PubMed] [Google Scholar]
- 15.Verheyen E M, Cooley L. Development (Cambridge, UK) 1994;120:717–728. doi: 10.1242/dev.120.4.717. [DOI] [PubMed] [Google Scholar]
- 16.Carlsson L, Nystrom L E, Sundkvist I, Markey F, Linderg U. J Mol Biol. 1977;115:465–483. doi: 10.1016/0022-2836(77)90166-8. [DOI] [PubMed] [Google Scholar]
- 17.Pantaloni D, Carlier M F. Cell. 1993;75:1007–1014. doi: 10.1016/0092-8674(93)90544-z. [DOI] [PubMed] [Google Scholar]
- 18.Lassing I, Lindberg U. Nature (London) 1985;314:472–474. doi: 10.1038/314472a0. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka M, Shibata H. Eur J Biochem. 1985;151:291–287. doi: 10.1111/j.1432-1033.1985.tb09099.x. [DOI] [PubMed] [Google Scholar]
- 20.Mullins R D, Kelleher J F, Xu J, Pollard T D. Mol Biol Cell. 1998;9:841–852. doi: 10.1091/mbc.9.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welch M D, Rosenblatt K, Skoble J, Portnoy D A, Mitchison T J. Science. 1998;281:105–108. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- 22.Goldschmidt-Clermont P J, Kim J W, Machesky L M, Rhee S G, Pollard T D. Science. 1991;251:1231–1233. doi: 10.1126/science.1848725. [DOI] [PubMed] [Google Scholar]
- 23.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 24.Deng W P, Nickoloff J A. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 25.Mayboroda O, Schluter K, Jockusch B M. Cell Motil Cytoskeleton. 1997;37:166–177. doi: 10.1002/(SICI)1097-0169(1997)37:2<166::AID-CM9>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 26.Purich D L, Southwick S. Biochem Biophys Res Commun. 1997;231:686–691. doi: 10.1006/bbrc.1997.6158. [DOI] [PubMed] [Google Scholar]
- 27.Kang F, Laine R O, Bubb M R, Southwick F S, Purich D L. Biochemistry. 1997;36:8384–8392. doi: 10.1021/bi970065n. [DOI] [PubMed] [Google Scholar]
- 28.Ren R, Mayer B J, Cicchetti P, Baltimore D. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- 29.Chien C-T, Bartel P L, Strenglanz R, Fields S. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rothkegal M, Mayboroda O, Rohde M, Wucherpfennig C, Valenta R, Jockusch B M. J Cell Sci. 1996;109:83–90. doi: 10.1242/jcs.109.1.83. [DOI] [PubMed] [Google Scholar]
- 31.Feng S, Chen J K, Yu H, Schreiber S L. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 32.Yu H, Rosen M K, Shin T B, Seidel D C, Brugge J S, Schreiber S L. Science. 1992;258:1665–1668. doi: 10.1126/science.1280858. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe N, Maduale P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Joskusch B M, Narumiya S. EMBO J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suetsugu S, Hiroaki M, Takenawa T. EMBO J. 1998;17:6516–6526. doi: 10.1093/emboj/17.22.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buss F, Temm-Grove C, Henning A, Jockusch B M. Cell Motil Cytoskeleton. 1992;22:51–61. doi: 10.1002/cm.970220106. [DOI] [PubMed] [Google Scholar]
- 36.Moldovan N I, Milliken E E, Irani K, Chen J, Sohn R H, Finkel T, Goldschmidt-Clermont P J. Curr Biol. 1996;7:24–30. doi: 10.1016/s0960-9822(06)00024-8. [DOI] [PubMed] [Google Scholar]
- 37.Bar-Sagi D, Rotin D, Batzer A, Mandiyan V, Schlessinger J. Cell. 1993;74:83–91. doi: 10.1016/0092-8674(93)90296-3. [DOI] [PubMed] [Google Scholar]
- 38.Gout I R, Dhand I D, Hiles M J, Fry G, Panayotou P, Das O, Waterfield M D. Cell. 1993;75:25–36. [PubMed] [Google Scholar]
- 39.Simon M A, Dodson G S, Rubin G M. Cell. 1993;73:169–177. doi: 10.1016/0092-8674(93)90169-q. [DOI] [PubMed] [Google Scholar]
- 40.Weng S, Thomas S J, Rickles R J, Taylor J A, Brauer A W, Siedel-Dugan C, Michael W M, Dreyfuss G, Brugge J S. Mol Cell Biol. 1994;14:4509–4521. doi: 10.1128/mcb.14.7.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]