Abstract
We describe a novel interaction between HIV-1 Rev and microtubules (MTs) that results in the formation of bilayered rings that are 44–49 nm in external diameter, 3.4–4.2 MD (megadaltons) in mass, and have 28-, 30-, or 32-fold symmetry. Ring formation is not sensitive to taxol, colchicine, or microtubule-associated proteins, but requires Mg2+ and is inhibited by maytansine. The interaction involves the NH2-terminal domain of Rev and the face of tubulin exposed on the exterior of the MTs. The NH2-terminal half of Rev has unexpected sequence similarity to the tubulin-binding portion of the catalytic/motor domains of the microtubule-destabilizing Kin I kinesins. We propose a model wherein binding of Rev dimers to MTs at their ends causes segments of two neighboring protofilaments to peel off and close into rings, circumferentially containing 14, 15, or 16 tubulin heterodimers, with Rev bound on the inside. Rev has a strong inhibitory effect on aster formation in Xenopus egg extracts, demonstrating that it can interact with tubulin in the presence of normal levels of cellular constituents. These results suggest that Rev may interact with MTs to induce their destabilization, a proposition consistent with the previously described disruption of MTs after HIV-1 infection.
Keywords: acquired immunodeficiency syndrome, HIV-1 Rev, kinesin, microtubules, tubulin
Introduction
HIV-1 is the causative agent of human acquired immune deficiency syndrome. It has been shown that HIV-1 infection of human epidermal keratinocytes and intestinal epithelial cells induces cellular ultrastructural changes and massive disruption of microtubules (MTs). These changes have been attributed in part to the activities of HIV-1 gp120 (Cenacchi et al. 1996; Delezay et al. 1997; Malorni et al. 1997). Numerous changes in the vimentin and actin cytoskeleton organizations are also observed after HIV-1 infection and these have been ascribed to interactions with the viral proteins Vif, Vpr, and Nef (Kaminchik et al. 1994; Macreadie et al. 1995; Karczewski and Strebel 1996; Fackler et al. 1997; Gu et al. 1997).
MTs comprise one of the major cytoskeletal systems of the cell and play an essential role in cytoplasmic organization and cell division. MTs interact with a variety of proteins, including the kinesins, a group of plus- and minus-end–directed motor proteins that travel along MTs to accomplish intracellular transport (Titus and Gilbert 1999). MTs either can be stable structures, as in ciliary axonemes, or dynamic ones, as in mitotic spindles, where they alternately undergo growth and shrinkage. This dynamic instability is regulated by several proteins that either stabilize MTs or induce their disassembly. The stabilization of MTs is affected by a variety of microtubule-associated proteins (MAPs) such as MAP2 and MAP4 (Drewes et al. 1999). Conversely, the destabilization of MTs is caused by proteins such as the katanins, which break MTs internally to initiate disassembly (Hartman and Vale 1999), or by proteins that catalyze end-based disassembly either from one end, such as Stathmin/Oncoprotein 18 (Wallon et al. 2000), or from both ends, such as XKCM1 and XKIF2 (Desai et al. 1999). Together, these proteins regulate MT dynamics, and, as illustrated by the antagonistic activities of XKCM1 and XMAP215, may act as regulatory pairs (Tournebize et al. 2000).
Rev is a key HIV-1 regulatory protein produced early in infection that enables the export of unspliced and partially spliced mRNA from the nucleus, thereby inducing the switch to the late phase of the viral replication cycle. Rev binds to these transcripts via a specific interaction between its arginine-rich motif and the phosphate groups in the Rev response element on the mRNA (Hammarskjold 1977; Cole and Saavedra 1997; Cullen 1998; Frankel and Young 1998; Izaurralde and Adam 1998; Mattaj and Englmeier 1998; Pollard and Malim 1998; Fukumori et al. 1999). Rev also interacts with various host proteins (Bogerd et al. 1995; Fisher et al. 1995; Miyazaki et al. 1995; Stutz et al. 1995; Fritz and Green 1996; Bevec et al. 1996; Henderson and Percipalle 1997; Farjot et al. 1999). Because Rev has both a nuclear export signal and a nuclear localization signal it is able to function as a nucleocytoplasmic shuttle protein that conveys the viral mRNAs out through the nuclear pore in a manner that avoids default splicing (Hammarskjold 1977; Cole and Saavedra 1997; Cullen 1998; Frankel and Young 1998; Izaurralde and Adam 1998; Mattaj and Englmeier 1998; Pollard and Malim 1998; Fukumori et al. 1999).
Rev is a small (13-kD), basic (pI = 9.2) protein with a high affinity for RNA (Heaphy et al. 1991; Wingfield et al. 1991; Daly et al. 1993; Zemmel et al. 1996). In addition to its cognate RNAs, Rev also binds strongly to other polyanions such as poly G, poly dG, and even polyglutamate, presumably via the arginine-rich motif. In fact, Rev, which polymerizes in vitro into long and very stable tubes (Heaphy et al. 1991; Wingfield et al. 1991; Watts et al. 1998), is effectively depolymerized by all of these polyanions. On the other hand, tubulin is a heterodimer composed of two 50-kD acidic monomers (pI = 4.8 and 5.2, respectively), positioned in MTs in such a way that the COOH-terminal polyglutamate tracts of the monomers are all presented on the outside of the structure (Sackett 1995a; Nogales et al. 1998, Nogales et al. 1999). Such an arrangement suggests a priori that Rev and tubulin may interact via the highly charged arginine-rich motif on the former and the polyglutamate tract on the latter.
In this paper we describe a strong interaction between Rev and MTs in both defined in vitro and cell extract conditions. We have observed in vitro that Rev filaments react readily with MTs to form Rev–tubulin toroidal (RTT) complexes with a mean external diameter of 47 nm. These structures are somewhat reminiscent of those formed by GDP-tubulin (Diaz et al. 1994), or those formed when MTs are either disassembled by cooling (Melki et al. 1989; Mandelkow et al. 1991) or exposed to antineoplastic drugs such as Dolastatin-10 (Bai et al. 1995). These complexes form even in the absence of the tubulin polyglutamate tail. Rev also has unexpected sequence similarity to the tubulin-binding domain of the microtubule-destabilizing kinesin-like proteins XKCM1 and XKIF2, indicating a mode of interaction other than a simple attraction between the glutamate and arginine tracts. Xenopus egg extract assays, used to assess the effect of Rev on MT polymerization in an intracellular milieu, demonstrate the probability of an in vivo interaction between Rev and MTs. Taken together, these results demonstrate the potential for Rev to interact with tubulin to alter MT dynamics in HIV-1–infected cells.
Materials and Methods
Proteins
HIV-1 Rev.
The expression, purification, and polymerization of Rev protein has been described previously (Wingfield et al. 1991; Watts et al. 1998). In brief, Rev was expressed in Escherichia coli and purified by a combination of ion exchange and gel filtration steps in the presence of urea. Rev was refolded and polymerized by dialysis against 50 mM sodium phosphate, 600 mM ammonium sulfate, 150 mM sodium chloride, 50 mM sodium citrate, pH 7.0, and then exhaustively dialyzed against 20 mM Hepes, 100 mM sodium chloride, 50 mM sodium citrate, pH 7.0. Filaments were concentrated by pelleting and resuspension in a minimal volume of the same buffer. Protein concentrations, corrected for light scattering as described (Wingfield et al. 1991), were typically 100 mg/ml. Rev was kept at 4°C at all times. For some experiments a nonpolymerizing NH2-terminal construct of Rev (Rev 1–59) was also used.
Tubulin.
Rat brain tubulin was purified from MT protein (Sackett et al. 1991) by differential polymerization as previously described (Wolff et al. 1996). Purified tubulin was stored in 25 mg/ml stocks in 100 mM MES, 1 mM EGTA, 1 mM MgCl2, pH 6.9 (MEM), drop-frozen in liquid nitrogen, and subsequently thawed only once before use within 6 h. Cow brain tubulin was prepared similarly. Subtilisin-cleaved tubulin, in which the acidic COOH-terminal peptide is removed from both α- and β-tubulin (Bhattacharyya et al. 1985; Sackett 1995a), was prepared as previously described (Knipling et al. 1999). Tubulin was either polymerized into MTs (2 mM GTP and 50 μM Taxol present in the buffer) or maintained as dimers and small oligomers (2 mM GDP, 50 μM colchicine).
Formation of RTT and Tubulin–Drug Complexes
Rev stock, typically 100 mg/ml in 20 mM Hepes, 100 mM sodium chloride, and 50 mM sodium citrate, pH 7.0, was diluted just before use with 100 mM MES and 2 mM MgCl2, pH 6.9, to minimize aggregation of the protein due to reduction in the concentration of citrate ion. Tubulin stock, typically 25 mg/ml in MEM, was thawed, warmed to 37°C, and diluted with MEM also just before use, in this case to avoid time-dependent denaturation of the protein. In most experiments, Rev was at 0.52 mg/ml and tubulin at 2 mg/ml. Mixing in equal volume (and equimolar monomer) ratios at room temperature resulted in the immediate formation of ring-like complexes. Note: it is conventional to refer to tubulin concentrations relative to the α/β tubulin heterodimer; however, this convention does not apply to Rev. Consequently, for purposes of clarity and simplicity, all protein stoichiometries in this communication refer to molecular monomers, e.g., Rev/tubulin 1:1 means 1 Rev polypeptide chain per 1 tubulin polypeptide chain. Ring-like complexes involving Rev 1–59 were prepared in a similar fashion. Ring-like complexes of tubulin were also induced with the antineoplastic drug Dolastatin-10. Tubulin at 1 mg/ml (20 μM monomer) in MEM and drug (15 μM) were incubated for 1 h at room temperature and then diluted 10-fold with MEM for electron microscopy.
Rev–Tubulin Complexes; Masses and Stoichiometries
Stoichiometry of Rev–Tubulin in Soluble Complexes.
Rev–tubulin complex formation is Mg2+ dependent; if no Mg2+ ion is present (buffer is 100 mM MES, 1 mM GDP, and 1 mM EGTA, pH 6.9) or chelated by excess citrate then no structures are visible in the electron microscope and only small soluble complexes are formed. Gel filtration on a Superdex-200 column equilibrated with 100 mM MES, pH 6, gave a single broad peak with the retention time of a 110-kD particle, a value close to 113 kD, which corresponds to a Rev/tubulin 1:2 complex. However, no protein stoichiometries were determined by SDS-PAGE due to difficulties in resolving unique species in the above peak.
Stoichiometry of Rev–Tubulin in RTT.
Rev and tubulin were mixed as described above and RTT formation was confirmed by electron microscopy. RTTs were resolved from unassembled protein and smaller complexes by gel filtration on a Sephacryl S-500 (Amersham Pharmacia Biotech) column equilibrated with MEM. RTTs were recovered in the void peak, as confirmed by electron microscopy. Particles were concentrated by ultrafiltration, precipitated with acetone, and analyzed by reducing SDS-PAGE. Coomassie-stained bands were photographed and quantified by optical scanning.
Analytical Ultracentrifugation and Dynamic Light Scattering.
RTTs were analyzed by analytical ultracentrifugation and dynamic light scattering to determine their mass and hence the stoichiometries of the constituent proteins. Sedimentation velocity analysis was performed at 20°C using a Beckman XLA analytical ultracentrifuge (Beckman Coulter) equipped with absorption optics. Sedimentation coefficients were derived using software provided by Beckman Coulter. Dynamic light scattering was performed on a Brookhaven Instruments Corporation laser light scattering spectrometer equipped with an argon ion laser and a BI2030AT digital correlator. Diffusion coefficients were obtained from fits to the autocorrelation functions using software provided by Brookhaven Instruments Corporation. Mass values for Dolastatin-10–induced rings were also determined.
Mass Determinations by Scanning Transmission Electron Microscope.
The mass values of RTTs were determined by scanning transmission electron microscopy (STEM) and for this purpose the specimens were prepared in a volatile buffer. Tubulin was diluted to 1 mg/ml in 100 mM ammonium acetate and 2 mM MgCl2, pH 6.8. Rev was diluted to 18 mg/ml with 100 mM ammonium acetate and 25 mM sodium citrate, pH 6.8. Both protein solutions were frozen in liquid nitrogen and stored on dry ice until ready for microscopy at the Brookhaven STEM facility (Thomas et al. 1994; Wall et al. 1998). RTTs were formed fresh (from frozen constituents) because freezing and thawing was found to be disruptive to the preformed complexes. Immediately after thawing and warming to room temperature, 68 μl of tubulin was mixed with 1 μl of Rev (equimolar quantities), incubated 0.5 h at room temperature, diluted 10-fold with the ammonium acetate/magnesium buffer, and prepared for microscopy in the STEM. Inclusion of 100 mM NaCl in the dilution buffer alleviated the slight aggregation that occurred upon initial mixing of the proteins. Tobacco mosaic virus included in the preparations served as a mass standard (131.4 kD/nm). Mass determinations of RTTs were subsequently made from the micrographs using interactive PIC routines (Trus et al. 1996).
The mass values of the ring-like complexes induced by Dolastatin-10 were also determined by STEM. In this case tubulin, prepared as above but at 2 mg/ml, was mixed in a 1:1 volume ratio with the drug (100 μM in ammonium acetate/magnesium buffer). Rings induced by Dolastatin-10 were more fragile than those formed by Rev and tended to fragment during grid preparation. Consequently, only mass-per-unit-length values were determined.
Conventional Electron Microscopy
Specimens at a concentration of 0.1 mg/ml were adsorbed onto freshly air glow–discharged carbon-coated grids, rinsed with MEM, negatively stained with 1% uranyl acetate, and air dried. Micrographs were recorded with a Zeiss EM902 (Carl Zeiss, Inc.) at a nominal magnification of ×30,000. Magnifications were calibrated with reference to the 4.0-nm axial spacing of MTs (Metoz and Wade 1997), as measured by Fourier analysis of micrographs, and cross-calibrated with reference to the 4.06-nm axial spacing of bacteriophage T4 tails (Booy et al. 1996), measured in the same way.
Image Processing
Micrographs of negatively stained RTTs were digitized on a SCAI microdensitometer (Carl Zeiss, Inc.) at 7 μm per pixel and then binned twofold, thereby corresponding to 4.6 Å per pixel at the specimen. Particle images were selected using X3D Preprocess, a semiautomated procedure that also normalizes the data and corrects for any background gradients (Conway et al. 1993), and then processed with the PIC suite of programs (Trus et al. 1996). In brief, 423 particles were selected, and care was taken to pick only those that were well stained and nondeformed. Because the particles varied slightly in diameter, they were sorted into two size classes, and then translationally aligned by cross-correlation methods, using a centered, azimuthally averaged particle as reference (one for each class). Particles were centered to within 0.5 pixel. These data were then analyzed for the presence of symmetries using the algorithms described by Kocsis et al. 1995, which found 28-, 30-, and 32-fold symmetries to be statistically significant. The particles were then sorted according to whether the 28-, 30-, or 32-fold component of their power spectrum predominated in the appropriate radial zone. The images in each final data set were combined by correlation averaging (n = 37, 62, and 19 for the 28-, 30-, and 32-fold symmetric particles, respectively). The power spectrum of each averaged image was calculated, and the appropriate order of rotational symmetry was imposed. As an additional confirmation of the classification, alternative symmetrization of the correlation averages was imposed (e.g., 28-fold symmetrization of the 30-fold average), which produced a rotationally symmetric image essentially devoid of any azimuthal contrast. For side views, 100 particles were selected, aligned by cross-correlation, and then symmetrized along both axes.
Sequence Alignment
Sequence alignments were done with Genetics Computer Group (GCG) software using the Bestfit routine (version 10.0). To assess the significance of the alignments, the XKCM1 sequence was randomized 50 times (using the −RAN parameter) and aligned with Rev each time. For every isolate of Rev, the quality of the alignment, as defined in the GCG documentation, was reduced to a statistically significant degree.
Xenopus Egg Extract Assays
Preparation of sperm chromatin and 9,000-g extracts of ovulated eggs were prepared as described previously (Lohka and Masui 1983; Miller et al. 1999). Cytoplasmic extracts were supplemented with 10 μg/ml cytochalasin B, 10 μg/ml leupeptin, 10 μg/ml pepstatin, 10 μg/ml chymostatin, 2 mM ATP, 9 mM creatine phosphate, and 150 μg/ml creatine phosphokinase. Glycerol was added to a final volume of 5%. Extracts were frozen by dropwise addition into liquid nitrogen. The frozen beads were stored below −70°C until use.
Induction of MT asters by addition of sperm chromatin to the Xenopus egg extracts was used to test for an interaction between Rev and tubulin in a complex mixture containing cellular constituents. Assays were performed as described (Murray 1991; Moreau and Way 1999). In brief, 9 μl of thawed extract was mixed with 0.3 μl of Rhodamine-labeled tubulin (Cytoskeleton) and 1 μl of Rev (1 mg/ml final concentration) or Rev buffer (20 mM Hepes, 100 mM sodium chloride, and 50 mM sodium citrate, pH 7.0). After mixing, 1 μl of diluted sperm chromatin was added and mixed and polymerization was allowed to continue for 10–20 min at room temperature. Samples were mounted in a fixative containing 4′,6-diamidino-2-phenylindole dihydrochloride, and examined in a fluorescence microscope. 100 single sperm heads were photographed and scored in both the presence and absence of Rev.
Results and Discussion
During attempts to find solution conditions for Rev suitable for structural studies, we observed that the filaments could be depolymerized effectively by 5 mM decaglutamate. Under these conditions, Rev retained a significant amount of helical secondary structure as shown by circular dichroism (data not shown). The recent structure determination of tubulin by electron crystallography has highlighted the disposition of the COOH-terminal polyglutamate tracts on the exterior of the MTs (Nogales et al. 1999), confirming earlier biochemical evidence for an exterior location of the COOH termini of both the α- and β-subunits (Sackett and Wolff 1986; Sackett 1995a). Taken together, these observations suggested a structural basis for an interaction between Rev and MTs.
Conditions for Rev–Tubulin Complex Formation
Preliminary investigations were made by simply mixing Rev filaments with MTs and observing the results by electron microscopy. Mixing resulted in the immediate formation of toroidal complexes (Fig. 1, A–D). At the macroscopic level, a slight transient turbidity can be observed, suggesting that large initial aggregates form which then dissociate into small nonscattering rings. Simple kinetic experiments measuring turbidity at 340 nm by mixing reactants in a cuvette showed that the reaction occurred almost instantaneously. Inclusion of 100 mM NaCl suppressed this turbidity but did not affect the formation of the rings.
Figure 1.
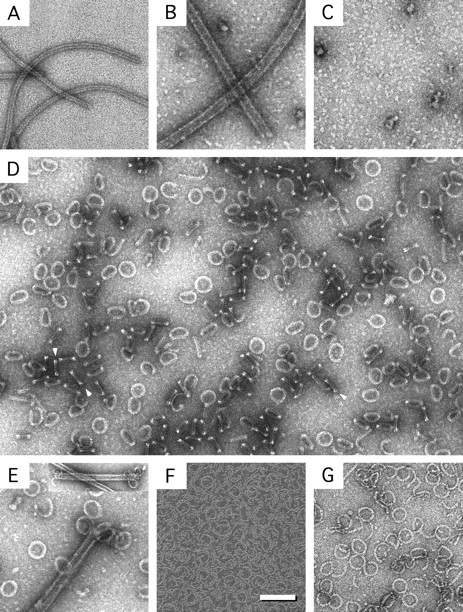
Rev–tubulin rings and related complexes. Shown are Rev filaments (A), MTs (B), colchicine depolymerized tubulin (C), RTTs (D), RTTs forming at the ends of MTs (E), Rev 1–59–tubulin rings (F), and Dolastatin-10–tubulin rings (G). RTT double-bars are visible in D (arrows). The inset in E shows MTs depolymerizing at both ends. Bars: (A–G) 100 nm; (inset) 200 nm.
Reciprocal titrations showed that the excess protein remained in the filamentous form, indicating that depolymerization and RTT formation were not a buffer effect. Moreover, no persistent turbidity appeared at any ratio of Rev/tubulin so that all solutions of RTT were completely clear. When MTs were added in molar excess, the ends occasionally appeared curled and in some instances RTT could be seen, apparently in the process of forming (Fig. 1 E). The appearance of the Rev-promoted rings at the ends of the MTs is similar to that of MTs depolymerizing in the cold (Melki et al. 1989; Mandelkow et al. 1991) or in the presence of Kin I kinesins (Desai et al. 1999).
Experiments were performed to investigate the basis of the Rev–tubulin interaction, and in particular to determine whether this was a simple charge interaction between the respective arginine and glutamate tracts. Inclusion of up to 200 mM NaCl had no apparent effect on RTT formation, but at 500 mM NaCl rings were no longer observed although both Rev filaments and MTs had been depolymerized, as judged by electron microscopy. These experiments were repeated at pH 6, 7, and 8, and also at 4°C and 25°C, again with no adverse effect on RTT formation. These observations suggest that the association is more complex than a simple electrostatic interaction, because charge shielding or neutralization of the polyglutamate tract would be expected to be altered when varying these conditions (Sackett 1995a; Wolff et al. 1995). More significantly, the complexes were also formed from subtilisin-treated tubulin, in which the COOH-terminal glutamate tract has been removed, further supporting the view that the interaction is more specific than a simple electrostatic one.
However, formation of RTTs was Mg2+-dependent. Chelation of Mg2+ by citrate ion introduced along with the Rev protein, for which citrate is required to prevent lateral aggregation (Watts et al. 1998), prevented RTT formation unless either supplemental Mg2+ was included or the Rev was introduced in a suitably small volume. Increasing the citrate concentration yielded progressively fewer RTTs, addition of a 1–2 mM excess of Mg2+ ion over citrate restored RTT formation, and addition of a slight excess of EDTA over Mg2+ again gave fewer RTTs. In the absence of Mg2+, a soluble Rev–tubulin complex was formed, with a median M r of ∼110 kD, as determined by gel filtration (data not shown), while in the presence of 1–2 mM Mg2+, large RTT complexes were formed. The stoichiometry of Rev/tubulin monomers in the latter complexes was 1:1 (recall that protein stoichiometries refer to individual polypeptide chains), as shown by SDS-PAGE of material purified by gel filtration (data not shown). Mg2+ is known to be required for tubulin stability per se (Menendez et al. 1998). Here it may also be required for formation of the RTT complex itself.
RTTs were formed by both rat and bovine brain tubulin, which shows that the phenomenon is not limited to a particular species. RTTs were formed from taxol-stabilized MTs as well as from a preformed colchicine–tubulin complex, demonstrating that RTTs can form from both polymerized and nonpolymerized tubulin. RTTs also formed in the presence of MT-associated proteins that stabilize MTs. However, RTT formation was completely blocked by prior incubation of tubulin with maytansine equimolar to tubulin dimer (data not shown). Because maytansine is known to bind to the β-tubulin subunit at the vinca site (Sackett 1995b), this result suggests that Rev may bind at or near this location.
These results demonstrate that the Rev–tubulin interaction is specific. Other basic proteins can interact with tubulin, including histones, lysozyme, polylysines, polyarginines, and other polyamines (Mithieux et al. 1984; Multigner et al. 1992). In those cases, however, the products of interaction were large insoluble precipitates that produced high turbidity. Some of these, like lysozyme, are quite sensitive to salt addition (Mithieux et al. 1984). Others, like histones, yield very different results depending on the ratio to tubulin and whether or not the tubulin is polymerized (Mithieux et al. 1984; Multigner et al. 1992). In contrast, the Rev–tubulin interaction results in clear solutions of ring polymers regardless of the initial state of tubulin polymerization, or the presence of added salt, or of significant changes in pH or temperature. Furthermore, RTT formation is prevented by micromolar concentrations of maytansine and proceeds through an intermediate that forms only at the ends of the MTs.
Rev Domain Specificity
Preliminary experiments to define the binding site on Rev using selected oligopeptides from various regions of Rev were all negative. However, an NH2-terminal construct of Rev, comprising residues 1–59, which essentially consists of the two predicted α-helical domains (Auer et al. 1994; Thomas et al. 1997) and shows high helical content as judged by circular dichroism (data not shown), also forms large numbers of thin-rimmed rings when mixed in a 1:1 molar ratio with tubulin (Fig. 1 F). Ring induction by Rev 1–59 is not prevented when MTs are assembled by GTP alone, but, unlike wild-type Rev, is completely blocked by taxol, showing that the affinity of the truncated Rev is less than that of the full-length molecule. Together, these observations point to a specific mode of association, possibly involving helix 2 of the NH2-terminal helix-turn-helix domain of Rev and the exterior face of tubulin at or near the vinca site. Sequence similarity considerations (below) support this proposition.
Mass Determinations of RTTs and Drug-induced Rings
To determine the masses of RTT, measurements were made from dark-field STEM micrographs of unstained, frozen-dried specimens (Thomas et al. 1994; Wall et al. 1998). Freshly prepared specimens were imaged at the Brookhaven STEM facility. The data were calibrated against an internal standard (tobacco mosaic virus). The histogram of mass measurements is shown in Fig. 2. It yields an average mass of 3.8 MD, but the breadth of the distribution and the standard deviation, which at ±0.43 MD is high for a complex of this size, suggest that the particles are somewhat heterogeneous (Fig. 2 and Table ).
Figure 2.
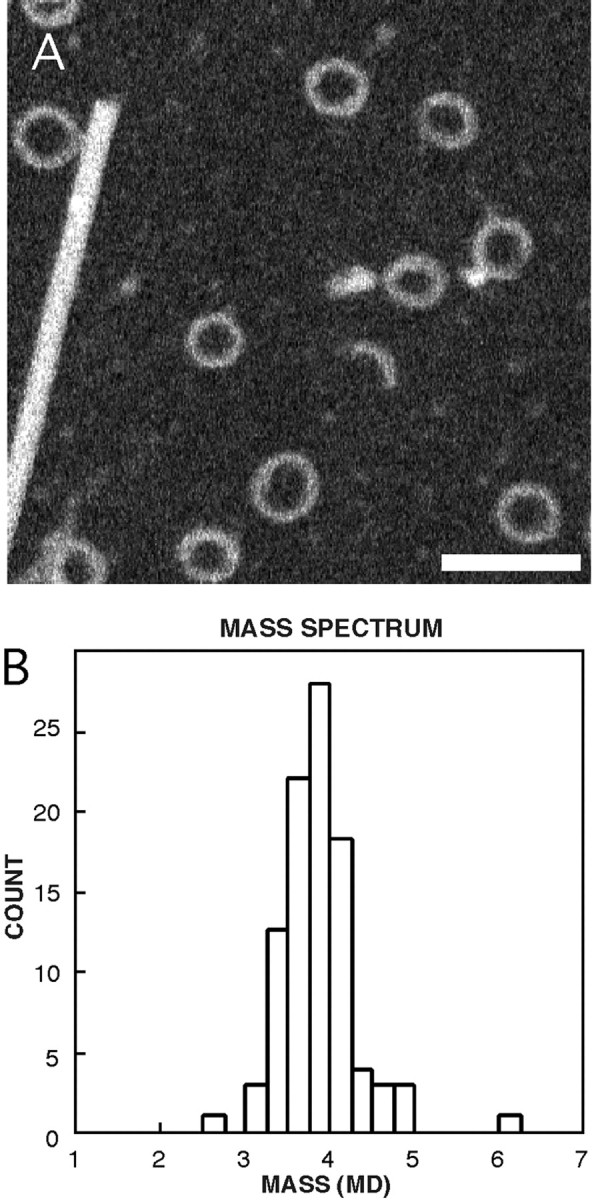
Mass distribution of RTTs determined by STEM. Shown are a field of RTTs and a TMV particle (A), and the mass histogram (B). Bar, 100 nm.
Table 1.
Dimensions and Masses of RTT and Dolastatin-10–Tubulin Rings
| Complex | Diameter | Width | Mass | Mass/length | Mass |
|---|---|---|---|---|---|
| nm | nm | MD | kD/nm | MD | |
| Rev, 28-fold | 37.4, n = 37 | 10.1, n = 37 | – | – | – |
| 30-fold | 41.0, n = 62 | 9.9, n = 62 | 3.8 ± 0.43, n = 99 | 30.1 | 4.15 ± 0.2 |
| 32-fold | 42.2, n = 19 | 9.8, n = 19 | – | – | – |
| Dolastatin-10 | 44.6 ± 1.2, n = 41 | 5.2 ± 0.4, n = 41 | – | 12.9 ± 2.2, n = 55 | 1.4 ± 0.1 |
Mass determinations were also made by analytical ultracentrifugation and light scattering which gave a consistent but slightly higher mass of 4.15 ± 0.2 MD (Table ). The sedimentation coefficient of 49.5 S may be compared with the value of 42 S reported for cold-induced tubulin rings with 26 ± 3 tubulin dimers (Frigon and Timasheff 1975).
RTTs were also compared with similar ring-like aggregates induced in tubulin by the antineoplastic drug Dolastatin-10 (Bai et al. 1995). Compared with the drug complexes, the RTTs are visually denser and have a higher mass (Fig. 1D and Fig. G; Table ). In RTTs, the cross-section of the torus is greater than in the tubulin–drug complexes although the overall diameter is similar. The drug-induced rings are also more fragile and tended to fragment despite the rather gentle application onto prewetted grids used during preparation for the STEM (Thomas et al. 1994). The mass-per-unit-length values, as well as the diameters of the Dolastatin-10 rings, suggest that these rings are probably composed of a single protofilament (expected mass-per-unit-length >11.9 kD/nm). The ratio of the mass-per-unit-length of the RTTs (30.1 kD/nm) to that of the drug-induced rings (12.9 kD/nm) is 2.3, in good agreement with the value of 2.5 obtained from the ratio of the molecular weight of the Rev/tubulin 2:2 complex (126 kD) and the tubulin monomer (50 kD). This indicates that the drug-induced rings are probably composed of a single protofilament, whereas the RTTs are formed from Rev/tubulin 2:2 complexes. As discussed below, RTTs are composed of two stacked rings, with each ring a copolymer of Rev and tubulin. Consequently, RTTs have the mass of two 50-kD tubulin monomers and two 13-kD Rev monomers over the same length that Dolastatin-10 rings have only one 50-kD tubulin monomer.
Image Analysis of RTT Reveals Polymorphism
Initial visual inspection of negatively stained RTTs (e.g., Fig. 1) revealed a faint periodicity, particularly around the periphery. To investigate this observation further, the images were analyzed computationally for the presence of rotational symmetry. All complexes that were unbroken and were not visibly flattened or otherwise distorted, were aligned translationally. This data set was analyzed quantitatively by a statistical procedure for rotational symmetry detection (Kocsis et al. 1995). In essence, this procedure uses two statistical measures to test whether any orders of symmetry are represented in the image set more strongly than in a similar set of background images (which are assumed to be nonsymmetric). One measure is Student's t test. The other is expressed as a product of ratios between corresponding Fourier components for the experimental and background images. Each radial zone is analyzed separately. The method is capable of detecting multiple symmetries in polymorphic mixtures of particles, provided that each component is sufficiently represented in the overall population (Kocsis et al. 1995).
The order of symmetry most frequently represented in the data set is 30-fold, which was detected between radii of 20 and 24 nm, peaking at 22 nm (Fig. 3). Also statistically significant are 28-fold symmetry at somewhat lower radii (19–22 nm), and 32-fold symmetry at somewhat higher radii (22–24 nm). It appears that only even symmetries are present, as no 29- or 31-fold symmetries were detected as significant.
Figure 3.
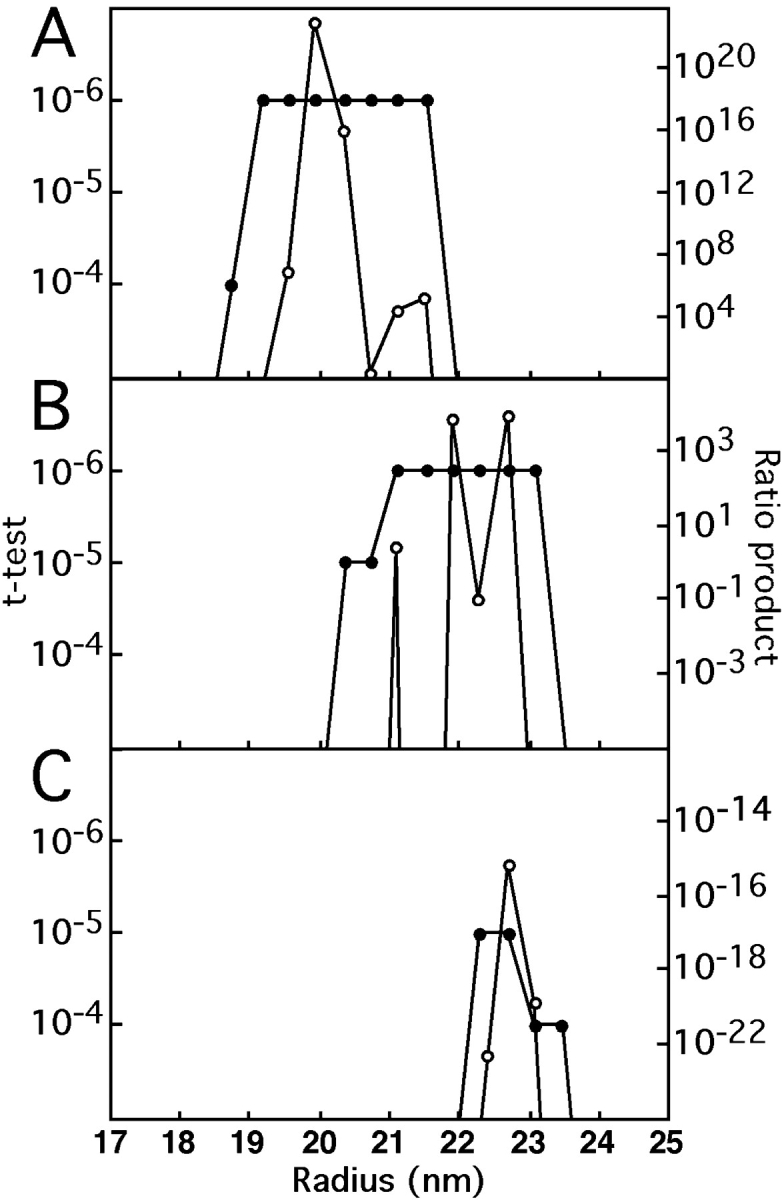
Detection of rotational symmetries in RTTs as a function of radius. Two measures of significance, Student's t test and the ratio product (Kocsis et al. 1995), were applied to two subsets of images, comprising 167 smaller rings and 256 larger rings, respectively. For polar-sampled images, the t test assesses at each radius and for each potential order of symmetry, whether that symmetry is significantly represented in the data set as a whole. The only significant symmetries detected were 28-fold (A) in the smaller set, and 30-fold (B), and 32-fold (C) in the larger set. For instance, in A, 28-fold symmetry is detected at the P < 10−6 level at radii of 18–22 nm. N-fold symmetry at radius R corresponds to an azimuthal spacing of 2πR/N in the specimen. For a given candidate symmetry at a given radius, the ratio product is the product of the corresponding Fourier amplitudes from all the images, normalized against the average amplitude from the background images. This product converges rapidly to zero except when a certain symmetry is significantly present. The relative proportions of particles with different symmetries may be estimated from the product ratios when both saturate the t test–calculated limit of P < 10−6. In this case, it may be estimated that 30-fold rings are three- to fourfold more abundant in the large subset than are 32-fold rings.
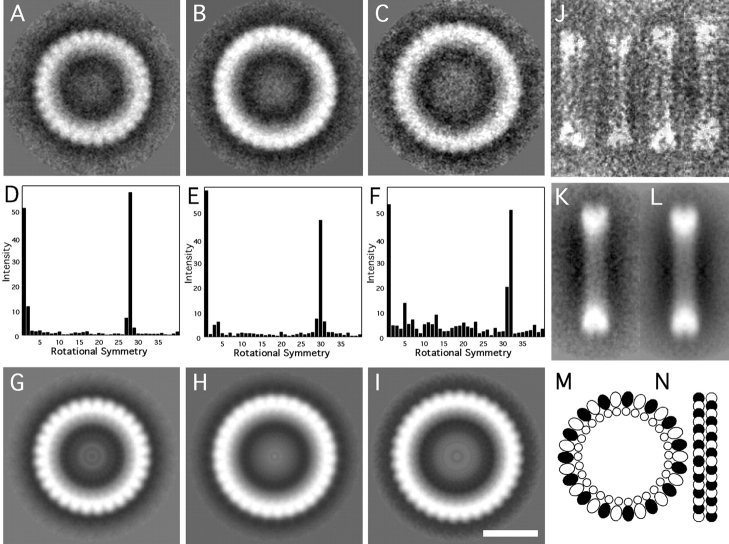
Having determined that these symmetries were present, the particles were sorted according to whether their 28-, 30-, or 32-fold Fourier components were strongest in the radial zone of interest. The set of 30-fold particles was largest (53%). To confirm this classification, the three sets of sorted images were analyzed separately with the symmetry detection algorithm; only the expected symmetry was found to be statistically significant in each case. At this point, the images in each of the three sets were subjected to several cycles of correlation averaging. The results are shown in Fig. 4A–C. Interestingly, although the mean outside diameter is 47 nm, the three averaged complexes vary in diameter in proportion to their respective orders of symmetry. Their outer diameters are 44.0 nm (28-fold), 46.8 nm (30-fold), and 49.1 nm (32-fold), respectively. The symmetries are most strongly expressed around their outer edges. The power spectra of the three averaged images, shown in Fig. 4D–F, further confirm their respective orders of symmetry. At this point, rotational symmetrization could be considered a valid operation, and was performed for each of the three averaged images, resulting in a further reduction in noise (Fig. 4, G–I).
Figure 4.
Image analysis and a molecular model of RTTs. Shown are the cross-correlation averages of 28-, 30-, and 32-fold symmetric particles viewed axially (A–C), the corresponding power spectra (D–F), and the final symmetrized images (G–I). Also shown are selected side-views of rings (J), the correlation average (K), and a symmetrized image (L). A molecular model of the 30-fold RTT is shown in M and N. In the model, Rev monomers are located on the inside and tubulin monomers on the outside (M). The side-view (N) shows RTTs as composed of bi-layered rings. The α- and β-tubulin monomers are differentiated as black and white. Bar, 20 nm.
RTT Rings Are Bilayered
Electron micrographs of negatively stained preparations show considerable numbers of what appear to be side-views of the RTT complexes. In this view, the particles are bars, ∼8 nm wide, and of somewhat variable length. Usually, they are divided into two parallel striations (Fig. 4, J–L). Therefore, it appears that these complexes are double rings. The variability in length may represent, to some extent, differential flattening, but the above analysis indicates it is also due in part to polymorphism. Similar bars have been observed previously (Voter and Erickson 1979; Melki et al. 1989; Mandelkow et al. 1991).
Stoichiometry of RTT Complexes
The observations that the RTT rings contain equimolar amounts of Rev and tubulin monomers are bilayered, and are 28-, 30-, or 32-fold symmetric in axial projection, agree well with the STEM mass data. Assuming that they contain 28, 30, and 32 Rev/tubulin 2:2 complexes, respectively, their corresponding predicted masses are 3.53, 3.78, and 4.03 MD, in excellent agreement with the observed distribution of particle masses (Fig. 2 B). We conclude that the three subpopulations are not resolved in this distribution, although their polymorphism contributes to its spread.
A Molecular Model for RTT Complexes
In principle, tubulin heterodimers may be arranged in either of two ways, i.e., with their long axis directed either circumferentially or axially. The former alternative is supported by the fact that only even orders of symmetry are observed, since they correspond to integral numbers of heterodimers (14, 15, or 16, respectively). However, the distinction between α- and β-subunits is not discernible at the current resolution.
A molecular model for the 30-fold rings is shown in Fig. 4M–N. The model is based on the following considerations: because tubulin subunits are approximately four times more massive than Rev subunits, they must contribute the majority of the stain-excluding material seen in the complexes, i.e., the outer ring. Its thickness, 6.5 nm, matches the known dimensions of the ellipsoidal tubulin monomer (4.6 × 4.0 × 6.5 nm; width, height, and depth, respectively), as determined by electron crystallography (Nogales et al. 1999). Moreover, the spacing of subunits around the ring (4.3 nm at a radius of 21 nm) agrees well with the width of the monomer and the previously reported 4–4.3-nm spacings of tubulin in rings (Voter and Erickson 1979; Diaz et al. 1994; Nicholson et al. 1999).
Since the outer edge of the RTT ring shows marked serrations (Fig. 4 H), it is likely to correspond to the inner surface of an MT (Nicholson et al. 1999). This disposition puts the outer surface of the MT on the inside. The inner part of the complex is lined with a 3–4-nm-thick ring of lower density, which we ascribe to Rev molecules. Periodicity is not evident in this inner ring (Fig. 4, G–I), but given the equimolarity of Rev and tubulin, we would expect this inner ring to consist of 30 monomers of Rev, in one ring, of the 30-fold RTT. The calculated spacing of 3.3 nm at a radius of 15.5 nm compares well with one of the spacings (3.8 nm) with which Rev dimers are packed in filaments (Watts et al. 1998), and with the 3.5-nm calculated molecular diameter of Rev (Wingfield et al. 1991).
Formation of RTTs
The starting materials for RTT formation were Rev filaments and MTs, and in some experiments colchicine–tubulin (Fig. 1, A–C). Rev filaments are quite stable and normally depolymerize very slowly but are nevertheless in dynamic equilibrium with subunits, probably free dimers (Wingfield et al. 1991). MTs in MEM with 50 μM taxol and 2 μM GTP at room temperature are also stable structures with no obvious curling at the termini. When mixed together, however, both filaments depolymerized and, in the presence of 2 mM Mg2+, formed RTTs.
How do RTT form? Various curved aggregates of tubulin alone have been described (Mandelkow et al. 1991; Lobert et al. 1993; Diaz et al. 1994; Bai et al. 1995, Bai et al. 1996, Bai et al. 1999; Menendez et al. 1998). GDP-tubulin is thought to be in a “tensioned” state in MTs and to relax into a lower-energy curved conformation when the MTs depolymerize (Mandelkow et al. 1991; Diaz et al. 1994). Curved oligomers are thought to form when a single protofilament curls inside out tangential to the MTs (Mandelkow et al. 1991). In fact, this may be the usual mode of MT shortening during events such as chromosome-to-pole migration during anaphase (Mandelkow et al. 1991). In the current situation, Rev appears to bind to tubulin to form a curved complex wherein the tubulin is, presumably, located on the outside and Rev on the inside of the ring. However, the occurrence of RTT in colchicine–tubulin preparations shows that intact MTs are not strictly required for RTTs to form.
Sequence Similarity of HIV-1 Rev to the Kin I Kinesins
The in vivo assembly of MTs is controlled by the nucleotide state of tubulin and by several regulatory proteins that either stabilize the MTs or cause their rapid disassembly. Stabilization of MTs is affected by a variety of structural MAPs such as MAP2 and MAP4 (Drewes et al. 1999), and, in Xenopus, XMAP215 (Tournebize et al. 2000), whereas destabilization is induced by katanins, which break MTs along their length and expose GDP–tubulin ends susceptible to depolymerization (Hartman and Vale 1999); Stathmin, which causes plus-end disassembly (Steinmetz et al. 2000; Wallon et al. 2000); and the Kin I kinesins, which induce disassembly at both ends (Desai et al. 1999). The Kin I kinesins XKCM1 and XKIF2 are thought to induce a destabilizing conformational change in GTP–tubulin subunits at the MT ends with subsequent release of an XKCM1/XKIF2–tubulin complex. ATP hydrolysis is later used to dissociate the XKCM1/XKIF2–tubulin complex (Desai et al. 1999).
Kinesins may be classified as N-, I-, or C-type, according to the position of the mechanochemical motor domain in the polypeptide chain. Rat brain kinesin is an N-type kinesin with the motor located between residues 1 and 335, whereas XKCM1 (730 amino acids) is an I-type kinesin (also known as M-type, from the internal location of the motor domain) with the motor domain located between residues 257 and 598 (Swissprot documentation). The motor/catalytic domains of kinesins are very similar in structure, consisting of a central 8-stranded β-sheet with three α-helices located on either side (Mandelkow and Hoenger 1999). Fitting the rat brain kinesin X-ray structure into the cryo-EM envelope of MTs decorated with kinesin has shown that it binds to MTs via helices 4 and 5 (Mandelkow and Hoenger 1999).
Unlike kinesin, the structure of Rev is not known. The protein appears to have two domains (Fig. 5 A): an NH2-terminal domain (residues 1–60) with two α-helices in a helix-turn-helix arrangement (Auer et al. 1994; Thomas et al. 1997), and a COOH-terminal domain that is predicted to be low in regular secondary structure (Watts et al. 1998). Rev binds to its target RNAs via an arginine-rich motif locate between residues 33 and 46 in the second of the two predicted helices. The RNA-binding domain is flanked on both sides by multimerization domains by which Rev oligomerizes along its cognate mRNA (Pollard and Malim 1998). Residues I52, I55, and I59 have been identified as forming a hydrophobic contact patch on helix 2 involved in formation of the helix-turn-helix structure (Auer et al. 1994; Thomas et al. 1997). Together, the RNA-binding domain and the COOH-terminal multimerization domain extend from residue 33 to at least residue 59, and probably somewhat beyond. Beyond that Rev has a leucine-rich region, known as the activation domain, whereby it interacts with cellular proteins.
Figure 5.
Domain structure and amino acid sequence similarity of HIV-1 Rev to kinesin XKCM1. A shows the locations of Helix 1, the polyproline sequence, Helix 2 (the RNA-binding region with flanking multimerization domains M1 and M2), and the activation domain of Rev. Key residues and positions are numbered above. B shows the XKCM1 motor/catalytic domain sequence from residues 500–550, and aligned Rev sequences below. In Rev, residues are identical (red), very similar (green), or similar (blue) to XKCM1. These residues are bold in XKCM1. Specific residues that are highly conserved in both Rev and the kinesins and that have identified structural and/or functional roles are marked by asterisks. Rev isolates are identified on the right. Rev isolate P04616 (carats) was used in this study.
Rev has sequence similarity to a portion of the catalytic/motor domain of XKCM1 (Fig. 5 B). In HIV-1 Rev P04616, the isolate used in this study, residues 34–70 correspond to residues 506–543 in XKCM1 (the specific residue numbers vary slightly depending on the Rev isolate). This sequence corresponds to helix 4 and the turn leading to helix 5 in kinesins, the specific portion of the motor whereby rat brain kinesin (PDB identifier 3kin; Mandelkow and Hoenger 1999) and KIF1A (Kikkawa et al. 2000) bind to tubulin. The highly conserved proline at the ends of the Rev similarity regions occurs just before helix 5. The same results were obtained when Rev was aligned with XKIF2, which is 87% identical overall with XKCM1 (data not shown).
Certain residues, indicated by asterisks in Fig. 5 B, are highly conserved in Rev isolates from HIV-1, HIV-2, and Simian Immunodeficiency Virus (SIV), as well as in XKCM1 and XKIF2 and other kinesins. Of these, residues T34, R39, and R42 have been shown by nuclear magnetic resonance (NMR) to be involved in Rev binding to the RRE (Battiste et al. 1996), whereas mutational studies show that residues I52, I55, and I59 form a hydrophobic helix–helix contact patch in Rev (Thomas et al. 1997). Alanine scanning and proteolysis studies have been applied to kinesin to determine how kinesin binds to tubulin. However, the exact residues involved remain uncertain (Mandelkow and Hoenger 1999).
Rev, like XKCM1 (Desai et al. 1999), causes disassembly of MTs from both ends (Fig. 1 E, inset). Also like XKCM1, Rev can depolymerize MTs into spirals in the presence of the nonhydrolyzeable GTP analogue GMPCPP (data not shown). The sequence similarities described above suggest a similar mode of binding to tubulin for both Rev and the Kin I kinesins. How the remaining NH2- and COOH-terminal sequences of Rev are involved is not clear. Certainly, the COOH-terminal domain of Rev stabilizes the complex because ring formation by the Rev 1–59 construct was prevented by taxol, whereas full-length Rev was able to form rings in the presence of taxol. In the kinesins, the flanking regions determine whether the protein translocates along MTs (conventional kinesin) or depolymerizes them (Kin I kinesins). In the Kin I kinesins, the released XKCM1/XKIF2-tubulin complex is dissociated by ATP hydrolysis (Desai et al. 1999). This has not been determined for Rev. However, Rev is subject to phosphorylation at a number of sites (Meggio et al. 1996; Fouts et al. 1997). Whether any of these phosphorylation events are involved in complex formation or whether other factors are required for complex dissociation is not known.
Rev–Tubulin Interactions in Cell Extracts
To test the hypothesis that Rev might interact with MTs in vivo, Xenopus egg extract aster formation assays were done (Fig. 6). It was found that Rev inhibited virtually all (94%) of the aster formation on sperm heads, whereas in the control condition (no Rev) almost all (92%) of the sperm heads formed asters. Moreover, the few asters that did form in the presence of Rev were distinctly smaller than in the control (Fig. 6 F). These results show that Rev strongly inhibits aster induction by sperm chromatin and demonstrate that Rev and tubulin can interact in the presence of cellular constituents including high concentrations of RNA. Whether RTTs actually occur in HIV-infected cells is not known, but the above results demonstrate that the affinity of Rev for tubulin persists in a cytoplasmic environment.
Figure 6.
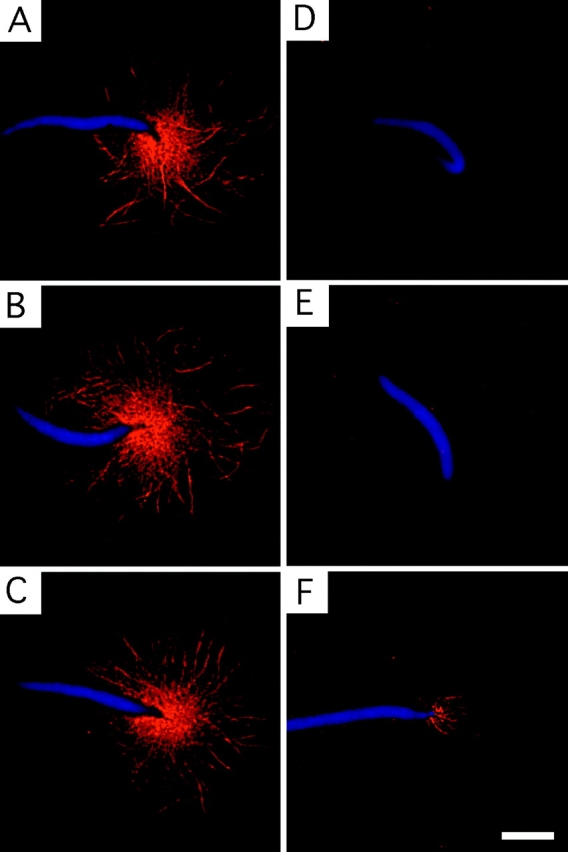
Inhibition of MT polymerization by Rev in Xenopus egg extracts. Shown is aster formation in the absence (A–C) and presence (D–F) of Rev. Sperm chromatin fluoresces blue (4′,6-diamidino-2-phenylindole dihydrochloride), and Rhodamine-tubulin fluoresces red. Bar, 10 μm.
A Rev titration series in the egg extract assay showed a concentration dependence. Although 1 mg/ml Rev almost totally suppressed aster formation, at <0.3–0.1 mg/ml (25–8 μM) Rev it was difficult to assess the inhibition of aster formation. Estimates of tubulin dimer concentration in the cell vary but are on the order of 20 μM (Gard and Kirschner 1987). These values are compatible with the formation of a Rev/tubulin 1:1 complex. Rev concentrations in infected and transformed cells appear to be difficult to determine but are generally believed to be low—certainly significantly lower than an abundant protein such as tubulin—and quite variable, both between cellular compartments and between cells (Kalland et al. 1994; Szilvay et al. 1995; Bøe et al. 1998) and over time. Consequently, it might seem unlikely that Rev would disrupt the MT cytoskeleton on a large scale, as is observed during HIV infection (Delezay et al. 1997). However, by analogy to the Kin I kinesins (Desai et al. 1999), Rev may only need to induce a destabilizing conformational change in the GTP–tubulin subunits at the plus-ends of MTs to cause their catastrophic collapse. Furthermore, the pronounced inhibitory effect on aster formation in egg extracts, even under conditions expressly designed to promote MT polymerization, suggests that Rev may localize to mitotic spindles to modulate the mitotic process. In fact, it has been shown that transient expression of Rev may slow or prevent the progression through mitosis (Miyazaki et al. 1995).
The results presented here suggest that expression of Rev may have consequences in addition to Rev's established role in mediating export of mRNA from the nucleus to the cytoplasm. These consequences probably involve alteration of MT dynamics due to destabilization of the polymerized state. This could be due to active destabilization of MT ends, as is seen with Stathmin/Oncoprotein 18 and especially the Kin I kinesins (Desai et al. 1999; Howell et al. 1999). Resolving the mechanism of Rev-induced MT depolymerization and understanding how HIV-1 infection might exploit altered MT dynamics will be the subjects of further investigation.
Acknowledgments
The authors wish to thank Drs. Martha Simon and Joe Wall for providing STEM data; Drs. James Conway, and Eva Kocsis for providing computer resources and programs; Dr. Benes Trus for guidance with image processing; Joshua Kaufman and Ira Palmer for expert technical assistance; and Dr. Adam Zlotnick for early discussions.
This work was supported in part by the National Institutes of Health (NIH) Intramural AIDS Targeted Antiviral Program (IATAP). The Brookhaven STEM is an NIH Supported Resource Center, NIH P41-RR01777, with additional support provided by the Department of Energy and the Office of Biological and Environmental Research.
Footnotes
Abbreviations used in this paper: MAP, microtubule-associated protein; MD, megadalton; MEM, 100 mM MES, 1 mM EGTA, 1 mM MgCl2, pH 6.9; MT, microtubule; RTT, Rev tubulin toroidal complex; STEM, scanning transmission electron microscope.
References
- Auer M., Gremlich H.-U., Seifert J.-M., Daly T.J., Parslow T.G., Casari G., Gstach H. Helix-loop-helix motif in HIV-1 Rev. Biochemistry. 1994;33:2988–2996. doi: 10.1021/bi00176a031. [DOI] [PubMed] [Google Scholar]
- Bai R., Taylor G.F., Schmidt J.M., Williams M.D., Kepler J.A., Pettit G.R., Hamel E. Interaction of dolastatin 10 with tubulininduction of aggregation and binding and dissociation reactions. Mol. Pharmacol. 1995;47:965–976. [PubMed] [Google Scholar]
- Bai R., Schwartz R.E., Kepler J.A., Pettit G.R., Hamel E. Characterization of the interaction of cryptophycin 1 with tubulinbinding in the Vinca domain, competitive inhibition of dolastatin 10 binding, and an unusual aggregation reaction. Cancer Res. 1996;56:4398–4406. [PubMed] [Google Scholar]
- Bai R., Durso N.A., Sackett D.L., Hamel E. Interactions of the sponge-derived antimitotic tripeptide hemiasterlin with tubulincomparison with dolastatin 10 and cryptophycin 1. Biochemistry. 1999;38:14302–14310. doi: 10.1021/bi991323e. [DOI] [PubMed] [Google Scholar]
- Battiste J.L., Mao H.Y., Rao N.S., Tan R.Y., Muhandiram D.R., Kay L.E., Frankel A.D., Williamson J.R. Alpha helix-RNA major groove recognition in an HIV-1 Rev peptide RRE RNA complex. Science. 1996;273:1547–1551. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- Bevec D., Jaksche H., Oft M., Wöhl T., Himmelspach M., Pacher A., Schebesta M., Koettnitz K., Dobrovnik M., Csonga R. Inhibition of replication in lymphocytes by mutants of the Rev cofactor eIF-5A. Science. 1996;271:1858–1860. doi: 10.1126/science.271.5257.1858. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya B., Sackett D.L., Wolff J. Tubulin, hybrid dimers, and tubulin S. Stepwise charge reduction and polymerization. J. Biol. Chem. 1985;260:10208–10216. [PubMed] [Google Scholar]
- Bøe S.O., Bjorndal B., Røsok B., Szilvay A.M., Kalland K.H. Subcellular localization of human immunodeficiency virus type 1 RNAs, Rev, and the splicing factor SC-35. Virology. 1998;244:473–482. doi: 10.1006/viro.1998.9110. [DOI] [PubMed] [Google Scholar]
- Bogerd H.P., Fridell R.A., Madore S., Cullen B.R. Identification of a novel cellular co-factor for the Rev/Rex class of retroviral proteins. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- Booy F.P., Trus B.L., Davison A.J., Steven A.C. The capsid architecture of channel catfish virus, an evolutionarily distant herpesvirus, is largely conserved in the absence of discernible sequence homology with herpes simplex virus. Virology. 1996;215:134–141. doi: 10.1006/viro.1996.0016. [DOI] [PubMed] [Google Scholar]
- Cenacchi G., Guiducci G., Pasquinelli G., Re M.C., Ramazzotti E., Furlini G., Malorni W., DeLuca M., Martinelli G.N. Early ultrastructural changes of human keratinocytes after HIV-1 contactan in vitro study. Eur. J. Dermatol. 1996;6:213–218. [Google Scholar]
- Cole C.N., Saavedra C. Regulation of the export of RNA from the nucleus. Semin. Cell Dev. Biol. 1997;8:71–78. doi: 10.1006/scdb.1996.0124. [DOI] [PubMed] [Google Scholar]
- Conway J.F., Trus B.L., Booy F.P., Newcomb W.W., Brown J.C., Steven A.C. The effects of radiation-damage on the structure of frozen hydrated HSV-1 capsids. J. Struct. Biol. 1993;111:222–233. doi: 10.1006/jsbi.1993.1052. [DOI] [PubMed] [Google Scholar]
- Cullen B.R. Posttranscriptional regulation by the HIV-1 Rev protein. Semin. Virol. 1998;8:327–334. [Google Scholar]
- Daly T.J., Doten R.C., Rennert P., Auer M., Jaksche H., Donner A., Fisk G., Rusche J.R. Biochemical characterization of binding of multiple HIV-1 Rev monomeric proteins to the Rev response element. Biochemistry. 1993;32:10497–10505. doi: 10.1021/bi00090a028. [DOI] [PubMed] [Google Scholar]
- Delezay O., Yahi N., Tamalet C., Baghdiguian S., Boudier J.A., Fantini J. Direct effect of type 1 human immunodeficiency virus (HIV-I) on intestinal epithelial cell differentiationrelationship to HIV-I enteropathy. Virology. 1997;238:231–242. doi: 10.1006/viro.1997.8829. [DOI] [PubMed] [Google Scholar]
- Desai A., Verma S., Mitchison T., Walczak C.E. Kin 1 kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- Diaz J.F., Pantos E., Bordas J., Andreu J.M. Solution structure of GDP-tubulin double rings to 3 nm resolution and comparison with microtubules. J. Mol. Biol. 1994;238:214–223. doi: 10.1006/jmbi.1994.1282. [DOI] [PubMed] [Google Scholar]
- Drewes G., Ebneth A., Mandelkow E.-M. MAPs, MARKs and microtubule dynamics. Trends Biol. Sci. 1999;272:307–311. doi: 10.1016/s0968-0004(98)01245-6. [DOI] [PubMed] [Google Scholar]
- Fackler O.T., Kienzle N., Kremmer E., Boese A., Schramm B., Klimkait T., Kucherer C., Mueller-Lantzsch N. Association of human immunodeficiency virus Nef protein with actin is myristoylation dependent and influences its subcellular localization. Eur. J. Biochem. 1997;247:843–851. doi: 10.1111/j.1432-1033.1997.00843.x. [DOI] [PubMed] [Google Scholar]
- Farjot G., Sergeant A., Mikaelian I. A new nucleoporin-like protein interacts with both HIV-1 Rev nuclear export signal and CRM-1. J. Biol. Chem. 1999;274:17309–17317. doi: 10.1074/jbc.274.24.17309. [DOI] [PubMed] [Google Scholar]
- Fisher U., Huber J., Boelens W.C., Mattaj I.W., Lührman R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Fouts D.E., True H.L., Cengel K.A., Celander D.W. Site-specific phosphorylation of the human immunodeficiency virus type-1 Rev protein accelerates formation of an efficient RNA-binding conformation. Biochemistry. 1997;36:13256–13262. doi: 10.1021/bi971551d. [DOI] [PubMed] [Google Scholar]
- Frankel A.D., Young J.A.T. HIV-1fifteen proteins and an RNA. Annu. Rev. Biochem. 1998;67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- Frigon R.P., Timasheff S.N. Magnesium-induced self-association of calf brain tubulin. I. Stoichiometry. Biochemistry. 1975;14:4559–4566. doi: 10.1021/bi00692a001. [DOI] [PubMed] [Google Scholar]
- Fritz C.C., Green M.R. HIV Rev uses a conserved cellular protein export pathway for the nucleocytoplasmic transport of viral RNAs. Curr. Biol. 1996;6:848–854. doi: 10.1016/s0960-9822(02)00608-5. [DOI] [PubMed] [Google Scholar]
- Fukumori T., Kagawa S., Iida S., Oshima Y., Akari H., Koyama A.H., Adachi A. Rev-dependent expression of three species of HIV-1 mRNAs. Int. J. Mol. Med. 1999;3:297–302. doi: 10.3892/ijmm.3.3.297. [DOI] [PubMed] [Google Scholar]
- Gard D.L., Kirschner M.W. Microtubule assembly in cytoplasmic extracts of Xenopus oocytes and eggs. J. Cell Biol. 1987;105:2191–2201. doi: 10.1083/jcb.105.5.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J.R., Emerman M., Sandmeyer S. Small heat shock protein suppression of Vpr-induced cytoskeletal defects in budding yeast. Mol. Cell Biol. 1997;17:4033–4042. doi: 10.1128/mcb.17.7.4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarskjold M.L. Regulation of retroviral RNA export. Semin. Cell Dev. Biol. 1977;8:83–90. doi: 10.1006/scdb.1996.0127. [DOI] [PubMed] [Google Scholar]
- Hartman J.J., Vale R.D. Microtubule disassembly by ATP-dependent oligomerization of the AAA enzyme katanin. Science. 1999;286:782–785. doi: 10.1126/science.286.5440.782. [DOI] [PubMed] [Google Scholar]
- Heaphy S., Finch J.T., Gait M.J., Karn J., Singh M. Human immunodeficiency virus type 1 regulator of virion expression, Rev, forms nucleo-protein filaments after binding to a purine-rich “bubble” located within the rev-response region of viral RNA. Proc. Natl. Acad. Sci. USA. 1991;88:7366–7370. doi: 10.1073/pnas.88.16.7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson B.R., Percipalle P. Interactions between HIV Rev and nuclear import and export factorsthe Rev nuclear localisation signal mediates specific binding to human importin-beta. J. Mol. Biol. 1997;274:693–707. doi: 10.1006/jmbi.1997.1420. [DOI] [PubMed] [Google Scholar]
- Howell B., Larsson N., Gullberg M., Cassimeris L. Dissociation of the tubulin-sequestering and microtubule catastrophe-promoting activities of oncoprotein 18/stathmin. Mol. Biol. Cell. 1999;10:105–118. doi: 10.1091/mbc.10.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E., Adam S. Transport of macromolecules between the nucleus and the cytoplasm. RNA. 1998;4:351–364. [PMC free article] [PubMed] [Google Scholar]
- Kalland K.H., Szilvay A.M., Brokstad K.A., Sætrevik W., Haukenes G. The human immunodeficiency virus type I Rev protein shuttles between the cytoplasm and nuclear compartments. Mol. Cell Biol. 1994;14:7436–7444. doi: 10.1128/mcb.14.11.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminchik J., Margalit R., Yaish S., Drummer H., Amit B., Sarver N., Gorecki M., Panet A. Cellular-distribution of HIV type-1 Nef proteinidentification of domains in Nef required for association with membrane and detergent-insoluble cellular matrix. AIDS Res. Hum. Retroviruses. 1994;10:1003–1010. doi: 10.1089/aid.1994.10.1003. [DOI] [PubMed] [Google Scholar]
- Karczewski M.K., Strebel K. Cytoskeleton association and virion incorporation of the human immunodeficiency virus type 1 Vif protein. J. Virol. 1996;70:494–507. doi: 10.1128/jvi.70.1.494-507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa M., Okada Y., Hirokawa N. 15 angstrom resolution model of the monomeric kinesin motor, KIF1A. Cell. 2000;100:241–252. doi: 10.1016/s0092-8674(00)81562-7. [DOI] [PubMed] [Google Scholar]
- Knipling L., Hwang J., Wolff J. Preparation and properties of pure tubulin S. Cell Motil. Cytoskel. 1999;43:63–71. doi: 10.1002/(SICI)1097-0169(1999)43:1<63::AID-CM7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Kocsis E., Cerritelli M.E., Trus B.L., Cheng N., Steven A.C. Improved methods for determination of rotational symmetries in macromolecules. Ultramicroscopy. 1995;60:219–228. doi: 10.1016/0304-3991(95)00070-2. [DOI] [PubMed] [Google Scholar]
- Lobert S., Hennington B.S., Correia J.J. Multiple sites for subtilisin cleavage of tubulineffects of divalent-cations. Cell Motil. Cytoskel. 1993;25:282–297. doi: 10.1002/cm.970250308. [DOI] [PubMed] [Google Scholar]
- Lohka M., Masui Y. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science. 1983;220:719–721. doi: 10.1126/science.6601299. [DOI] [PubMed] [Google Scholar]
- Macreadie I.G., Castelli L.A., Hewish D.R., Kirkpatrick A., Ward A.C., Azad A.A. A domain of human immunodeficiency virus type-1 Vpr containing repeated H(S/F)RIG amino-acid motifs causes cell-growth arrest and structural defects. Proc. Natl. Acad. Sci. USA. 1995;92:2770–2774. doi: 10.1073/pnas.92.7.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malorni W., Guiducci G., Pasquinelli G., Rivabene R., Re M.C., Ramazzotti E., DeLuca M., LaPlaca M., Cenacchi G. HIV-type 1 induces specific cytoskeleton alterations in human epithelial cells in culture. Eur. J. Dermatol. 1997;7:263–269. [Google Scholar]
- Mandelkow E., Hoenger A. Structures of kinesin and kinesin-microtubule interactions. Curr. Opin. Cell Biol. 1999;11:34–44. doi: 10.1016/s0955-0674(99)80005-2. [DOI] [PubMed] [Google Scholar]
- Mandelkow E.M., Mandelkow E., Milligan R.A. Microtubule dynamics and microtubule caps - a time-resolved cryoelectron microscopy study. J. Cell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I.W., Englmeier L. Nucleocytoplasmic transportthe soluble phase. Annu. Rev. Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Meggio F., D'Agostino D.M., Ciminale V., Chieco-Bianchi L., Pinna L.A. Phosphorylation of HIV-1 Rev proteinimplication of protein kinase CK2 and pro-directed kinases. Biochem. Biophys. Res. Commun. 1996;226:547–554. doi: 10.1006/bbrc.1996.1392. [DOI] [PubMed] [Google Scholar]
- Melki R., Carlier M.F., Pantaloni D., Timasheff S.N. Cold depolymerization of microtubules to double ringsgeometric stabilization of assemblies. Biochemistry. 1989;28:9143–9152. doi: 10.1021/bi00449a028. [DOI] [PubMed] [Google Scholar]
- Menendez M., Rivas G., Diaz J.F., Andreu J.M. Control of the structural stability of the tubulin dimer by one high affinity bound magnesium ion at nucleotide N-site. J. Biol. Chem. 1998;273:167–176. doi: 10.1074/jbc.273.1.167. [DOI] [PubMed] [Google Scholar]
- Metoz F., Wade R.H. Diffraction by helical structures with seamsmicrotubules. J. Struct. Biol. 1997;118:128–139. [Google Scholar]
- Miller M.W., Caracciolo M.R., Berlin W.K., Hanover J.A. Phosphorylation and glycosylation of nucleoporins. Arch. Biochem. Biophys. 1999;367:51–60. doi: 10.1006/abbi.1999.1237. [DOI] [PubMed] [Google Scholar]
- Mithieux G., Alquier C., Roux B., Rousset B. Interactions of tubulin with chromatin proteinsH1 and core histones. J. Biol. Chem. 1984;259:15523–15531. [PubMed] [Google Scholar]
- Miyazaki Y., Takamatsu T., Nosaka T., Fujita S., Martin T.E., Hatanaka M. The cytotoxicity of human immunodeficiency virus type 1 Revimplications for its interaction with the nucleolar protein B23. Exp. Cell Res. 1995;219:93–101. doi: 10.1006/excr.1995.1209. [DOI] [PubMed] [Google Scholar]
- Moreau V., Way M. In vitro approaches to study actin and microtubule dependent cell processes. Curr. Opin. Cell Biol. 1999;11:152–158. doi: 10.1016/s0955-0674(99)80019-2. [DOI] [PubMed] [Google Scholar]
- Multigner L., Gagnon J., Van Dorsselaer A., Job D. Stabilization of sea urchin flagellar microtubules by histone H1. Nature. 1992;360:33–39. doi: 10.1038/360033a0. [DOI] [PubMed] [Google Scholar]
- Murray A.W. Cell-cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Nicholson W.V., Lee M., Downing K.H., Nogales E. Cryo-electron microscopy of GDP-tubulin rings. Cell Biochem. Biophys. 1999;31:175–183. doi: 10.1007/BF02738171. [DOI] [PubMed] [Google Scholar]
- Nogales E., Wolf S.G., Downing K.H. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- Nogales E., Whittaker M., Milligan R.A., Downing K.H. High-resolution model of the microtubule. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- Pollard V.W., Malim M.H. The HIV-1 Rev protein. Annu. Rev. Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- Sackett D.L. Structure and function in the tubulin dimer and the role of the acidic carboxyl terminus Subcell. Biochem. 24 1995. 255 302a [DOI] [PubMed] [Google Scholar]
- Sackett D.L. Vinca site agents induce structural changes in tubulin different from and antagonistic to changes induced by colchicine site agents Biochemistry. 34 1995. 7010 7019b [DOI] [PubMed] [Google Scholar]
- Sackett D.L., Wolff J. Proteolysis of tubulin and the substructure of the tubulin dimer. J. Biol. Chem. 1986;261:9070–9076. [PubMed] [Google Scholar]
- Sackett D.L., Knipling L., Wolff J. Isolation of microtubule protein from mammalian brain frozen for extended periods of time. Prot. Expr. Purif. 1991;2:390–393. doi: 10.1016/1046-5928(91)90099-5. [DOI] [PubMed] [Google Scholar]
- Steinmetz M.O., Kammerer R.A., Jahnke W., Goldie K.N., Lustig A., van Oostrum J. Op18/stathmin caps a kinked protofilament-like tubulin tetramer. EMBO (Eur. Mol. Biol. Organ.) J. 2000;19:572–580. doi: 10.1093/emboj/19.4.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F., Neville M., Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- Szilvay A.M., Brokstad K.A., Kopperud R., Haukenes G., Kalland K.H. Nuclear export of the human immunodeficiency virus type I nucleocytoplasmic shuttle protein Rev is mediated by its activation domain and is blocked by transdominant negative mutants. J. Virol. 1995;69:3315–3323. doi: 10.1128/jvi.69.6.3315-3323.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D., Schultz P., Steven A.C., Wall J.S. Mass analysis of biological macromolecular complexes by STEM. Biol. Cell. 1994;80:181–192. doi: 10.1111/j.1768-322x.1994.tb00929.x. [DOI] [PubMed] [Google Scholar]
- Thomas S., Hauber J., Casari G. Probing the structure of the HIV-1 trans activator protein by functional analysis. Prot. Eng. 1997;10:103–107. doi: 10.1093/protein/10.2.103. [DOI] [PubMed] [Google Scholar]
- Titus M.A., Gilbert S.P. The diversity of molecular motorsan overview. Cellular and Molecular Life Sciences. 1999;56:181–183. doi: 10.1007/s000180050420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize R., Popov A., Kinoshita K., Ashford A.J., Rybina S., Pozniakovsky A., Mayer T.U., Walczak C.E., Karsenti E., Hyman A.A. Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat. Cell Biol. 2000;2:13–19. doi: 10.1038/71330. [DOI] [PubMed] [Google Scholar]
- Trus B.L., Kocsis E., Conway J.F., Steven A.C. Digital image processing of electron micrographsthe PIC system-III. J. Struct. Biol. 1996;116:61–67. doi: 10.1006/jsbi.1996.0011. [DOI] [PubMed] [Google Scholar]
- Voter W.A., Erickson H.P. Tubulin ringscurved filaments with limited flexibility and two modes of association. J. Supramol. Struct. 1979;10:419–431. doi: 10.1002/jss.400100405. [DOI] [PubMed] [Google Scholar]
- Wall J.S., Hainfeld J.F., Simon M.N. Scanning transmission electron microscopy of nuclear structures. Methods Cell Biol. 1998;53:139–164. doi: 10.1016/s0091-679x(08)60878-x. [DOI] [PubMed] [Google Scholar]
- Wallon G., Rappsilber J., Mann M., Serrano L. Model for stathmin/OP18 binding to tubulin. EMBO (Eur. Mol. Biol. Organ.) J. 2000;19:213–222. doi: 10.1093/emboj/19.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts N., Misra M., Wingfield P.W., Stahl S., Cheng N., Trus B., Williams R., Steven A.C. Three-dimensional structure of HIV-1 Rev protein polymers. J. Struct. Biol. 1998;121:41–52. doi: 10.1006/jsbi.1998.3964. [DOI] [PubMed] [Google Scholar]
- Wingfield P.T., Stahl S.J., Payton M.A., Venkatesan S., Misra M., Steven A.C. HIV-1 Rev expressed in recombinant Escherichia colipurification, polymerization, and conformational properties. Biochemistry. 1991;30:7527–7534. doi: 10.1021/bi00244a023. [DOI] [PubMed] [Google Scholar]
- Wolff J., Knipling L., Sackett D.L. Charge-shielding and the “paradoxical” stimulation of tubulin polymerization by guanidine hydrochloride. Biochemistry. 1995;35:5910–5920. doi: 10.1021/bi9527395. [DOI] [PubMed] [Google Scholar]
- Wolff J., Sackett D.L., Knipling L. Cation selective promotion of tubulin polymerization by alkali metal chlorides. Prot. Sci. 1996;5:2020–2028. doi: 10.1002/pro.5560051008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemmel R.W., Kelley A.C., Karn J., Butler P.J.G. Flexible regions of RNA structure facilitate co-operative Rev assembly on the Rev-response element. J. Mol. Biol. 1996;258:763–777. doi: 10.1006/jmbi.1996.0285. [DOI] [PubMed] [Google Scholar]