Abstract
Programmed cell death or apoptosis leads to the activation of the caspase-activated DNase (CAD), which degrades chromosomal DNA into nucleosomal fragments. Biochemical studies revealed that CAD forms an inactive heterodimer with the inhibitor of caspase-activated DNase (ICAD), or its alternatively spliced variant, ICAD-S, in the cytoplasm. It was initially proposed that proteolytic cleavage of ICAD by activated caspases causes the dissociation of the ICAD/CAD heterodimer and the translocation of active CAD into the nucleus in apoptotic cells. Here, we show that endogenous and heterologously expressed ICAD and CAD reside predominantly in the nucleus in nonapoptotic cells. Deletional mutagenesis and GFP fusion proteins identified a bipartite nuclear localization signal (NLS) in ICAD and verified the function of the NLS in CAD. The two NLSs have an additive effect on the nuclear targeting of the CAD–ICAD complex, whereas ICAD-S, lacking its NLS, appears to have a modulatory role in the nuclear localization of CAD. Staurosporine-induced apoptosis evoked the proteolysis and disappearance of endogenous and exogenous ICAD from the nuclei of HeLa cells, as monitored by immunoblotting and immunofluorescence microscopy. Similar phenomenon was observed in the caspase-3–deficient MCF7 cells upon expressing procaspase-3 transiently. We conclude that a complex mechanism, involving the recognition of the NLSs of both ICAD and CAD, accounts for the constitutive accumulation of CAD/ICAD in the nucleus, where caspase-3–dependent regulation of CAD activity takes place.
Keywords: apoptosis, chromosomal DNA degradation, caspase-activated DNase, nuclear targeting, caspase-3
Introduction
Apoptosis is an essential process that controls cell numbers during development and participates in the elimination of cells that have undergone irreparable genomic damage (Jacobson et al. 1997; Nagata 1997). Apoptotic cells show distinctive morphological perturbations, including membrane blebbing, cytoskeletal changes, chromatin condensation, and DNA fragmentation (Jacobson et al. 1997; Wyllie 1980). The pathways and the nucleases involved in the progressive destruction of the genomic DNA have remained elusive until the recent cloning of the heterodimeric DNA fragmentation factor (ICAD/CAD or DFF; Liu et al. 1997; Enari et al. 1998).
The DNA fragmentation factor consists of two subunits, the 40-kD caspase-3–activated DNase (CAD or DFF40) and the 45-kD inhibitor of CAD (ICAD or DFF45; Liu et al. 1997, Liu et al. 1998; Enari et al. 1998; Sakahira et al. 1998). Biochemical isolation of the ICAD–CAD complex from the cytosolic fraction led to the cloning of the mouse and human orthologues of ICAD and CAD (Liu et al. 1997, Liu et al. 1998; Enari et al. 1998; Halenbeck et al. 1998). While the NH2-terminal CIDE-N domain, identified in both ICAD and CAD, appears to account for heterodimerization (Inohara et al. 1998), a putative nuclear localization signal (NLS) was identified at the COOH terminus of CAD based on sequence alignment to the SV-40 large T antigen signal (Enari et al. 1998). CAD is expressed exclusively as an ICAD/CAD heterodimer, leading to the suggestion that NLS is masked in nonapoptotic cells (Enari et al. 1998).
Heterodimerization of ICAD/CAD is obligatory not only to prevent chromosomal DNA degradation in growing cells, but also to ensure co- or posttranslational folding of CAD in the cytoplasm (Liu et al. 1997, Liu et al. 1998; Enari et al. 1998; Halenbeck et al. 1998; Samejima et al. 1998). According to this early model, activation of the apoptotic signaling cascade leads to the proteolysis of ICAD by effector caspases, permitting the dissociation of the ICAD–CAD complex, and the subsequent nuclear uptake of activated CAD (Enari et al. 1998). Besides the full-length ICAD (or ICAD-L), alternatively spliced variants, ICAD-S, lacking the last 66 and 62 amino acid residues at the COOH terminus have been identified in mice and humans, respectively (Gu et al. 1999; Sakahira et al. 1999). These ICAD variants can also dimerize with CAD (Gu et al. 1999), but their function is obscure.
While compelling evidence has characterized the endonuclease activity of mouse CAD (mCAD) and its human orthologue (hCAD; Enari et al. 1998; Halenbeck et al. 1998; Inohara et al. 1999; Janicke et al. 1998; Liu et al. 1998; Mukae et al. 1998; Tang and Kidd 1998), neither the colocalization of endogenous ICAD and CAD in the cytoplasm nor the nuclear translocation of activated CAD has been demonstrated. However, immunolocalization data, obtained on cells overexpressing epitope-tagged hCAD or hICAD, suggested that both ICAD and CAD are nuclear (Liu et al. 1998). In addition, a fusion protein, consisting of mICAD and the green fluorescent protein (GFP), has been found to associate with the nuclei of a variety of transfected cells. Based on these observations, it was proposed that the nuclear import of GFP-ICAD was accomplished by association with either CAD or polypeptides actively targeted into the nucleus (Samejima and Earnshaw 1998). The heterodimerization of ICAD and CAD was not directly ensured in these experiments, thus, the controversy regarding the subcellular localization of the ICAD–CAD complex has remained largely unresolved.
To better understand the subcellular compartmentalization of the apoptotic DNase, our objectives were to establish the cellular distribution and to elucidate the targeting determinants of the CAD–ICAD complex. Immunolocalization of the endogenous ICAD with two polyclonal antibodies provides the direct evidence for the constitutive nuclear targeting of ICAD. Using deletional mutagenesis and fusion proteins, we have identified an NLS at the COOH terminus of ICAD and confirmed the function of the NLS of CAD. The two NLSs appear to contribute in an additive manner to the nuclear targeting of the ICAD–CAD heterodimer, implying that the caspase-3–dependent activation of CAD takes place in the nucleus.
Materials and Methods
Cell Lines and Transfection
HeLa, COS-1, MCF7, HTE, PANC, MDCK, CHO, and BHK-21 cells were grown in α-modified Eagle's or DME medium supplemented with 10% FCS at 37°C under an atmosphere of 5% CO2. Transient transfections were performed with the calcium phosphate precipitation method, Effecten (QIAGEN), or Fugene (Roche) on 60–70% confluent cells. Cells were harvested after 48 h of transfection.
To generate HeLa cells stably expressing hICAD-C-myc, cells were transfected with the pcDNA3-hICAD-C-myc expression vector as calcium phosphate precipitates, and selected in the presence of 0.5 mg/ml G418 (Geneticin; GIBCO BRL). Clones were screened by indirect immunostaining using the mouse monoclonal anti-myc (9E10, Covance Research Products Inc.) antibody.
Plasmid Constructions
The cDNA of human ICAD was isolated by PCR cloning, using a cDNA library prepared from Caco-2 cells (American Type Culture Collection, accession number HTB37) as a template. The cDNAs encoding the mCAD and hCAD were provided by Dr. S. Nagata (Osaka University Medical School, Osaka, Japan) and Dr. R. Halenbeck (Chiron Corp., Emeryville, CA), respectively. The membrane-targeted EGFP construct, encoding EGFP and the ras farnesylation site (EGFPF) and the cDNA of procaspase-3 were the gift of Dr. W. Jiang (Jiang 1998) and Dr. V. Dixit, respectively. Both hICAD and mCAD were subcloned into the expression plasmid pcDNA3 or into a modified version, incorporating in-frame fusions of the coding sequences for HA, myc or flag epitopes, at either the COOH or the NH2 terminus. Deletion mutants ICADΔ265-330 (ICAD-S), ICADΔ306-331 (ICADΔNLS), mCADΔ329-344 (CADΔNLS), and hCADΔ329-338 (hCADΔNLS) were generated by PCR mutagenesis. cDNA of fusion proteins, comprising EGFP (Cormack et al. 1996) and the NLS of hICAD (EGFP-ICAD(306-331) or mCAD (EGFP-CAD(326-344)), were obtained by inserting the corresponding cDNA fragments (amino acids 306–331 from ICAD and 326–344 from CAD) into the EcoRI and ApaI sites of the pEGFP-C1 (CLONTECH Laboratories, Inc.). The plasmid encoding GST-hICAD fusion protein was constructed by insertion of the full-length coding region of hICAD into the EcoRI and XhoI sites of pGEX-4T1 plasmid. All constructs were verified by dideoxy chain termination DNA sequencing.
Bacterial Expression of hICAD and mCAD
To generate polyclonal anti-ICAD antibody, the full-length coding region of hICAD was fused in-frame with GST in the pGEX-4T1 vector and transformed in HB101 bacteria. Production of the fusion protein was induced with 0.1 mM isopropyl β-d-thiogalactopyranoside. The bacteria suspension was lysed by sonication in 0.5 M NaCl, 20 mM Hepes, 10% glycerol, 0.1 mM EDTA, and 1 mM DTT, pH 7.5. GST-hICAD was purified from the soluble fraction using glutathione Sepharose 4B (Sigma Chemical Co.), eluted with sonication buffer supplemented with 10 mM reduced glutathione, and further purified with SDS-PAGE. Gel slices, containing GST-hICAD were crushed for immunization of rabbits. Recombinant hICAD, hICAD-His6, and mCAD-His6 were expressed in BL21(DE3) cells using the pET15b (Novagen) expression plasmid and purified according to the supplier's recommendations using metal affinity chromatography.
Polyclonal Antibody Production
Purified GST-hICAD fusion protein was sent to Harlam Bioproducts for Science for inoculation into rabbits. Immunization was achieved with four boost of injections (0.5 mg protein/rabbit). The specificity of the rabbit antibodies was determined by comparing the activity of the immune and preimmune serum. For immunoblotting and immunofluorescence, the antibody was used at 1:1,000–1:3,000 dilution, respectively.
Immunofluorescence Microscopy
Fluorescence staining of transfected and nontransfected cells was carried out on glass coverslips after fixing (4% paraformaldehyde for 20 min) and permeabilizing (0.2% Triton X-100 in PBS for 5 min) the cells as previously described (Lechardeur et al. 1999). Primary antibodies were as follows: affinity-purified polyclonal goat anti-ICAD (K-17), anti-myc (monoclonal and polyclonal; Santa Cruz Biotechnology, Inc.), anti-HA (monoclonal 16B12; Covance Research Products Inc., and polyclonal, Santa Cruz Biotechnology, Inc.), and anti-Flag (M2-monoclonal; Sigma Chemical Co.). Secondary anti-mouse and anti-rabbit antibodies were conjugated to fluorescein or rhodamine (Jackson ImmunoResearch Laboratories). Double immunostaining was performed with pairs of polyclonal and monoclonal primary antibodies. No significant spillover of the fluorescence signal between the FITC and rhodamine filter sets was detected. Immunofluorescence micrographs were captured on a Zeiss Axiovert 100 inverted fluorescence microscope (63×/1.4 Planachromat objective) and a Contax camera as described previously (Lechardeur et al. 1999). Color slides (1600 ASA Kodachrome) were scanned by a Polaroid slide scanner, and the images were manipulated with Adobe Photoshop 5.0 software on a Power Macintosh. Exposure times and background substraction were held constant for the documentation of the time course of ICAD distribution. Laser confocal fluorescence microscopy and image processing was performed using the Zeiss LSM 510 imaging system with Plan neofluor 40×/1.3 objective.
Immunoblotting and Coimmunoprecipitation
For immunoblottings, cells were washed with ice-cold PBS and lysed in RIPA buffer (150 mM NaCl, 20 mM Tris-HCl, 1% Triton X-100, 0.1% SDS, and 0.5% sodium deoxycholate, pH 8.0) containing 10 μg/ml of leupeptin and pepstatin, 10 mM iodoacetamide, and 1 mM PMSF for 20 min at 4°C. Nuclei and unbroken cells were removed by centrifugation (15,000 g for 15 min at 4°C). Soluble proteins were denatured in Laemmli sample buffer, separated with SDS-PAGE, and transferred to a nitrocellulose membrane. Immunoblotting, using anti-HA, anti-myc, or anti-hICAD primary antibodies and enhanced chemiluminescence Western blot kit (Amersham), were performed as previously described (Lukacs et al. 1994). The CAD-N-HA/ICAD-C-myc heterodimer was isolated with immunoprecipitation, using anti-HA antibody from cell lysates prepared in TNT buffer (0.2% Triton X-100, 150 mM NaCl, 20 mM Tris-HCl, pH 7.2). The immunoprecipitate was separated with SDS-PAGE, transferred to nitrocellulose, and detected with anti-myc antibody.
Detection of Apoptosis
TUNEL assay was performed as previously described (Lechardeur et al. 1999). Annexin V staining and determination of caspase-3 activity of HeLa cell extract were carried out with the ApoAlert annexin V and caspase-3 fluorescent assay kits (CLONTECH Laboratories, Inc.), respectively.
Online Supplemental Materials
Supplemental Figure 1.
Exogenous hICAD is confined to the nucleus at a low level of expression. HeLa cells were transiently transfected with hICAD-C-myc and immunostained with anti-myc and polyclonal anti-ICAD antibodies. (Available at http://www.jcb.org/cgi/content/full/150/2/321/DC1)
Supplemental Figure 2.
Expression level of ICAD-S and ICAD-L. (a and b) The expression level of ICAD-S relative to ICAD-L was determined with quantitative immunoblotting. (c) The weak cytosolic immunostaining of endogenous ICAD is eliminated in HeLa cells expressing exogenous mCAD-N-HA, presumably because of the nuclear translocation of the hICAD-S/mCAD complex. (Available at http://www.jcb.org/cgi/content/full/150/2/321/DC1)
Supplemental Figure 3.
The NLSs of both hCAD and hICAD contribute to the nuclear accumulation of the ICAD–CAD complex. Coimmunolocalization of full-length or truncated hCAD-N-HA and hICAD-C-myc was carried out as depicted on Fig. 7 a. (Available at http://www.jcb.org/cgi/content/full/150/2/321/DC1)
Figure 7.
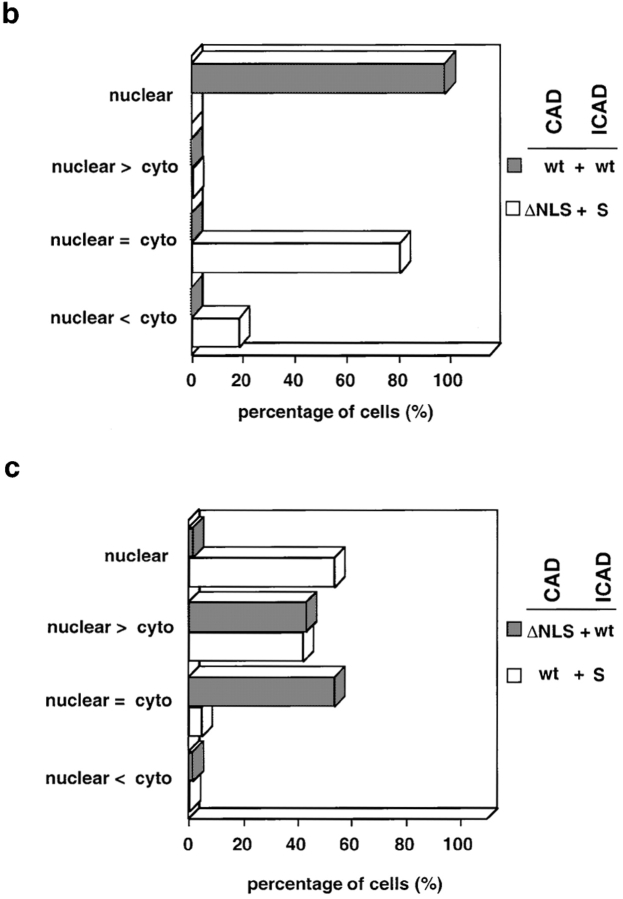
The NLSs of both mCAD and hICAD contribute to the nuclear accumulation of the ICAD–CAD complex. Coimmunolocalization of full-length or truncated mCAD-N-HA and hICAD-C-myc was carried out as on Fig. 6. (a) HeLa cells were cotransfected with the indicated expression constructs encoding for the full-length or truncated mCAD-N-HA and hICAD-C-myc and visualized by indirect immunostaining. Identical fields are shown for ICAD (red) and CAD (green) staining. (b and c) Estimation of the nucleocytoplasmic concentration gradient of the exogenous ICAD–CAD complex, based on the subcellular distribution of mCAD-N-HA or mCADΔNLS-N-HA in the presence of hICAD-C-myc or hICAD-S-C-myc. According to the nucleocytoplasmic concentration gradient of CAD-N-HA or CADΔNLS-N-HA, the distribution pattern of transfected cells was classified into four groups: (1) mCAD was exclusively nuclear (nuclear); (2) mCAD concentration was higher in the nucleus than in cytoplasm; (3) no significant concentration difference could be recognized between the nucleus and the cytoplasm; and (4) the concentration of mCAD in the cytoplasm was higher than in the nucleus. The composition of the exogenous ICAD–CAD complex is indicated on the right: wt, mCAD-N-HA or hICAD-C-myc; ΔNLS, mCADΔNLS-N-HA; and S, hICAD-S-C-myc. For each transfection, >400 cells were scored in two to three independent experiments. Bar, 10 μm.
Supplemental Figure 4.
Distribution of endogenous and exogenous hICAD in HeLa cells after biochemical fractionation. The ICAD content of nuclear (nu), microsomal (mi), and cytosolic (cy) fractions of HeLa cells, obtained by differential centrifugation, were determined with immunoblot analysis. (Available at http://www.jcb.org/cgi/content/full/150/2/321/DC1)
Results
Localization of ICAD in Nonapoptotic Cells
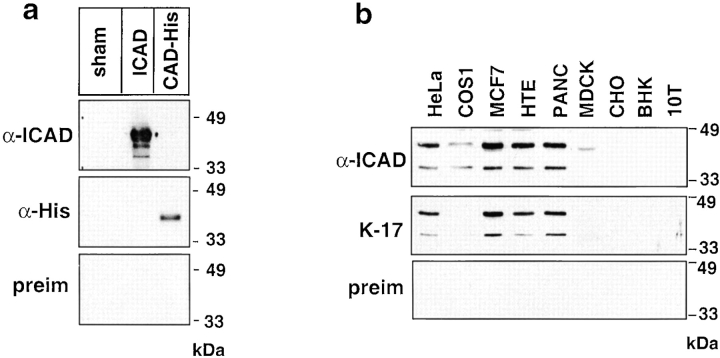
The subcellular localization of endogenous ICAD was examined using a polyclonal anti-hICAD antibody, developed by immunizing rabbits with purified GST-hICAD fusion protein. Western blot analysis showed that the rabbit anti-hICAD immune serum, but not the preimmune serum, recognizes recombinant hICAD with an apparent molecular mass of ∼45 kD (Fig. 1 a), corresponding to the predicted molecular mass of hICAD (Liu et al. 1997). No immuno cross-reactivity was observed with recombinant mCAD-(His)6 (apparent molecular mass ∼40 kD), despite the presence of the conserved CIDE-N domains in both CAD and ICAD (Inohara et al. 1998) (Fig. 1 a).
Figure 1.
Characterization of the anti-hICAD antibody. (a) The polyclonal rabbit anti-ICAD immune serum recognizes recombinant hICAD on immunoblot. Full-length hICAD and mCAD-(His)6 were expressed in BL21(D3) cells, and the bacterial lysates (∼10 μg protein/lanes) were separated with SDS-PAGE, transferred to nitrocellulose, and polypeptides were visualized by enhanced chemiluminescence using rabbit polyclonal anti-hICAD antiserum (α-ICAD), mouse monoclonal anti-His (α-His) primary antibody, or preimmune serum with the corresponding HRP-conjugated secondary antibody. Lysate obtained from sham-transformed bacteria were used as negative controls. (b) Western blot analysis of endogenous hICAD expression. Equal amounts of protein (50 μg) of the indicated cell lysate were separated with SDS-PAGE and subjected to immunoblotting as described in Materials and Methods. Two prominent immunoreactive polypeptides, with an apparent molecular mass of ∼45 and ∼36 kD, corresponding to the full-length ICAD (ICAD-L) and the alternatively spliced variant ICAD-S, respectively, are recognized with the rabbit (α-ICAD) and goat (K-17) anti-hICAD antibodies but not with the rabbit preimmune serum (preim).
Two major polypeptides were recognized by the anti-hICAD immune serum, but not by the preimmune serum, in whole cell lysates of HeLa, COS-1, MCF7, HTE, and PANC cells, all of human or primate origin (Fig. 1 b). The slower migrating band, with an apparent molecular mass of ∼45 kD, corresponds to the predicted mass of the full-length hICAD (ICAD-L). The less abundant polypeptide, with an apparent molecular mass of ∼36 kD, is consistent in size with the alternatively spliced variant, hICAD-S (Sakahira et al. 1998, Sakahira et al. 1999; Gu et al. 1999), and was not detected with a polyclonal anti-ICAD antibody, specific for the COOH terminus of ICAD-L (data not shown). A comparable pattern of ICAD expression was revealed with the affinity-purified goat polyclonal K-17 anti–ICAD antibody (Fig. 1 b). Thus, the rabbit and goat anti-hICAD immune serums recognize human ICAD, but cannot recognize hamster (CHO and BHK), mouse (10T), and dog (MDCK) orthologues (Fig. 1 b).
The subcellular distribution of endogenous hICAD was established with indirect immunofluorescence microscopy in five cell lines, including HeLa, HTE, PANC, COS-1, and MCF7, using two independent anti-ICAD antibodies. The representative fluorescence micrographs of HeLa and HTE cells show that hICAD resides predominantly within the nucleus and is excluded from the nucleolus, with a consistently weak fluorescence signal in cytoplasm (Fig. 2, a and b). No nuclear staining was detectable with the rabbit preimmune serum or after the adsorption of the specific antibody to recombinant ICAD-His6 (Fig. 2 a). Importantly, the strong nuclear staining of ICAD could be observed with the affinity-purified goat polyclonal anti-ICAD antibody, as illustrated in the HeLa cells (Fig. 2 c). These data indicate that hICAD constitutively accumulates in the nucleus in a variety of cells with the maintenance of a reduced level in the cytoplasm.
Figure 2.
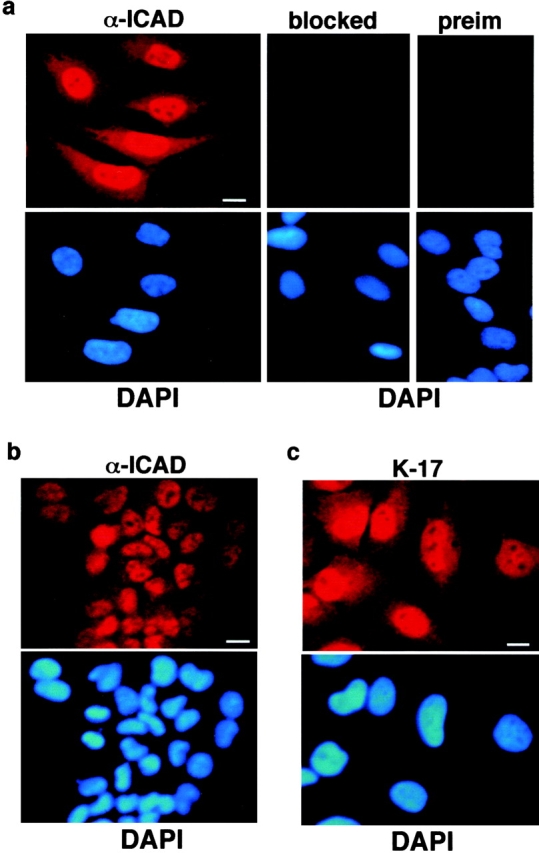
Indirect immunolocalization of endogenous hICAD with fluorescence microscopy. HeLa (a and c) or human tracheal epithelia (b) cells were fixed, permeabilized, and immunostained with rabbit (a and b) or goat K-17 (c) polyclonal anti-hICAD primary and tetramethylrhodamine-conjugated secondary antibody. Chromosomal DNA was visualized with DAPI (4′,6′-diaminidino-2-phenylindole), and identical fields are shown for hICAD and DNA staining. Adsorption of the rabbit immune serum to recombinant ICAD-His6 prevented the immunostaining of the nuclei (a, blocked). The preimmune serum does not recognize any specific signal (a, preim). Bars: 10 μm.
The Nuclear Localization Signal of ICAD
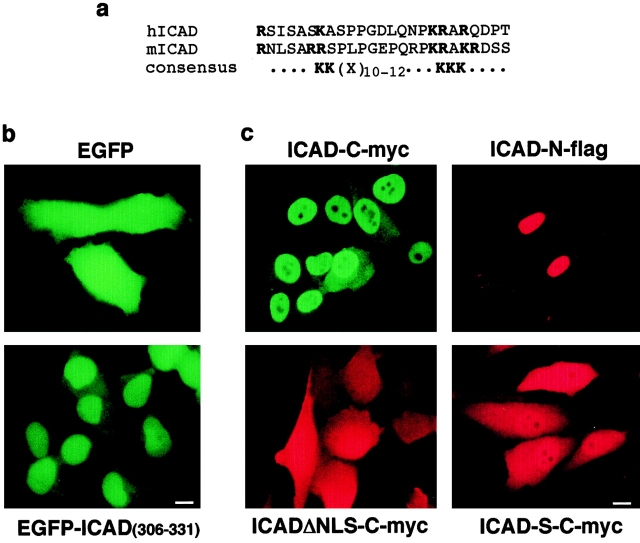
The nuclear import ICAD could be achieved passively in association with CAD or another carrier molecule. Alternatively, an unrecognized NLS residing in ICAD could mediate nuclear uptake. Primary sequence analysis of the mouse and human ICAD revealed two series of basic residues separated by a 10–amino acid spacer at the extreme COOH terminus, which is reminiscent of the consensus sequence of the bipartite NLS of nucleoplasmin (Fig. 3 a; Dingwall et al. 1988; Robbins et al. 1991). Although the alignment to consensus of the bipartite NLS was less striking for hICAD, compared with its mouse counterpart, (Dingwall and Laskey 1991; Jans and Hubner 1996; Fig. 3 a), the functional significance of the NLS was tested using a fusion protein, comprising the enhanced green fluorescent protein (EGFP) and the last 26–amino acid residues of hICAD (EGFP-ICAD306-331). The transiently expressed EGFP-ICAD306-331 was clearly nuclear (Fig. 3 b, bottom), in contrast to the EGFP, which was found uniformly distributed throughout the cells (Fig. 3 b, top). Thus, the COOH-terminal tail of hICAD is sufficient to confer nuclear import capacity to EGFP, and may be implicated in the nuclear localization of ICAD. During the revision of this manuscript similar results were reported by Samejina and Earnshaw (Samejina and Earnshaw 2000).
Figure 3.
Nuclear localization signals of hICAD. (a) The COOH-terminal amino acid residues of the human (h) and mouse (m) ICAD are shown aligned. The positively charged amino acid residues, representing the putative recognition signals in the bipartite NLS, are indicated in bold. (b) The COOH terminus of hICAD directs EGFP into the nucleus. HeLa cells were transiently transfected with the expression plasmid encoding the EGFP or the EGFP-ICAD306-331 chimera and observed with fluorescence microscopy. (c) The COOH terminus confers nuclear targeting to hICAD. HeLa cells were transiently transfected with the expression vectors encoding hICAD-C-myc, hICAD-N-flag, hICADΔNLS-C-myc, and hICAD-S-C-myc, as indicated. Localization of ICAD was monitored by indirect immunofluorescence using the corresponding anti-epitope antibodies. Bars: 10 μm.
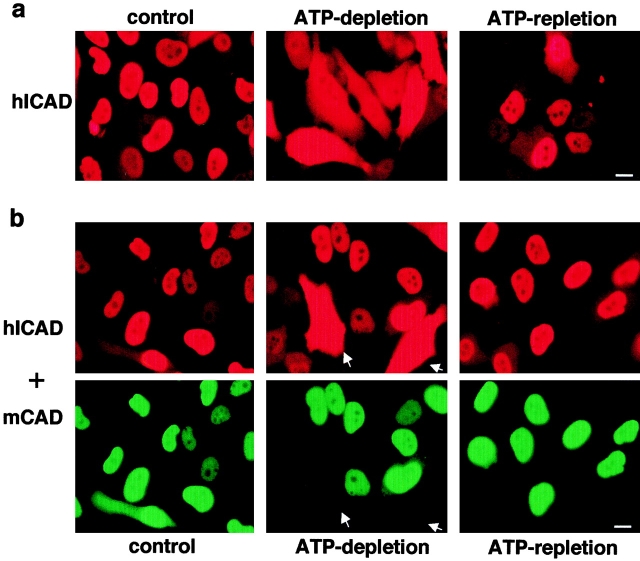
Overexpressed hICAD consistently appeared largely nuclear, with a low level of cytosolic expression in different expression systems (HeLa, COS-1, and BHK), regardless of the epitope (c-myc, flag, or HA [influenza hemagglutinin]) introduced at the COOH or NH2 termini (Fig. 3 c, top and data not shown). Several lines of evidence indicate that NLS-dependent active uptake accounts for the nuclear localization of overexpressed hICAD. First, the nuclear accumulation of hICAD was clearly compromised without NLS. Strong cytosolic immunostaining was detected for hICADΔNLS-C-myc, a recombinant version of hICAD prepared by deleting the last 26–amino acid residues, comprising the NLS consensus (Fig. 3 c, bottom). Further, prominent cytosolic retention was observed for the epitope-tagged ICAD-S, representing the alternatively spliced isoform, which lacks the last 66 amino acid residues (Fig. 3 c, bottom right), suggesting that the nuclear import of exogenous hICAD cannot be attributed to association with CAD. Secondly, both stably and transiently transfected hICAD-C-myc was predominantly nuclear at expression levels, which were comparable to that of endogenous ICAD in HeLa cells (Fig. 4 a, left, and Figure S1 [available at http://www.jcb.org/cgi/content/full/150/2/321/ DC1]). Finally, depletion of the cellular ATP content by metabolic inhibitors dissipated the nucleocytosolic gradient of hICAD-C-myc (Fig. 4 a, middle), supporting the hypothesis that nuclear accumulation of hICAD involves active transport. Irreversible deterioration of the nuclear envelope could not account for this phenomenon, since hICAD-C-myc reconcentrated in the nuclei upon the recovery of the cellular ATP content (Fig. 4 a, right). These observations are consistent with the activity of a previously unrecognized NLS at the COOH terminus of hICAD. Furthermore, they also suggest that the weak cytosolic immunostaining, obtained with the polyclonal anti-ICAD antibodies (Fig. 2), is, most likely, because of the presence of low levels of monomeric ICAD-S, confined to the cytoplasm (Figure S2, a and b [available at http://www.jcb.org/cgi/content/full/150/2/321/DC1]). Supporting this notion, the cytosolic immunostaining of ICAD-S was virtually abolished upon transient expression of CAD-N-HA, with a concomitant increase in the nuclear immunostaining of ICAD (Figure S2, c).
Figure 4.
The effect of reversible energy depletion on the distribution of hICAD and mCAD. ATP depletion induces the cytosolic release of overexpressed hICAD in the absence (a) but not in the presence of mCAD (b). Indirect immunolocalization of hICAD and mCAD was carried out on HeLa cells stably expressing hICAD-C-myc (hICAD) (a), or transiently overtransfected with mCAD-N-HA (b, hICAD + mCAD). The epitopes were visualized as described in Materials and Methods. ATP depletion was achieved by incubating the cells in the presence of metabolic inhibitors (4 μg/ml antimycin A, 10 mM deoxy-d-glucose, and 10 μM carbonyl cyanide p-chlorophenyl hydrazone; ATP-depletion) for 2 h at 37°C. ATP repletion was attained by extensive rinses and incubating the cells in normal medium for 3 h (ATP-repletion). Export of ICAD from the nuclei into the cytoplasm upon ATP depletion is evident in cells without mCAD expression (arrowheads). Bars: 10 μm.
The ICAD/CAD Heterodimer Is Nuclear in Nonapoptotic Cells
It has been established previously that co- or posttranslational heterodimerization of CAD and ICAD is necessary for the expression of CAD. The possible impact of heterodimerization on the subcellular targeting of ICAD was assessed by transient coexpression of epitope-tagged human ICAD and mouse CAD.
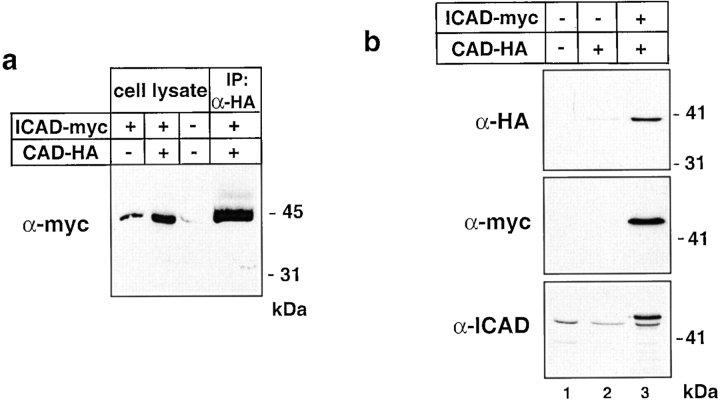
Three lines of evidence indicated that epitope-tagged human ICAD, a highly conserved orthologue of mouse ICAD (Liu et al. 1997; Sakahira et al. 1998), dimerizes efficiently with mouse CAD. First, physical interaction between mCAD and hICAD was demonstrated by coimmunoprecipitation of the hICAD-C-myc/mCAD-N-HA complex with anti-HA antibody and subsequent immunoblotting the precipitate with anti-myc antibody (Fig. 5 a). Second, the expression level of mCAD-N-HA was substantially higher (more than sixfold) in the presence of exogenous hICAD-C-myc, as demonstrated by quantitative immunoblot analysis (Fig. 5 b, lanes 2 and 3). As a corollary, the level of expression of the exogenous hICAD-C-myc was significantly higher than that of the endogenous form, which was detected by the rabbit anti-hICAD antibody (Fig. 5 b, lanes 2 and 3). Finally, the release of hICAD-C-myc, from the nucleus into the cytosol, was virtually undetectable upon ATP depletion from those nuclei that expressed both hICAD-C-myc and mCAD-N-HA (Fig. 4 b), but not hICAD-C-myc alone (Fig. 4 a). This may be explained by the diffusional barrier of the nuclear pore complex for polypeptides >45 kD, such as the hICAD–mCAD complex (molecular mass ∼85 kD; Dingwall and Laskey 1991). Collectively, these experiments indicate that human ICAD can substitute its mouse counterpart as a chaperone and as an inhibitor of mCAD, as reported for the heterodimerization of monkey or avian ICAD with mCAD (Enari et al. 1998; Samejima et al. 1998). Since similar data were obtained with coexpressing human CAD and ICAD, we confirmed that expression of both mCAD and hCAD is strictly limited by the abundance of the endogenous ICAD and increased level of CAD through heterologous expression can be achieved by introducing and binding of exogenous ICAD.
Figure 5.
Expression and interaction of mCAD with hICAD. (a) hICAD associates with mCAD. HeLa cells were transiently transfected with hICAD-C-myc and mCAD-N-HA expression constructs. Immunoprecipitation of the ICAD–CAD complex was performed with monoclonal mouse anti-HA antibody. Whole cell lysates and immunoprecipitate (IP) were separated with SDS-PAGE and probed with anti-myc antibody. (b) Heterologous expression of mCAD-N-HA is limited by the endogenous hICAD. HeLa cells were transiently transfected with the plasmid encoding the indicated constructs and carrier DNA (pcDNA3). 2 d after transfection, cells were lysed and equal amounts of protein (50 μg) were separated with SDS-PAGE and transferred to nitrocellulose. Polypeptides were visualized with enhanced chemiluminescence using anti-HA (α-HA), anti-myc (α-myc), or anti-ICAD antibodies.
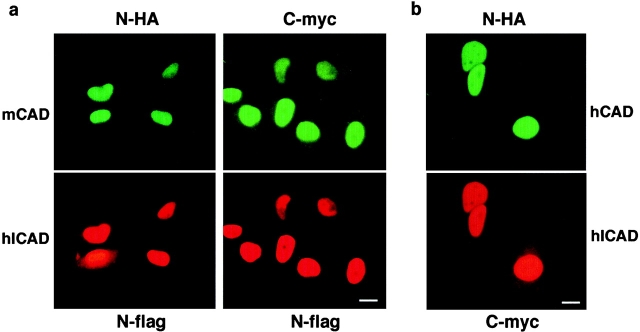
The subcellular distribution of hICAD in the ICAD–CAD complex was investigated by immunofluorescence staining of HeLa cells, transiently cotransfected with epitope-tagged hICAD and mCAD. The nearly exclusive nuclear expression of heterologous hICAD and mCAD was independent of the epitope (Fig. 6 a), which is consistent with the localization of endogenous hICAD (Fig. 2). Similar subcellular distribution was observed in COS-1 and BHK cells (data not shown). Association or binding to nuclear constituents is unlikely to play a major role in the nuclear retention of ICAD/CAD. In contrast to the nuclear proteins, which are resistant to detergent extraction because of their association with the nucleoskeleton or chromosomal DNA (e.g., lamina-associated polypeptide 2α, (Lim and Li 1996)), a low concentration of Triton X-100 was sufficient to remove both mCAD-N-HA and hICAD-C-myc from the nucleus (unpublished observation). As a corollary, monomeric hICAD appears to move freely in both directions across the nuclear pore complex.
Figure 6.
Colocalization of epitope-tagged CAD and ICAD. (a) Nuclear colocalization of epitope-tagged hICAD and mCAD and (b) hICAD and hCAD. HeLa cells were transiently transfected with the expression vectors encoding mCAD or hCAD with the indicated epitope. Immunostaining was performed with a combination of rabbit polyclonal anti-HA and mouse monoclonal anti-Flag or anti-myc antibodies, and the corresponding fluorescein- and rhodamine-conjugated secondary antibodies and were detected by fluorescence microscopy. Identical fields are shown for ICAD and CAD distribution. Bars: 10 μm.
Nuclear accumulation of ICAD/CAD could not be attributed to activation of apoptosis, since neither increased chromosomal DNA fragmentation nor phosphatidylserine translocation into the outer leaflet of the plasma membrane was detectable by terminal deoxynucleotidyl transferase–mediated dUTP nick end-labeling (TUNEL) assay or annexin V staining, respectively. Finally, identical localization was demonstrated for the epitope-tagged hICAD/hCAD heterodimer (Fig. 6 b), supporting our hypothesis that nuclear targeting of ICAD/CAD is an inherent characteristic of the complex, rather than the consequence of heterologous expression, and dimerization ICAD does not impair its nuclear accumulation.
The Role of ICAD in the Nuclear Targeting of the ICAD–CAD Complex
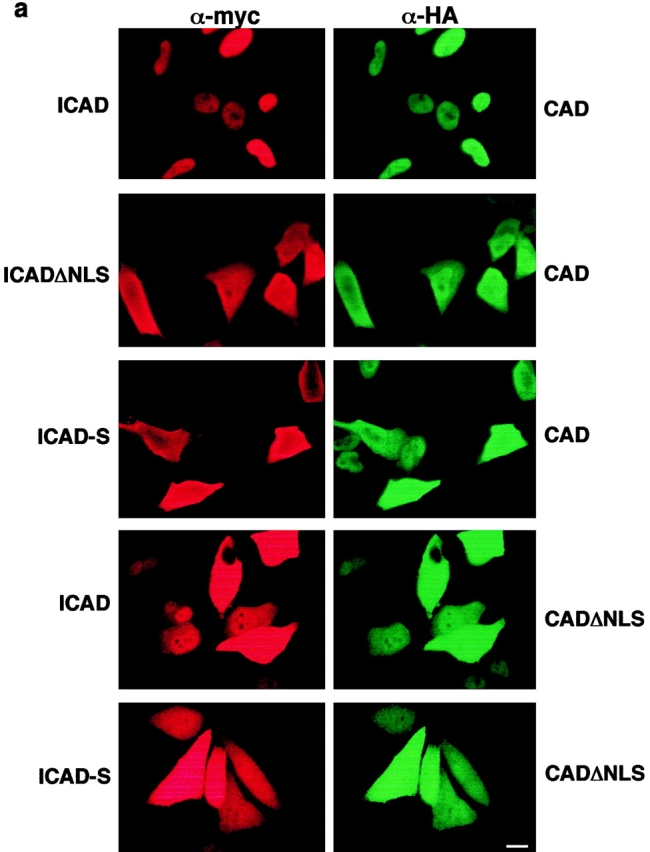
Polypeptides >45 kD require an NLS and interactions with nuclear transport receptors to be targeted specifically into the nucleus (Kalderon et al. 1984; Newmeyer and Forbes 1988; Dingwall and Laskey 1991). It is possible that nuclear accumulation of the ICAD–CAD complex, with a molecular mass of ∼85 kD, relies on either or both of the NLS motifs of its constituents. Given that the heterologously expressed ICAD–CAD complex is largely comprised of epitope-tagged components, the individual contribution of NLS of CAD and ICAD could be elucidated. The subcellular distribution of the ICAD–CAD complex was scored on the basis of the nucleocytoplasmic gradient of mCAD, since heterodimerization is obligatory for the expression of CAD (Enari et al. 1998; Halenbeck et al. 1998; Liu et al. 1998; Inohara et al. 1999). Data in the previous section and analysis of >400 transfected cells showed that hICAD–mCAD complex is essentially nuclear (Fig. 6 and Fig. 7, a and b, filled bars). In contrast, deletion of the ICAD NLS impeded the nuclear import of the complex, reflected by the cytosolic localization of mCAD-N-HA and hICADΔNLS-C-myc heterodimer (Fig. 7 a). Similar subcellular distribution of mCAD was observed in the presence of ICAD-S-C-myc (Fig. 7 a), suggesting that the NLS of hICAD is necessary for the efficient nuclear accumulation of the CAD–ICAD complex. Furthermore, these results also imply that nuclear uptake of CAD may be influenced by the expression level of ICAD-S. This latter possibility was verified by determining the subcellular distribution of mCAD in complex with hICAD-S. Nuclear accumulation of mCAD was observed only in 52% of the expressors and showed substantial cytosolic staining in the rest of the transfectants upon coexpression with hICAD-S (Fig. 7 c, empty bars).
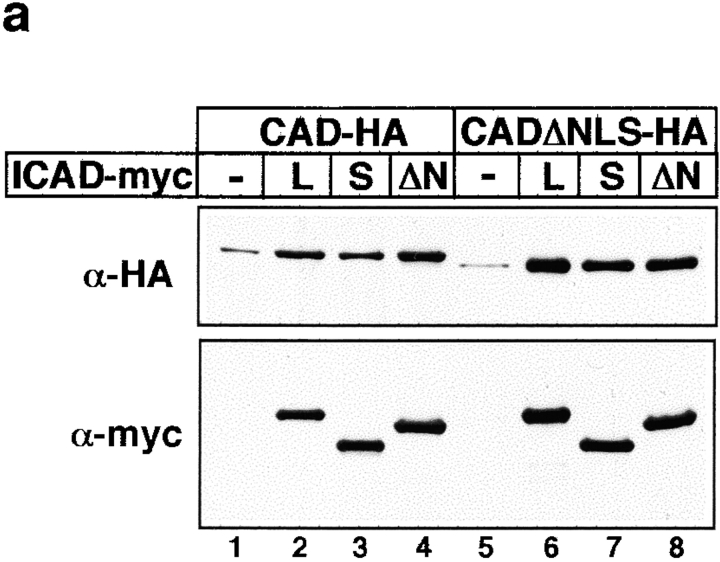
The possibility that hICADΔNLS or hICAD-S is unable to chaperone the folding of mCAD, leading to a nonnative, and perhaps mistargeted mCAD seems unlikely since the expression level of mCAD was augmented by either the full-length or truncated variants of hICAD more than six-fold, as measured by quantitative immunoblot analysis (Fig. 8 a, lanes 1–4). Similar distribution patterns were observed when human CAD-N-HA was coexpressed with the full-length or truncated hICADs (Figure S3 [available at http://www.jcb.org/cgi/content/full/150/2/321/DC1]), precluding the possibility that dimerization of hICAD with mCAD induced the exposure of a buried NLS. Hence, we conclude that the NLS of hICAD has an important role in the constitutive nuclear localization of the ICAD/CAD heterodimer.
Figure 8.
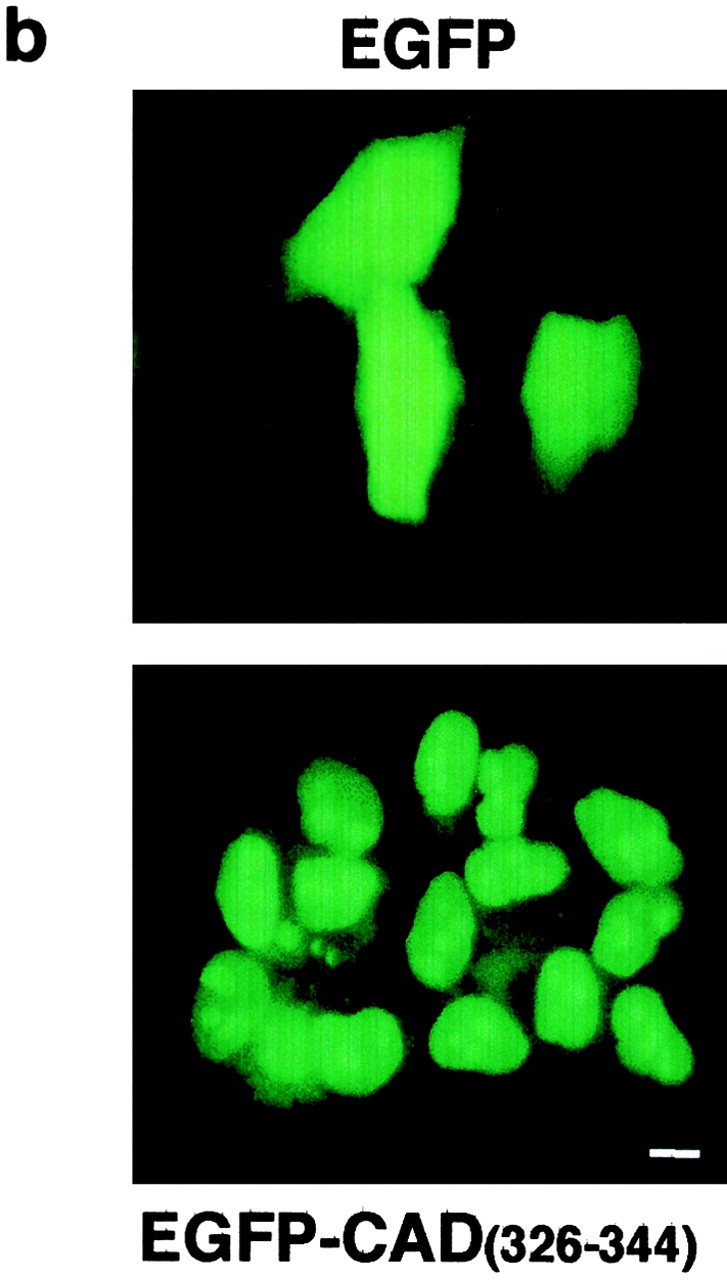
The importance of the NLS of CAD in its expression and nuclear targeting. (a) Heterologous expression of mCAD and mCADΔNLS requires the presence of either the full-length or the truncated variant of hICAD. The expression of mCAD-N-HA or mCADΔNLS-N-HA was determined in the absence or presence of hICAD-C-myc (L), hICAD-S-C-myc (S), or hICADΔNLS-C-myc (ΔN) in transiently transfected HeLa cells, using anti-HA antibody. The expression of ICAD was visualized with anti-myc antibody. (b) The COOH terminus of mCAD directs EGFP into the nucleus. HeLa cells were transiently transfected with the expression vector encoding EGFP or EGFP-CAD326-344 fusion, comprising the putative NLS of mCAD and observed with fluorescence microscopy. Bar, 10 μm.
The Role of CAD in the Nuclear Targeting of the ICAD–CAD Complex
To examine the possible role of CAD in the nuclear targeting of the ICAD–CAD complex, the function of its putative NLS was also evaluated. The COOH-terminal tail of mCAD, containing the positively charged amino acid cluster (326RRKQPARKKRPARKR344), was fused in-frame to EGFP. The transiently expressed EGFP-CAD326-344 chimeric polypeptide was nuclear in HeLa cells, indicating that the COOH terminus of mCAD is sufficient to confer nuclear import capacity to EGFP (Fig. 8 b).
The significance of the NLS in the context of mCAD was demonstrated by immunolocalization of the COOH terminally truncated mCAD lacking its NLS (CADΔNLS). Deletion of the NLS disrupted the nuclear import of the CADΔNLS-N-HA as well as the coexpressed ICAD-C-myc (Fig. 7 a). In contrast to the exclusive nuclear localization of mCAD and hICAD (Fig. 6 and Fig. 7, a and b, filled bar), the complex comprising the mCADΔNLS and hICAD remained cytosolic for >50% of the transfectants (Fig. 7 c, filled bar). The rest of the transfectants displayed incomplete nuclear accumulation with significant amount of cytosolic mCADΔNLS-NHA (Fig. 7 c). Importantly, deletion of the NLS in mCAD does not seem to interfere with its biosynthesis and folding, since the expression level of mCADΔNLS was comparable to that of mCAD, in the presence of either the full-length or the truncated hICADs (Fig. 8 a, lanes 6–8). Furthermore, deletion of the putative NLS of the human CAD caused a similar subcellular distribution of the ICAD–CADΔNLS complex (Figure S3). Thus, perturbation of the nuclear localization of the ICAD/CAD heterodimer by CADΔΝLS provides direct evidence for the role of CAD NLS. Therefore, we conclude that NLSs of both ICAD and CAD contribute to the constitutive nuclear import of the complex. Our results also imply that translocation of activated CAD from the cytoplasm into the nucleus is not occurring specifically to achieve fragmentation of chromosomal DNA during apoptosis.
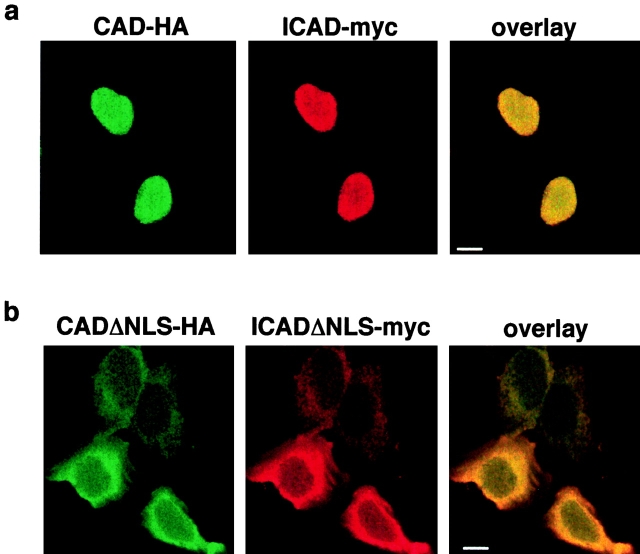
The NLSs of ICAD and CAD Contribute to the Nuclear Uptake of ICAD–CAD Complex in an Additive Manner
To assess whether the NLS of each constituent participates in the nuclear targeting of the ICAD–CAD complex, the subcellular distribution of the heterodimer composed of mCADΔNLS and ICADΔNLS was examined. In striking contrast to the nuclear colocalization of mCAD-N-HA and hICAD-C-myc (Fig. 9 a), the nuclear targeting of the complex lacking both NLSs was virtually abolished, as illustrated by the laser confocal fluorescent micrographs (Fig. 9 b). Comparable results were obtained with the mCADΔNLS-N-HA/hICAD-S-C-myc complex (Fig. 7 a). Nuclear accumulation of the mCADΔNLS-N-HA/hICAD-S-C-myc complex could not be recognized in 81% of the transfected cells, and appeared to be excluded from the nucleus in 19% of the expressors (Fig. 7 b, empty bar). Since the cytosolic expression of the ICAD/CAD was substantially more pronounced when both NLSs were absent, in contrast to single NLS deletion, we conclude that the NLSs of both ICAD and CAD are required for efficient nuclear import of the heterodimer.
Figure 9.
Coimmunolocaliza- tion of wt- and NLS-deficient ICAD and CAD with laser confocal microscopy. (a) Immunofluorescence detection of mCAD-N-HA and hICAD-C-myc in transiently transfected HeLa cells using anti-HA and anti-myc antibodies. mCAD and hICAD colocalize throughout the nucleus. (b) Deletion of the NLSs results in cytosolic localization of the exogenous ICAD–CAD complex. Immunofluorescence detection of overexpressed mCADΔNLS-N-HA and hICADΔNLS-C-myc in transiently transfected HeLa cells using anti-HA and anti-myc antibodies. Bars: 10 μm.
Staurosporine-induced Apoptosis Is Associated with the Caspase-dependent Release of ICAD from the Nucleus
Immunolocalization of endogenous hICAD suggests that proteolytic activation of ICAD/CAD takes place in the nucleus, rather than in the cytoplasm as proposed previously (Enari et al. 1998). To test this prediction, the cellular distribution of endogenous and epitope-tagged hICAD was monitored upon staurosporine-induced apoptosis. According to the immunostaining, staurosporine evoked a time-dependent disappearance of hICAD from the nuclei of HeLa cells. The nuclear expression of the endogenous hICAD appears to be abolished in most of the cells after 3 h of incubation with staurosporine (Fig. 10 a). Nearly complete elimination of the fluorescence signal derived from the exogenous ICAD-C-myc was observed after 2 h of incubation with staurosporine (Fig. 10 b). The more pronounced diminution of the ICAD-C-myc could be attributed to an increased caspase susceptibility of the monomeric hICAD-C-myc, and/or the small size of c-myc epitope, as compared with multiple epitopes detected with the polyclonal anti-ICAD antibody.
Figure 10.
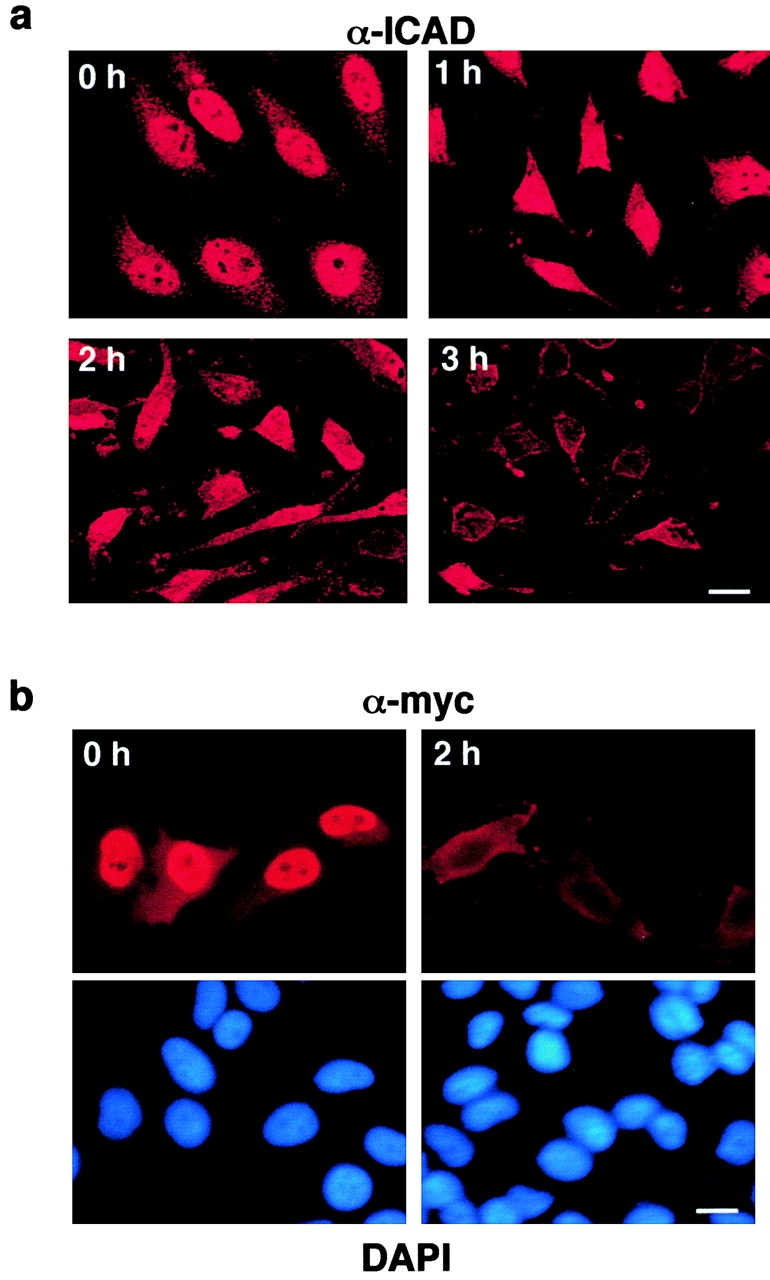
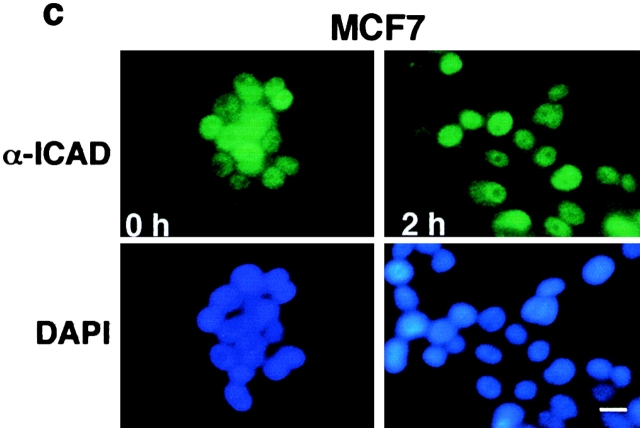
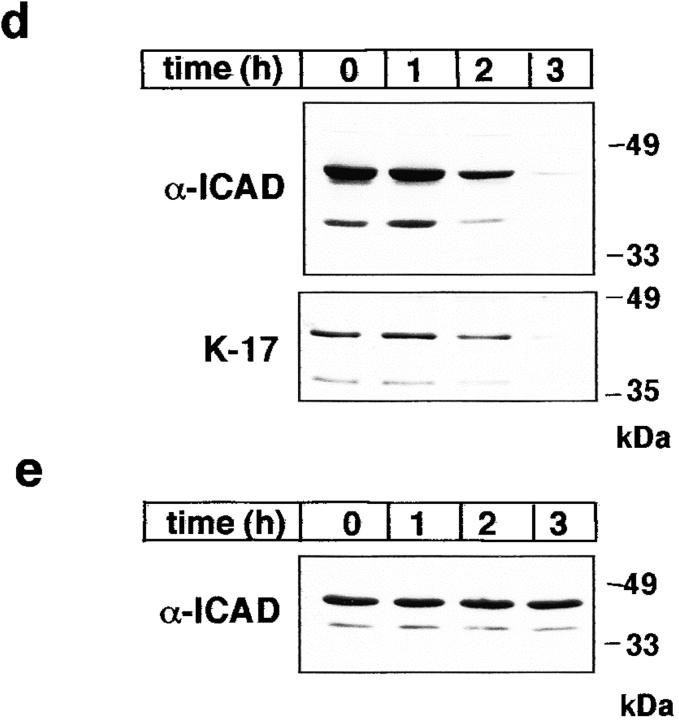
The effect of staurosporine on the localization and expression of hICAD. The subcellular distribution of endogenous hICAD (a and c) and transiently overexpressed hICAD-C-myc (b) were visualized in HeLa (a and b) and MCF7 (c) cells before and after staurosporine (2 μM) treatment for 2 h at 37°C. Cells were fixed, permeabilized, and hICAD and hICAD-C-myc were immunostained with the rabbit polyclonal anti-hICAD (α-ICAD) and anti-myc antibodies, respectively. Chromosomal DNA was visualized with DAPI, and identical fields are shown for hICAD and DNA staining. (d and e) The effect of staurosporine on the expression level of hICAD. HeLa (d) and MCF7 (e) cells were incubated in the presence of 2 μM staurosporine for the indicated time. Cellular proteins were solubilized in RIPA buffer, and equal amounts of protein (50 μg) were analyzed by immunoblotting using polyclonal rabbit (α-ICAD) and goat (K-17) anti-hICAD antibodies. Bars: 10 μm.
In sharp contrast to HeLa cells, the nuclear expression of the endogenous hICAD was preserved upon staurosporine treatment of MCF7 cells, a cell line which lacks functional caspase-3 (Janicke et al. 1998; Fig. 10 c). Consistent with the immunostaining results, a significant decrease in the total cellular pool of hICAD (and ICAD-S) was documented with immunoblot analysis of HeLa, but not MCF7 cells, upon induction of apoptosis with staurosporine (Fig. 10, d–e). The effect of staurosporine on ICAD degradation was sensitive to DEVD-CHO, an inhibitor of caspases-3, -6, and -7 (data not shown).
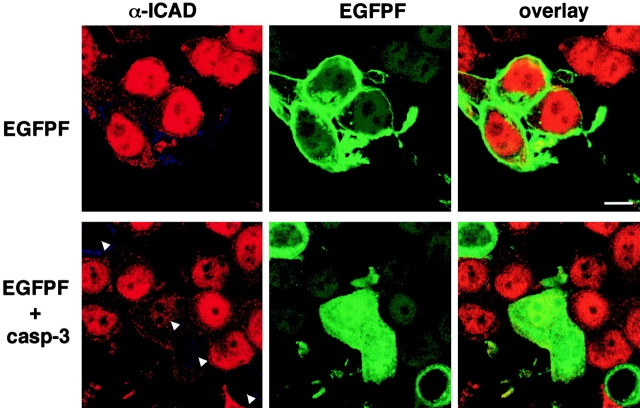
The lack of a staurosporine effect on the degradation of ICAD in MCF7 cells could not be explained by the absence of the upstream apoptotic signaling cascade, since hICAD could be eliminated from the nuclei after the complementation of the cells with procaspase-3. MCF7 cells were cotransfected with plasmids encoding procaspase-3 and membrane-targeted EGFP (EGFPF) at a molar ratio of 4:1. The inclusion of EGFPF facilitated the identification of procaspase-3 expressors. Activation of procaspase-3 with staurosporine caused the disappearance of endogenous ICAD from the nuclei of MCF7 cells expressing both EGFPF and caspase-3, but not EGFPF alone, as detected with immunostaining (Fig. 11). These in situ immunolocalization studies suggest that expression of caspase-3 is indispensable for proteolytic activation of ICAD/CAD, which takes place in the nucleus.
Figure 11.
The effect of staurosporine-induced apoptosis on the localization of hICAD in MCF7 cells. MCF7 cells were transiently transfected with plasmids encoding for (top) EGFPF and pcDNA3 or (bottom) procaspase-3 and EGFPF at a molar ration of 4:1. Immunostaining of endogenous hICAD was performed with rabbit anti-ICAD immune serum after exposing the cells to staurosporine (2 μM) for 2 h at 37°C and visualized with laser confocal microscopy. The arrowheads indicate those nuclei that have lost ICAD expression. Bar, 10 μm.
Discussion
Based on in vitro reconstitution of chromosomal DNA degradation, it was initially postulated that proteolytic cleavage of ICAD by caspase(s) would allow the dissociation of ICAD from CAD in the cytosol and unmasking the NLS of CAD, permitting the nuclear import of the active nuclease (Enari et al. 1998). However, recent reports of the nuclear localization of heterologously expressed epitope-tagged hICAD and GFP-mICAD fusion proteins were inconsistent with this activation mechanism, and suggested that ICAD may play a direct role in the nuclear targeting of CAD in nonapoptotic cells (Liu et al. 1998; Samejima and Earnshaw 1998). To resolve these conflicting observations and establish the mechanism of ICAD's action, immunolocalization of endogenous and heterologously expressed ICAD was performed in growing and apoptotic cells. Our results offer direct evidence for the constitutive nuclear targeting of the ICAD–CAD complex in nonapoptotic cells and document the caspase-3–dependent proteolytic cleavage of ICAD in the nucleus.
A number of observations indicate that the ICAD/CAD heterodimer resides, predominantly, in the nucleus of nonapoptotic cells. A panel of epitope-tagged ICAD and CAD variants was expressed in combinations or individually to demonstrate that they localize and colocalize in the nucleus. Nuclear localization was independent of the expression system or the epitope employed, and did not coincide with either DNA fragmentation or phosphatidylserine translocation (Fig. 4, Fig. 6, and Fig. 9). Moreover, inhibition of caspase activity with DEVD-CHO had no effect on the nuclear localization of exogenous ICAD/CAD or endogenous ICAD (data not shown). Finally and most importantly, we have demonstrated that endogenous hICAD is predominantly confined to the nucleus in diverse cell types with two independent polyclonal anti-hICAD antibodies (Fig. 2). Although we were unable to determine the subcellular localization of endogenous CAD, given the obligatory heterodimerization of the inactive CAD with ICAD, these experiments strongly suggest that the ICAD–CAD complex is targeted constitutively into the nucleus.
One of the most important findings of our study is the elucidation of the nuclear targeting signals of the ICAD–CAD complex. These experiments relied on the observation that coexpression of hICAD ensured a dramatic increase in the level of heterologous CAD, enabling investigations of the role of the NLSs without significant interference from the endogenous ICAD/CAD. In addition to identifying the NLS of ICAD, we have functionally verified the NLS in both mCAD and hCAD using two criteria. First, the NLSs of mCAD and hICAD were transferable. Both EGFP-ICAD306-331 and EGFP-CAD326-344 chimeras were targeted to the nucleus, in contrast to the homogenous cellular distribution of the EGFP (Fig. 3 b and 8 b). Second, the deletion of the bipartite NLS from the COOH terminus of hICAD manifested in the cytosolic accumulation of monomeric hICAD-S-C-myc and hICADΔNLS-C-myc as opposed to the full-length ICAD, which was predominantly nuclear (Fig. 3 c).
The role of the NLS of CAD was assessed in cells cotransfected with hICAD and truncated CAD. Incorporation of mCADΔNLS, or hCADΔNLS into the heterodimer partially disrupted its nuclear accumulation, similar to that observed in the presence of hICADΔNLS, implying that the NLS of CAD is also recognized by the nuclear import machinery in the complex. Importantly, when the NLSs of both ICAD and CAD were deleted, the nuclear exclusion of mCADΔNLS/hICAD-S and mCADΔNLS/hICADΔNLS became obvious (Fig. 9). The more pronounced cytosolic accumulation of the double mutants relative to that of single NLS deletion suggests that the two NLSs have an additive effect on the nuclear targeting efficiency of the ICAD–CAD complex. A cumulative effect of multiple nuclear localization signals on the targeting of soluble polypeptides is not without precedent. A number of proteins harbor two or more NLSs (e.g., c-myc, Matα2, and p53), which are required to achieve complete nuclear localization. Multiple copies of NLS apparently ensure more efficient targeting than do single copies (Dworetzky et al. 1988; Jans and Hubner 1996). The unique feature of ICAD/CAD targeting is that the two NLSs reside within distinct components of the complex.
In the light of the nuclear localization of the ICAD–CAD complex, it is reasonable to assume that the DNA fragmentation activity, which was isolated from the cytosol of apoptotic cells, was released from the nucleus during the preparation procedure, when ATP-dependent nuclear import was mitigated (Mitamura et al. 1998; Sabol et al. 1998). Perturbing the integrity of the nuclear envelope may have contributed to the cytosolic appearance of the ICAD–CAD complex during biochemical purification (Enari et al. 1998; Mitamura et al. 1998; Sabol et al. 1998). This speculation is supported by the findings that comparable amounts of ICAD and ICAD-C-myc/CAD-N-HA were associated with the nuclear and cytosolic fractions of HeLa cells, regardless of the homogenization technique used (nitrogen cavitation or freeze-thaw method, Figure S4 [available at http://www.jcb.org/cgi/content/full/150/2/321/DC1]). Furthermore, rapid loss of the exogenous ICAD–CAD complex was detected from the nuclei of NIH3T3 cell during digitonin fractionation (Dr. Robert Halenbeck, personal communication). These observations are in sharp contrast with the exclusive nuclear localization of the ICAD-C-myc/CAD-N-HA complex, as demonstrated by immunostaining, but are in line with previous reports documenting the loss of nuclear proteins during subcellular fractionation (Liu et al. 1998; Pain et al. 1983).
Activation of the ICAD/CAD Heterodimer Occurs in the Nucleus
The constitutive nuclear accumulation of ICAD/CAD dimer implies that regulation of CAD activity, most likely, takes place in the nucleus. How nuclear ICAD/CAD is activated upon apoptosis is an important question in understanding the mechanism of chromosomal DNA degradation. Convincing in vivo and in vitro data demonstrate that activation of CAD requires the proteolytic cleavage of ICAD by the caspase cysteine proteases (Mitamura et al. 1998; Sabol et al. 1998; Samejima et al. 1998; Tang and Kidd 1998; Sakahira et al. 1999). Our results provide some insights into this process in situ. Progressive disappearance of both the endogenous and the heterologously expressed hICAD was observed upon staurosporine-induced apoptosis from the nuclei of HeLa cells (Fig. 10). In contrast, staurosporine failed to promote ICAD degradation in caspase-3–deficient MCF7 cells, unless these cells were transiently transfected with the expression plasmid encoding procaspase-3 (Fig. 10 and Fig. 11), underlining the central role of caspase-3 in the regulation of CAD activity (Enari et al. 1998; Liu et al. 1998).
In the absence of evidence for the constitutive recycling of ICAD/CAD between the cytoplasm and the nucleus, the apoptotic signaling cascade must either trigger nuclear uptake of the heterotetrameric, activated caspase-3 or activate procaspase-3 within the nucleus to direct the proteolytic cleavage of the heterodimeric ICAD. While we are unable to distinguish between these possibilities at the present time, circumstantial and direct evidence suggest that nuclear translocation of both procaspases and active caspases can occur. Immunolocalization experiments have convincingly shown the nuclear import of caspase-9 in apoptotic neurons and suggested a similar process for caspase-3 in neuroblastoma cells (Nakagawara et al. 1997; Krajewski et al. 1999). Degradation of several caspase substrates, such as lamins, gelsolin, poly(ADP-ribose) polymerase, DNA-dependent protein kinase, and NUP153, is confined to the nucleus (Lazebnik et al. 1994, Lazebnik et al. 1995; Zheng et al. 1998; Buendia et al. 1999), implying that active caspases are present in the apoptotic nucleus. Finally, nuclear localization signals were revealed in the prodomain of caspase-1 and caspase-2, which appear to be required for the nuclear targeting (Colussi et al. 1998; Mao et al. 1998).
What are the biological advantages of the constitutive accumulation of ICAD/CAD in the nucleus? First, programmed cell death is often associated with a loss of mitochondrial membrane potential and impaired ATP production by the respiratory chain (Green and Reed 1998), which would be anticipated to compromise active nuclear import. Nuclear residence of ICAD/CAD renders the ATP-dependent nuclear import of CAD unnecessary in apoptotic cells. Second, since dimer formation is a prerequisite for the efficient nuclear targeting of CAD, this mechanism may play a role in ensuring that CAD remains enzymatically inactive in the nuclei of growing cells. On the other hand, the proximity of the DNase substrate would readily facilitate the cleavage of chromosomal DNA when the apoptotic enzyme cascade is activated. Finally, the nuclear targeting of ICAD/CAD may provide an additional mechanism for modulating the apoptotic tendency of nuclei. Since alterations in the ratio of ICAD-S/ICAD have been documented (Sabol et al. 1998), and ICAD-S appears to impair nuclear targeting of the heterodimer, as demonstrated by the cytosolic accumulation of ICAD-S–CAD complex (Fig. 7), we speculate that elevated expression of ICAD-S may decrease the apoptotic propensity of the nuclei. The apparently higher binding affinity of ICAD-S to CAD as compared with that of ICAD would further delay the caspase-dependent activation of the nuclease when the expression level of the alternatively spliced ICAD is elevated (Gu et al. 1999). However, the regulatory mechanisms, involved in the determination of the steady state level of ICAD-S, are presently not known.
In conclusion, the experimental data presented in this study indicate that two independent NLSs, identified in ICAD and CAD, are necessary and sufficient to render highly efficient, constitutive nuclear targeting of the ICAD/ CAD heterodimer. These observations, together with the nuclear accumulation of endogenous hICAD, and its redistribution on staurosporine-induced apoptosis in both HeLa and caspase-3 transfected MCF7 cells, imply that regulation of the apoptotic nuclease by the proteolytic cleavage of ICAD takes place in the nucleus.
Supplemental Material
Acknowledgments
We are grateful to Drs. S. Nagata, R. Halenbeck, V. Dixit, and W. Jiang for providing the cDNAs of mouse CAD, human CAD, procaspase-3, and EGFPF, respectively, and to Drs. S. Grinstein and M. Manolson for careful reading of the manuscript. We are indebted to Dr. R. Halenbeck for sharing unpublished results. We also thank Dr. H. O'Brodovich for his continuous support and Drs. D.W. Andrews and W. Trimble for helpful suggestions. The valuable assistance of K.-J. Sohn, J. So, and J. Sandu is highly appreciated.
This study was supported by the Medical Research Council (MRC) of Canada and the Canadian Cystic Fibrosis Foundation (CCFF; Sparx II Program). D. Lechardeur was supported, in part, by a CCFF Postdoctoral Fellowship. The instrumentation was partially covered by an Ontario Thoracic Society Block Term grant. J.M. Rommens and G.L. Lukacs are Scholars of MRC of Canada.
Footnotes
The online version of this article contains supplemental material.
Abbreviations used in this paper: CAD, caspase-3–activated DNase; DFF, DNA fragmentation factor; EGFP, enhanced green fluorescent protein; GST, glutathione-S-transferase; hICAD-L, full-length human ICAD; hICAD-S, alternatively spliced variant of hICAD; ICAD, inhibitor of caspase-activated DNase; NLS, nuclear localization signal.
References
- Buendia B., Santa-Maria A., Courvalin J.C. Caspase-dependent proteolysis of integral and peripheral proteins of nuclear membranes and nuclear pore complex proteins during apoptosis. J. Cell Sci. 1999;112:1743–1753. doi: 10.1242/jcs.112.11.1743. [DOI] [PubMed] [Google Scholar]
- Colussi P.A., Harvey N.L., Kumar S. Prodomain-dependent nuclear localization of the caspase-2(Nedd2) precursor. J. Biol. Chem. 1998;273:24535–24542. doi: 10.1074/jbc.273.38.24535. [DOI] [PubMed] [Google Scholar]
- Cormack B.P., Vladivia R.H., Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Robbins J., Dilworth S.M., Roberts B., Richardson W.D. The nucleoplasmin nuclear localization sequence is larger and more complex than that of SV-40 large T-antigen. J. Cell Biol. 1988;107:841–849. doi: 10.1083/jcb.107.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C., Laskey R.A. Nuclear targeting sequencesa consensus? Trends Biochem. Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Dworetzky S.I., Lanford R.E., Feldherr C.M. The effect of variations in the number and sequence of targeting signals on nuclear uptake. J. Cell Biol. 1988;107:1279–1287. doi: 10.1083/jcb.107.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enari M., Sakahira H., Yokoyama H., Okawa K., Iwamatsu A., Nagata S. A caspase-activated DNase degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- Green D.R., Reed J.C. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- Gu J., Dong R.-P., Zhang C., McLaughlin D.F., Wu M.X., Schlossman S.F. Functional interaction DFF35 and DFF45 with caspase-activated DNA fragmentation nuclease DFF40. J. Biol. Chem. 1999;274:20759–20762. doi: 10.1074/jbc.274.30.20759. [DOI] [PubMed] [Google Scholar]
- Halenbeck R., MacDonald H., Roulston A., Chen T.T., Conroy L., Williams L.T. CPAN, a human nuclease regulated by the caspase-sensitive inhibitor DFF45. Curr. Biol. 1998;8:537–540. doi: 10.1016/s0960-9822(98)79298-x. [DOI] [PubMed] [Google Scholar]
- Inohara N., Koseki T., Chen S., Wu X., Nunez G. CIDE, a novel family of cell death activators with homology to the 45 kDa subunit of the DNA fragmentation factor. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:2526–2533. doi: 10.1093/emboj/17.9.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N., Koseki T., Chen S., Benedict M.A., Nunez G. Identification of regulatory and catalytic domains in the apoptosis nuclease DFF40/CAD. J. Biol. Chem. 1999;274:270–274. doi: 10.1074/jbc.274.1.270. [DOI] [PubMed] [Google Scholar]
- Jacobson M.D., Weil M., Raff M.C. Programmed cell death in animal development. Cell. 1997;88:355–366. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- Janicke R.U., Sprengart M.L., Wati M.R., Porter A.G. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J. Biol. Chem. 1998;273:9357–9360. doi: 10.1074/jbc.273.16.9357. [DOI] [PubMed] [Google Scholar]
- Jans D.A., Hubner S. Regulation of protein transport to the nucleuscentral role of phosphorylation. Physiol. Rev. 1996;76:651–685. doi: 10.1152/physrev.1996.76.3.651. [DOI] [PubMed] [Google Scholar]
- Jiang W. Analysis of cell-cycle profiles in transfected cells using membrane-targeted GFP. Biotechnique. 1998;24:348–350. [PubMed] [Google Scholar]
- Kalderon D., Richardson W.D., Markham A.F., Smith A.E. Sequence requirements for nuclear localization of simian virus 40 large T-antigen. Nature. 1984;311:109–118. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- Krajewski S., Krajewska M., Ellerby L.M., Welsh K., Xie Z., Deveraux Q.L., Salvesen G.S., Bredesen D.E., Rosenthal R.E., Fiskum G., Reed J.C. Release of caspase-9 from mitochondria during neuronal apoptosis and cerebral ischemia. Proc. Natl. Acad. Sci. USA. 1999;96:5752–5757. doi: 10.1073/pnas.96.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazebnik Y.A., Kaufman S.H., Desnoyers S., Poirier G.G., Earnshaw W.C. Cleavage of poly(ADP-ribose) polymerase by a protease with properties like ICE. Nature. 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- Lazebnik Y.A., Takahashi A., Moir R., Goldman R., Poirier G.G., Kaufman S.H., Earnshaw W.C. Studies of the lamin proteinase reveal multiple parallel biochemical pathways during apoptotic execution. Proc. Nat. Acad. Sci. USA. 1995;92:9042–9046. doi: 10.1073/pnas.92.20.9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechardeur D., Sohn K.-J., Haardt M., Joshi P.B., Monck M., Graham R.W., Beatty B., Squire J., O'Brodovich H., Lukacs G.L. Metabolic instability of plasmid DNA in the cytosola potential barrier to gene therapy. Gene Ther. 1999;6:482–497. doi: 10.1038/sj.gt.3300867. [DOI] [PubMed] [Google Scholar]
- Lim A., Li B.F.L. The nuclear targeting and nuclear retention properties of a human DNA repair protein O6-methylguanine-DNA methyltransferase are both required for its nuclear localizationthe possible implications. EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:4050–4060. [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zou H., Slaughter C., Wang X. DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell. 1997;89:175–184. doi: 10.1016/s0092-8674(00)80197-x. [DOI] [PubMed] [Google Scholar]
- Liu X., Li P., Widlak P., Zou H., Luo X., Garrard W.T., Wang X. The 40-kDa subunit of DNA fragmentation factor induces DNA fragmentation and chromatin condensation during apoptosis. Proc. Natl. Acad. Sci. USA. 1998;95:8461–8466. doi: 10.1073/pnas.95.15.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs G.L., Mohamed A., Kartner N., Chang X.-B., Riordan J.R., Grinstein S. Conformational maturation of CFTR but not its mutant counterpart (ΔF508) occurs in the endoplasmic reticulum and requires ATP. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:6076–6086. doi: 10.1002/j.1460-2075.1994.tb06954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P.-L., Jiang Y., Wee B.Y., Porter A.G. Activation of caspase-1 in the nucleus requires nuclear translocation of pro-caspase-1 mediated by its prodomain. J. Biol. Chem. 1998;273:23621–23624. doi: 10.1074/jbc.273.37.23621. [DOI] [PubMed] [Google Scholar]
- Mitamura S., Ikawa H., Mizuno N., Kaziro Y., Itoh H. Cytosolic nuclease activated by caspase-3 and inhibited by DFF-45. Biochem. Biophys. Res. Commun. 1998;243:480–484. doi: 10.1006/bbrc.1998.8122. [DOI] [PubMed] [Google Scholar]
- Mukae N., Enari M., Sakahira H., Fukuda Y., Inazawa J., Toh H. Molecular cloning and characterization of human caspase-activated DNase. Proc. Natl. Acad. Sci. USA. 1998;95:9123–9128. doi: 10.1073/pnas.95.16.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S. Apotosis by death factor. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- Nakagawara A., Nakamura Y., Ikeda H., Hiwasa T., Kuida K., Su M.S., Zhao H., Cnaan A., Sakiyama S. High levels of expression and nuclear localization of interleukin-1 β converting enzyme (ICE)and CPP32 in favorable human neuroblastomas. Cancer Res. 1997;57:4578–4584. [PubMed] [Google Scholar]
- Newmeyer D.D., Forbes D.J. Nuclear import can be separated into distinct steps in vitronuclear pore binding and translocation. Cell. 1988;52:641–653. doi: 10.1016/0092-8674(88)90402-3. [DOI] [PubMed] [Google Scholar]
- Pain P.L., Austerberry C.F., Desjarlais L.J., Horovitz S.B. Protein loss during nuclear isolation. J. Cell Biol. 1983;97:1240–1242. doi: 10.1083/jcb.97.4.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J., Dilworth S.M., Laskey R.A., Dingwall C. Two independent basic domains in nucleoplasmin nuclear targeting sequenceidentification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- Sabol S.L., Li R., Lee T.Y., Abdul-Khalek R. Inhibition of apoptosis-associated DNA fragmentation activity in nonapoptotic cellsthe role of DNA fragmentation factor-45 (DFF45/ICAD) Biochem. Biophys. Res. Commun. 1998;253:151–158. doi: 10.1006/bbrc.1998.9770. [DOI] [PubMed] [Google Scholar]
- Sakahira H., Enari M., Nagata S. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature. 1998;391:96–99. doi: 10.1038/34214. [DOI] [PubMed] [Google Scholar]
- Sakahira H., Enari M., Nagata S. Functional differences of two forms of the inhibitor of caspase-activated DNase, ICAD-L and ICAD-S. J. Biol. Chem. 1999;274:15740–15744. doi: 10.1074/jbc.274.22.15740. [DOI] [PubMed] [Google Scholar]
- Samejima K., Earnshaw W.C. ICAD/DFF regulator of apoptotic nuclease is nuclear. Exp. Cell Res. 1998;243:453–459. doi: 10.1006/excr.1998.4212. [DOI] [PubMed] [Google Scholar]
- Samejina K., Earnshaw W.C. Differential localization of ICAD-L and ICAD-S in cells due to the removal of a COOH-terminal NLS from ICAD-L by alternative splicing. Exp. Cell. Res. 2000;255:314–320. doi: 10.1006/excr.2000.4801. [DOI] [PubMed] [Google Scholar]
- Samejima K., Shigenobu T., Kottke T.J., Enari M., Sakahira H., Cooke A.C., Durrieu F., Martins L.M., Nagata S., Kaufman S.H., Ernshaw W.C. Transition from caspase-dependent to caspase-independent mechanisms at the onset of apoptotic execution. J. Cell Biol. 1998;143:225–239. doi: 10.1083/jcb.143.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Kidd V.J. Cleavage of DFF-45/ICAD by multiple caspases is essential for its function during apoptosis. J. Biol. Chem. 1998;273:28549–28552. doi: 10.1074/jbc.273.44.28549. [DOI] [PubMed] [Google Scholar]
- Wyllie A.H. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature. 1980;284:555–556. doi: 10.1038/284555a0. [DOI] [PubMed] [Google Scholar]
- Zheng T.S., Schlosser S.F., Dao T., Hingorani R., Crispe I.N., Boyer J.L., Flavell R.A. Caspase-3 controls both cytoplasmic and nuclear events associated with Fas-mediated apoptosis in vivo. Proc. Natl. Acad. Sci. USA. 1998;95:13618–13623. doi: 10.1073/pnas.95.23.13618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.