Abstract
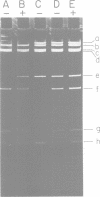
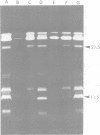
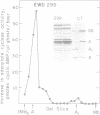
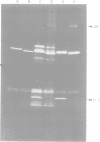
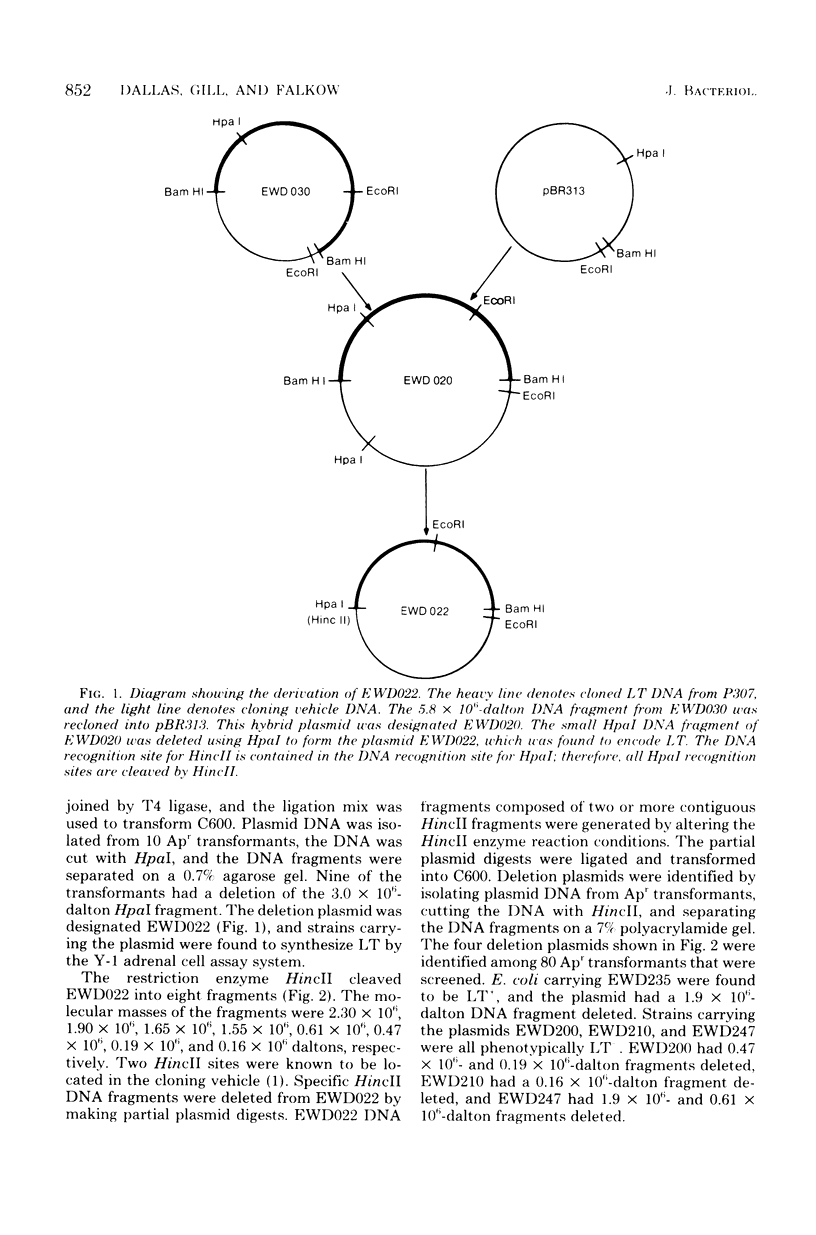
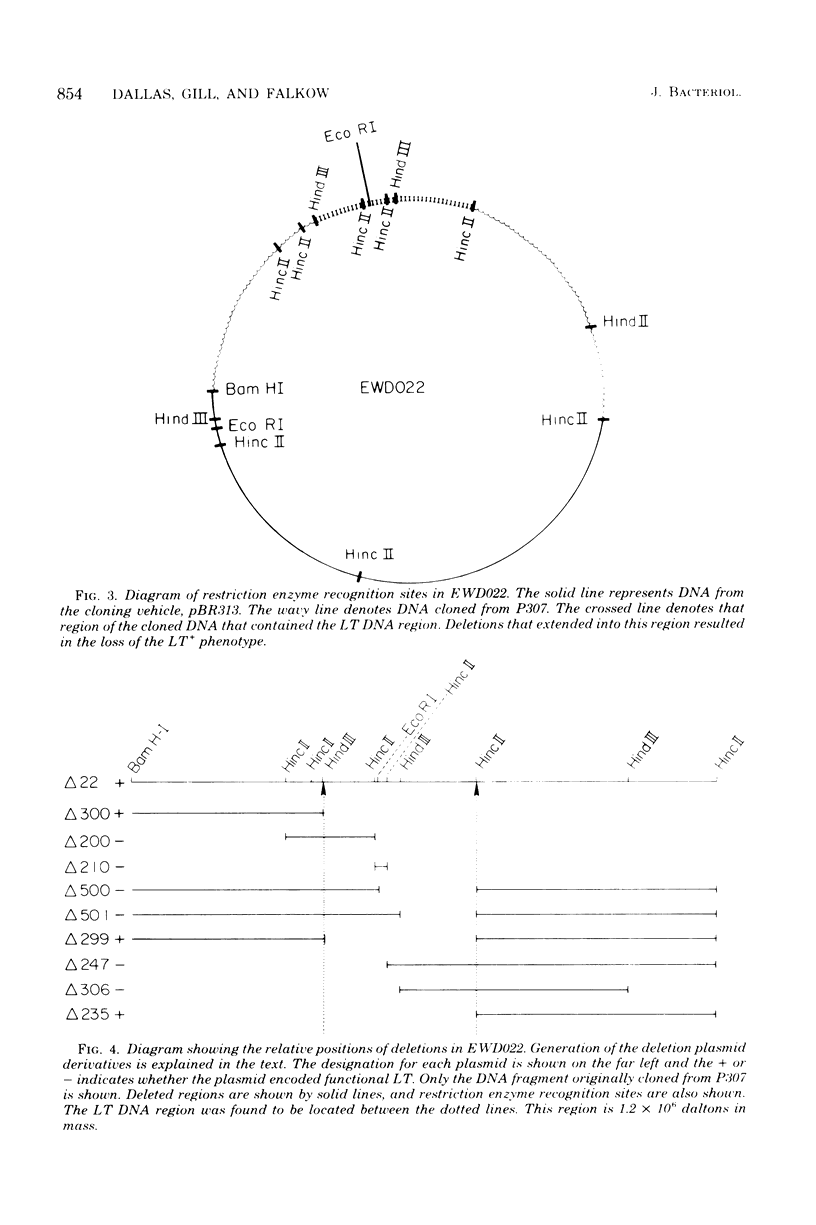
The structure and products of the two cistrons encoding the Escherichia coli heat-labile toxin (LT) were studied. The LT deoxyribonucleic acid (DNA) region had been isolated as part of a DNA fragment from the plasmid P307, and this fragment was joined to the cloning vector pBR313. Deletion mutations of various lengths were introduced into the LT DNA region and into the adjacent DNA sequences. Analysis of the deletions indicated that the maximum size of the LT DNA region was 1.2 x 10(6) daltons. Two proteins of 11,500 daltons and 25,500 daltons had been shown to be encoded by the LT DNA region. The functions of these LT gene products were investigated. The 11,500-dalton protein had an adsorption activity for Y-1 adrenal cells, and this protein was shown to form aggregates of four or five monomers. The 25,500-dalton protein was shown to have an adenylate cyclase-activating activity. The two cistrons encoding for each of the LT proteins have been located on a genetic map of the LT DNA region. Both cistrons are probably transcribed from the same promoter.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clements J. D., Finkelstein R. A. Demonstration of shared and unique immunological determinants in enterotoxins from Vibrio cholerae and Escherichia coli. Infect Immun. 1978 Dec;22(3):709–713. doi: 10.1128/iai.22.3.709-713.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Boyer H. W., Helling R. B. Construction of biologically functional bacterial plasmids in vitro. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer A. C., Curtiss R., 3rd Production, properties and utility of bacterial minicells. Curr Top Microbiol Immunol. 1975;69:1–84. doi: 10.1007/978-3-642-50112-8_1. [DOI] [PubMed] [Google Scholar]
- Gill D. M., King C. A. The mechanism of action of cholera toxin in pigeon erythrocyte lysates. J Biol Chem. 1975 Aug 25;250(16):6424–6432. [PubMed] [Google Scholar]
- Gill D. M., Rappaport R. S. Origin of the enzymatically active A1 fragment of cholera toxin. J Infect Dis. 1979 Jun;139(6):674–680. doi: 10.1093/infdis/139.6.674. [DOI] [PubMed] [Google Scholar]
- Gill D. M. The arrangement of subunits in cholera toxin. Biochemistry. 1976 Mar 23;15(6):1242–1248. doi: 10.1021/bi00651a011. [DOI] [PubMed] [Google Scholar]
- Kurosky A., Markel D. E., Peterson J. W., Fitch W. M. Primary structure of cholera toxin beta-chain: a glycoprotein hormone analog? Science. 1977 Jan 21;195(4275):299–301. doi: 10.1126/science.831277. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Parker R. C., Watson R. M., Vinograd J. Mapping of closed circular DNAs by cleavage with restriction endonucleases and calibration by agarose gel electrophoresis. Proc Natl Acad Sci U S A. 1977 Mar;74(3):851–855. doi: 10.1073/pnas.74.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgaramella V. Enzymatic oligomerization of bacteriophage P22 DNA and of linear Simian virus 40 DNA. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3389–3393. doi: 10.1073/pnas.69.11.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So M., Crosa J. H., Falkow S. Polynucleotide sequence relationships among Ent plasmids and the relationship between Ent and other plasmids. J Bacteriol. 1975 Jan;121(1):234–238. doi: 10.1128/jb.121.1.234-238.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]