Abstract
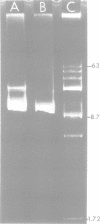
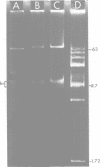
A 9.1 x 10(6)-dalton transposable deoxyribonucleic acid sequence resides within Pseudomonas aeruginosa plasmid R1033 and mediates resistance to gentamicin, streptomycin, sulfamethoxazole, chloramphenicol, and mercuric chloride. Transposability was demonstrated in Escherichia coli when this sequence, designated Tn1696, excised from R1033 and integrated into plasmid pMB8. Excision and insertion of Tn1696 occurred independently of the host Rec phenotype and may involve the 140-base pair, inverted deoxyribonucleic acid repeated region that flanks this sequence. Occurrence of a multiresistance transposon on a transferrable plasmid that has a broad host range may have serious epidemiological and therapeutic consequences.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Betlach M. C., Boyer H. W. Construction and characterization of new cloning vehicles. I. Ampicillin-resistant derivatives of the plasmid pMB9. Gene. 1977;2(2):75–93. doi: 10.1016/0378-1119(77)90074-9. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia P., Cairns J. Isolation of an E. coli strain with a mutation affecting DNA polymerase. Nature. 1969 Dec 20;224(5225):1164–1166. doi: 10.1038/2241164a0. [DOI] [PubMed] [Google Scholar]
- Dougan G., Sherratt D. The transposon Tn1 as a probe for studying ColE1 structure and function. Mol Gen Genet. 1977 Mar 7;151(2):151–160. doi: 10.1007/BF00338689. [DOI] [PubMed] [Google Scholar]
- Elwell L. P., Inamine J. M., Minshew B. H. Common plasmid specifying tobramycin resistance found in two enteric bacteria isolated from burn patients. Antimicrob Agents Chemother. 1978 Feb;13(2):312–317. doi: 10.1128/aac.13.2.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar W. E., Jr, O'dell N. M. beta-Lactamases and resistance to penicillins and cephalosporins in Serratia marcescens. J Infect Dis. 1976 Sep;134(3):245–251. doi: 10.1093/infdis/134.3.245. [DOI] [PubMed] [Google Scholar]
- Heffron F., Bedinger P., Champoux J. J., Falkow S. Deletions affecting the transposition of an antibiotic resistance gene. Proc Natl Acad Sci U S A. 1977 Feb;74(2):702–706. doi: 10.1073/pnas.74.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys G. O., Willshaw G. A., Anderson E. S. A simple method for the preparation of large quantities of pure plasmid DNA. Biochim Biophys Acta. 1975 Apr 2;383(4):457–463. doi: 10.1016/0005-2787(75)90318-4. [DOI] [PubMed] [Google Scholar]
- Jacoby G. A., Jacob A. E., Hedges R. W. Recombination between plasmids of incompatibility groups P-1 and P-2. J Bacteriol. 1976 Sep;127(3):1278–1285. doi: 10.1128/jb.127.3.1278-1285.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers J. A., Sanchez D., Elwell L. P., Falkow S. Simple agarose gel electrophoretic method for the identification and characterization of plasmid deoxyribonucleic acid. J Bacteriol. 1976 Sep;127(3):1529–1537. doi: 10.1128/jb.127.3.1529-1537.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price K. E., DeFuria M. D., Pursiano T. A. Amikacin, an aminoglycoside with marked activity against antibiotic-resistant clinical isolates. J Infect Dis. 1976 Nov;134(Suppl):S249–S261. doi: 10.1093/infdis/135.supplement_2.s249. [DOI] [PubMed] [Google Scholar]
- Rubens C., Heffron F., Falkow S. Transposition of a plasmid deoxyribonucleic acid sequence that mediates ampicillin resistance: independence from host rec functions and orientation of insertion. J Bacteriol. 1976 Oct;128(1):425–434. doi: 10.1128/jb.128.1.425-434.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahrabadi M. S., Bryan L. E., Van Den Elizen H. M. Further properties of P-2 R-factors of Pseudomonas aeruginosa and their relationship to other plasmid groups. Can J Microbiol. 1975 May;21(5):592–605. doi: 10.1139/m75-086. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Smith D. I., Lus R. G., Rubio Calvo M. C., Datta N., Jacob A. E., Hedges R. W. Third type of plasmid conferring gentamicin resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1975 Sep;8(3):227–230. doi: 10.1128/aac.8.3.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So M., Heffron F., Falkow S. Method for the genetic labeling of cryptic plasmids. J Bacteriol. 1978 Mar;133(3):1520–1523. doi: 10.1128/jb.133.3.1520-1523.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanisich V. A., Bennett P. M., Oritz J. M. A molecular analysis of transductional marker rescue involving P-group plasmids in Pseudomonas aeruginosa. Mol Gen Genet. 1976 Feb 2;143(3):333–337. doi: 10.1007/BF00269412. [DOI] [PubMed] [Google Scholar]