Abstract
The coordination of mitotic events is ensured through the spindle assembly checkpoint. BFA1 is required for this checkpoint in budding yeast because its disruption abolishes the mitotic arrest when spindle assembly is inhibited. Analysis of the genetic interaction of BFA1 with known mitotic checkpoint genes suggest that Bfa1 functions in the same pathway with Bub2 but not with Mad1 or Mad2. Both Bfa1 and Bub2 localize to spindle poles, and overexpression of Bfa1 arrests the cell cycle in anaphase. These findings suggest a bifurcation of the spindle assembly checkpoint: whereas one branch of the pathway, consisting of Mad1–3, Bub1 and 3, and Mps1, may prevent premature disjunction of sister chromosomes, the other, consisting of Bfa1 and Bub2, may function at spindle poles to prevent cytokinesis before the completion of chromosome segregation.
Cell cycle checkpoints ensure the fidelity of cell division by monitoring the completion of individual cell cycle events and controlling progression of the cell cycle through regulation of Cdk activity (reviewed in refs. 1 and 2). Inhibition of spindle assembly by microtubule drugs or mutations prevents cell exit from mitosis and entry into the next cell cycle (reviewed in refs. 3 and 4). Genetic analysis in budding yeast first led to the identification of several genes, BUB1, 2, and 3, MAD1, 2, and 3, and MPS1, which are required for the cell cycle arrest in response to inhibition of microtubule function (5–7). Homologs of several of these gene products have been identified in animal cells and shown to prevent mitotic progression in response to spindle defects (reviewed in refs. 3 and 4). Bub1, Bub3, Mad1, Mad2, and Mad3 homologs have been found to concentrate at the kinetochores in animal cells, suggesting a direct role for these proteins in monitoring kinetochore attachment to spindle microtubules (8–13). Ectopic expression of some of these proteins results in cell cycle arrest in metaphase in the absence of apparent spindle defects (14, 15). Thus, these proteins are thought to constitute a spindle assembly checkpoint that prevents the metaphase-to-anaphase transition before the chromosomes are properly attached to the spindle (reviewed in refs. 3 and 4).
Of the mitotic checkpoint proteins in budding yeast, Bub2 shows some functional differences from the rest. For example, the cell cycle delay caused by benomyl (a microtubule inhibitor) at a low dose that does not prevent spindle assembly but may interfere with kinetochore–microtubule interaction is abolished by mutations in the MAD genes and BUB1 and 3 but is not affected by mutations in BUB2 (16). In fission yeast, the Bub2 homolog, Cdc16, appears to be a negative regulator of cytokinesis: loss-of-function mutation of Cdc16 causes multiple septa to form during one cell cycle (17). Cdc16 interacts with Byr4, another negative regulator of cytokinesis (18), and stimulates the GTPase activity of Spg1, a positive regulator of cytokinesis (19, 20). A putative homolog of Byr4 has been found from the budding yeast genome database (18) and is now designated Bfa1 (Byr-four-alike). The sequence similarity between Byr4 and Bfa1 is confined to a COOH-terminal region that contains two imperfect repeats (18). In this report, I show that Bfa1 is a mitotic checkpoint protein in budding yeast and that it represents one of the two complementary branches of this checkpoint pathway, both required for the mitotic arrest in response to inhibition of spindle assembly.
MATERIALS AND METHODS
Media and Genetic Manipulations.
Yeast cell culture and genetic techniques were carried out by methods described by Sherman et al. (21). Yeast extract, peptone, dextrose (YPD) contained 2% glucose, 1% yeast extract, and 2% Bacto peptone. Yeast extract, peptone, galactose (YPG) contained 2% galactose, 1% yeast extract, and 2% Bacto peptone. Yeast extract, peptone, raffinose (YPR) contained 2% raffinose, 1% yeast extract, and 2% Bacto peptone.
Cloning, Plasmid and Strain Construction.
To generate a BFA1 disruption allele (Δbfa1), a 2.02-kb DNA fragment containing the BFA1 coding region (not including the stop codon) and 300-bp 5′ flanking sequence was amplified from yeast genomic DNA by using PCR and cloned between the XhoI and BamHI sites of pBluescript SK to yield pRL273. pRL273 was cut with PflMI and StuI, removing 76% of BFA1 coding region (amino acids 6–444), and blunted and ligated with a DNA fragment containing the HIS3 gene (22), yielding pRL280. To generate the Δbfa1 strain (RLY683), pRL280 was digested with XhoI and SpeI and transformed into RLY576, a wild-type strain in the W303 background. Gene disruption was confirmed by PCR analysis of the genomic DNA (data not shown). To generate a BUB2 disruption allele (Δbub2), a DNA fragment containing the BUB2 coding region was amplified from yeast genomic DNA by using PCR and cloned between the XhoI and XbaI sites of pBluescript SK to yield pRL275. pRL275 was cut with BamHI and StyI, removing 53% of BUB2 coding region (amino acids 90–252), and blunted and ligated with a DNA fragment containing the LEU2 gene (22), yielding pRL281. To generate the Δbub2 strain (RLY716), pRL281 was digested with XhoI and SpeI and transformed into RLY576. Gene disruption was confirmed by PCR analysis of the genomic DNA (data not shown). Δmad1 and Δmad2 alleles in the W303 background were obtained from Andrew Murray (University of California, San Francisco) and further backcrossed into the RLY576 W303 strain background.
A yeast integration plasmid expressing Mps1 under the GAL1 promoter (14) was introduced into RLY261 strain (Δbar1 in the same strain background as RLY576) to generate RLY700. The GAL1-MPS1∷URA3 allele was subsequently crossed into Δbfa1, Δbub2, Δmad1, or Δmad2 strains to generate the mutant strains bearing the GAL1-MPS1∷URA3 allele—RLY724, 727, 729, or 731, respectively. To generate a GAL1-BFA1 integration plasmid (pRL279), pRL273 was digested with StuI and BamHI, blunted, and ligated into the StuI site of pRL196, a vector for NH2-terminal (Myc)6 tagging of proteins under the GAL1 promoter. The fragment containing GAL1-(Myc)6-BFA1 was subcloned into pRS306 (23) for integration into the URA3 locus, yielding plasmid pRL283. pRL283 was linearized with XcmI and transformed into RLY576 to generate RLY690. The galactose-dependent expression of (Myc)6-Bfa1 was confirmed by immunoblot analysis (data not shown). The GAL1-(Myc)6-BFA1∷URA3 allele was subsequently crossed into Δbub2, Δmad1, or Δmad2 strains to generate the mutant strains bearing the GAL1-(Myc)6-BFA1∷URA3 allele—RLY688, 691, or 693, respectively.
To generate COOH-terminal green fluorescent protein (GFP)-tagged Bfa1, pRL273 was digested with XhoI and BamHI, and the Bfa1 contain fragment was ligated into the corresponding site of pRL73 (24). The Bfa1-GFP-containing fragment was subcloned into pRS306 for integration into the URA3 locus, yielding plasmid pRL282. pRL282 was linearized with XcmI and transformed into RLY261 to generate RLY603. To generate COOH-terminal GFP-tagged Bub2, a DNA fragment containing the BUB2 ORF (not including the stop codon) and 300 bp of the 5′ upstream sequence was amplified by using PCR and cloned between the XhoI-PstI sites of pRL73. The Bub2-GFP-containing fragment was subcloned into pRS306 for integration into the URA3 locus, yielding plasmid pRL288. pRL288 was linearized with XcmI and transformed into RLY716 to generate RLY735.
Immunofluorescence Staining.
Cells were fixed directly in growth media by addition of 37% formaldehyde to 5% final concentration. Immunofluorescence staining was carried out essentially as described (25). Rhodamine or FITC-conjugated secondary antibodies were purchased from Jackson ImmunoResearch.
Immunoblot Analysis.
Immunoblot analysis was carried out by using the enhanced chemiluminescence (ECL) detection kit (Amersham Pharmacia).
RESULTS
BFA1 Encodes a Mitotic Checkpoint Protein.
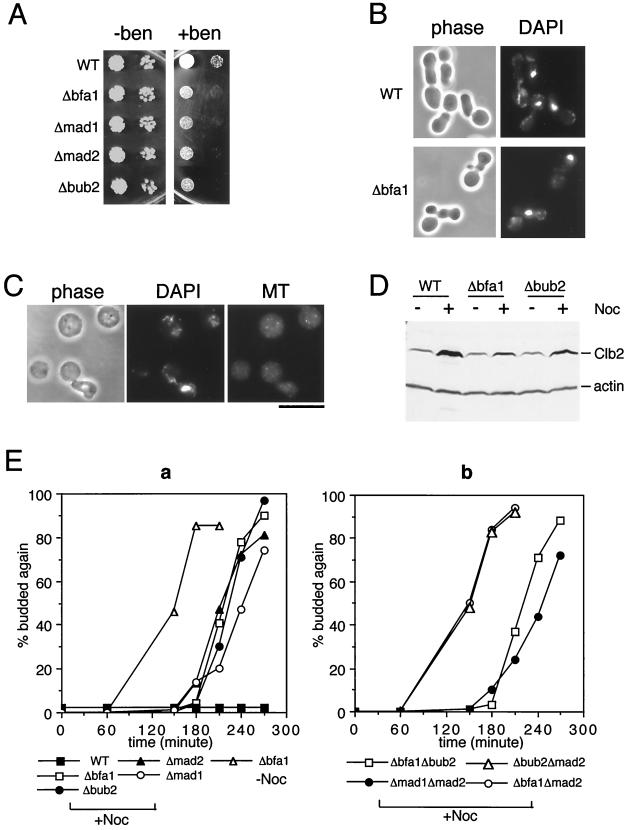
To determine the function of Bfa1 in budding yeast, a null allele was constructed in which 76% of the Bfa1 ORF was replaced by the HIS3 marker gene. Δbfa1 cells grow at a wild-type rate (Fig. 1A) and do not exhibit any apparent cell division defects, suggesting that unlike Byr4, Bfa1 is not an important regulator of cytokinesis during normal cell cycles. To determine whether Bfa1 functions in the mitotic checkpoint, Δbfa1 cells were tested for sensitivity to a sublethal concentration of benomyl. As shown in Fig. 1A, Δbfa1 cells exhibited the same degree of hypersensitivity to 10 μg/ml benomyl as mad1, mad2, and bub2 mutant cells.
Figure 1.
Bfa1 is a mitotic checkpoint protein acting in the same pathway as Bub2. (A) Cultures of RLY576 (WT, wild type), RLY683 (Δbfa1), RLY720 (Δmad1), RLY718 (Δmad2), and RLY716 (Δbub2) were grown to same density. Ten-fold serial dilutions were made and spotted onto YPD plates without (− ben) or with (+ ben) 10 μg/ml benomyl. Photographs were taken after 3 (− ben) and 5 (+ ben) days of growth at room temperature. (B) RLY576 and RLY683 cells were grown to 2 × 106 per ml and arrested with 5 μg/ml α-mating factor for 2.5 hours at room temperature. The cells were washed four times with water and resuspended in YPD containing 15 μg/ml nocodazol. After 5 hour growth at room temperature, cells were fixed and stained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize nuclear DNA. (C) The same fixed RLY683 cells were treated with 0.2 mg/ml zymolyase to remove the cell wall and double stained with DAPI and an anti-tubulin antibody as previously described (37). (D) Exponential cultures of RLY576, RLY683, and RLY716 cells were shifted to media with or without 15 μg/ml nocodazol (Noc) for 5 hours at room temperature. Total cell extracts were prepared and analyzed by immunoblotting using anti-Clb2 (38) and anti-actin (25) (as a loading control) antibodies. (E) RLY576, RLY683, RLY716, RLY718, RLY720, RLY684 (Δbfa1Δbub2), RLY686 (Δbfa1Δmad2), RLY736 (Δbub2Δmad2), and RLY737 (Δmad1Δmad2) cells were arrested in G1 with α-factor as described in B and released into media with or without 15 μg/ml nocodazol. For all strains used, >95% were arrested as unbudded cells after the α-factor treatment, and the rates at which budding occurred were identical after the release into media with or without nocodazol (budded cells always started to appear around 40 minutes after release and reached 85–90% at 60 min). Cells were fixed every 30 minutes and spotted on a microscope slide. The percentage of cells that had initiated a second round of budding was determined and plotted over time after release from the G1 arrest. The nuclear morphology was also examined for both wild-type and mutant cells obtained from the time points when the second round of budding had occurred. Most (>89%) of the mutant cells that had budded a second time in the presence of nocodazol had the morphology as shown in B (data not shown). In the absence of nocodazol, the newly budded mother cells stayed together with their former bud after cells were spotted into the glass slide. This was observed for both wild-type and various mutant strains of the W303 background, indicating a delay in cell separation or cell stickiness for this strain background. However, the nuclear morphology, as shown in B, was never observed in the absence of nocodazol (i.e., all cells with two buds had at least two distinct DAPI stained masses, data not shown). [Bar = 10 μm (C).]
The benomyl hypersensitivity of mitotic checkpoint mutants is due to their inability to arrest the cell cycle in mitosis when spindle assembly is inhibited (5, 6). To test whether Δbfa1 cells are also deficient in the mitotic arrest, cells were synchronized in G1 with α-mating factor and then released from the G1 arrest into media containing 15 μg/ml nocodazol (a more potent microtubule inhibitor than benomyl). Five hours later, as wild-type cells arrested in mitosis with a single large bud and an undivided nucleus, Δbfa1 cells, like the bub or mad mutants, had already initiated a second round of budding (Fig. 1B), indicating a failure in the mitotic arrest. Cell wall removal of Δbfa1 cells (but not wild-type cells; data not shown) fixed at the 5-hour time point gave rise to unbudded anucleate cells (3.7% before zymolyase treatment; 33% after the treatment), suggesting that cytokinesis had occurred in the absence of microtubules and nuclear division in Δbfa1 cells (Fig. 1C). When wild-type cells arrest in mitosis through the mitotic checkpoint, they maintain a high level of the mitotic cyclin Clb2 (14). Δbfa1 cells, however, like Δbub2 cells, failed to accumulate a high level of Clb2 under the same circumstances (Fig. 1D). The moderate increase in Clb2 level in Δbfa1 and Δbub2 cells grown in nocodazol was most likely caused by a short delay in the passage through mitosis (see below).
Bfa1 Functions in the Same Pathway with Bub2 but Not with Mad1 and Mad2.
An insight into the independent role of Bfa1 came from a close examination of the kinetics of mitotic progression in the presence of nocodazol. Although Δbfa1 cells failed to arrest, they still reproducibly delayed the passage through the cell cycle for 1–1.5 hours in comparison to cells released from the G1 arrest into media without nocodazol (Fig. 1E, a). A similar cell cycle delay was also observed in mad1, mad2, and bub2 null mutants released into media containing nocodazol (Fig. 1E, a). To determine whether the cell cycle delay of Δbfa1 cells in nocodazol is caused by the other mitotic checkpoint proteins present, double mutants were constructed between Δbfa1 and null alleles of mad2 and bub2. All of the double and single mutant strains were arrested to the same extent with α-factor and exited the G1 arrest with similar kinetics after release into media with or without nocodazol (see Fig. 1 legend). Pilot experiments were also carried out on cells released into media without nocodazol and showed that the timing of budding, nuclear division, and the second round of budding were virtually identical for all mutant and wild-type strains used (data not shown). The Δbfa1Δbub2 double-mutant cells exhibited the same cell cycle delay in the presence of nocodazol as either of the single mutants (Fig. 1E, b). The same delay was also observed with the Δmad1Δmad2 double mutant. The Δbfa1Δmad2 and Δbub2Δmad2 double-mutant cells, on the other hand, went through the cell cycle with the same kinetics as cells grown in the absence of nocodazol (Fig. 1E, b). This result suggests that Bfa1 and Bub2 are likely to act in a pathway separate from Mad1 and Mad2 in delaying the passage through mitosis.
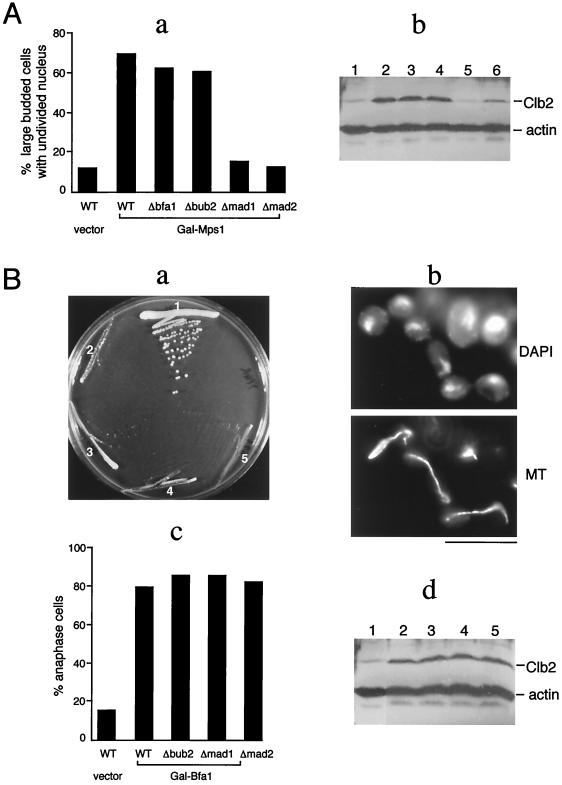
To further examine the independence of the Bfa1/Bub2 pathway from the Mad1/Mad2 pathway, a plasmid overexpressing the Mps1 protein under the GAL1 promoter was introduced into checkpoint mutant strains. It was shown previously that overexpression of Mps1 alone is sufficient to cause a mitotic delay that depends on the MAD and BUB genes, suggesting that Mps1 is an upstream component of the checkpoint pathway constituted by these genes (14). The data shown in Fig. 2A first confirmed that induction of Mps1 overexpression in galactose-containing media resulted in a mitotic delay in wild-type cells, as indicated by the accumulation of large budded cells with an undivided nucleus and an elevated level of Clb2. As expected, this delay was abolished in Δmad1 and Δmad2 cells. Overexpression of Mps1 in Δbfa1 and Δbub2 cells, by contrast, causes a mitotic delay similar to that observed in wild-type cells (Fig. 2A), suggesting that Bfa1 and Bub2 are unlikely to function downstream of Mps1 in the same way as do Mad1 and Mad2.
Figure 2.
(A) The mitotic delay induced by Mps1 overexpression is independent of Bfa1 and Bub2. RLY700 (WT), RLY724 (Δbfa1), RLY727 (Δbub2), RLY729 (Δmad1), and RLY731 (Δmad2) strains, which all contain the same integrated copy of Gal-Mps1 (14), were cultured overnight in media without galactose (YPR) and then shifted to media containing 2% galactose (YPG) for 6 hours. A control stain, RLY569, which contains the vector alone, was also included. (a) Cells were fixed, stained with DAPI, and the percentage of large budded cells (bud size greater than 2/3 of the mother size) with undivided DNA mass were determined and shown as histograms. (b) Total cell extracts were prepared and analyzed by immunoblotting using anti-Clb2 and anti-actin antibodies. Lane 1, RLY569; lane 2, RLY700; lane 3, RLY724; lane 4, RLY727; lane 5, RLY729; lane 6, RLY731. (B) Overexpression of Bfa1 induces an anaphase cell cycle arrest that is independent of the other mitotic checkpoint proteins. (a) Strains bearing vector alone (RLY569) or Gal-Bfa1 were streaked onto a plate containing 2% galactose. The photograph was taken after 3 days at 30°C. Sector 1, RLY569; sector 2, RLY690 (WT); sector 3, RLY688 (Δbub2); sector 4, RLY691 (Δmad1); sector 5, RLY693 (Δmad2). (b) The strains in B, a were cultured overnight in media without galactose (YPR) and then shifted to media containing 2% galactose (YPG) for 6 hours. Cells were fixed and stained with DAPI and an anti-tubulin (MT) antibody. The images show the arrest morphology of RLY690 cells. The same arrest morphology was also observed for the other Gal-Bfa1 bearing strains. (c) The percentages of cells exhibiting the arrest morphology, as shown in b, were determined for each of the strains in the experiment described in B, b. (d) Total extracts were prepared from each of the cell cultures in the experiment described in B, b and analyzed by immunoblotting by using anti-Clb2 and anti-actin antibodies. [Bar = 10 μm(B, b).]
Bfa1 Overexpression Causes a Cell Cycle Arrest in Anaphase.
Overexpression of Bfa1 under the Gal1 promoter in wild-type cells also blocked cell growth (Fig. 2B, a), but in this case, the cell cycle is blocked in anaphase where spindle elongation and chromosome segregation have already occurred (Fig. 2B, b and c), unlike the arrest point of Mps1 overexpression. The arrested cells also contained an elevated level of Clb2 (Fig. 2B, d). The anaphase arrest caused by Bfa1 overexpression was not affected by Δmad1, Δmad2, or Δbub2 mutations (Fig. 2B, c and d), suggesting that the Mad or Bub proteins do not function downstream of Bfa1 in activating anaphase arrest. Together, the above results support the idea that Bfa1 functions in a pathway separate from Mad1 and Mad2.
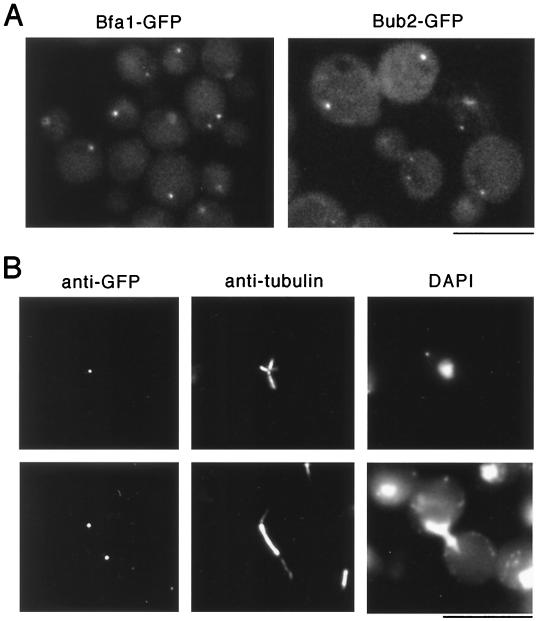
Bfa1 and Bub2 Proteins Localize to Spindle Poles.
The homologs of Mad1–3 and Bub1 and 3 in animal cells have been shown to localize to unattached kinetochores (8–13). In budding yeast, Mad1 exhibits a punctate nuclear staining pattern (26). To determine the localization of Bfa1 and Bub2, strains were constructed in which Bfa1 or Bub2, expressed under the BFA1 or BUB2 promoter, respectively, was tagged with GFP at the COOH terminus. The tagged constructs rescued the benomyl hypersensitivity of the corresponding null mutants (data not shown). Examination of cells expressing Bfa1-GFP or Bub2-GFP by fluorescence microscopy revealed that these proteins localized to one or two bright dots (Fig. 3A). Such localization was not seen in the control strain, which expresses the untagged proteins (data not shown), or strains expressing GFP-tagged proteins by using the same vector in other unrelated studies (24). Double immunofluorescence staining further showed that the Bfa1-GFP (Fig. 3B) or Bub2-GFP (data not shown) containing dots colocalized with the focal points of microtubule asters or spindle poles, suggesting that these proteins are localized to spindle pole bodies. Accumulation of these proteins inside the nucleus was never observed, even when they were overexpressed (data not shown).
Figure 3.
Localization of Bfa1 and Bub2 to the spindle pole bodies. Strains expressing Bfa1-GFP (RLY603) or Bub2-GFP (RLY735) at the endogenous level were constructed as described in Materials and Methods. (A) Live cells observed directly under a Nikon Eclipse E600 fluorescence microscopy equipped with an 100/1.40 oil DIC objective and a EXHQ450/50 DM480 LP/BA465LP GFP filter set. (B) Double immunofluroescence staining of fixed RLY603 cells by using a rat anti-tubulin antibody (37) and a rabbit anti-GFP antibody (a gift from P. Silver, Dana Farber Cancer Institute, Boston, MA). (Bar = 10 μm.)
DISCUSSION
A number of elegant studies in animal cells and in yeast have suggested that the signal that activates the mitotic checkpoint emanates from unattached kinetochores (reviewed in refs. 3, 4, and 27). Several of the spindle assembly checkpoint proteins have been found to localize selectively to the kinetochores of premetaphase chromosomes or misaligned chromosomes (8–13), suggesting that these proteins may play a direct role in monitoring the success of chromosome attachment. Mad2 has also been shown to act through Cdc20 to inhibit the anaphase promoting complex required for degradation of Pds1, a chromosome cohesion factor (reviewed in refs. 28 and 29). This is likely to be the key mechanism by which the mitotic checkpoint prevents premature disjunction of sister chromatids before kinetochore-spindle attachment.
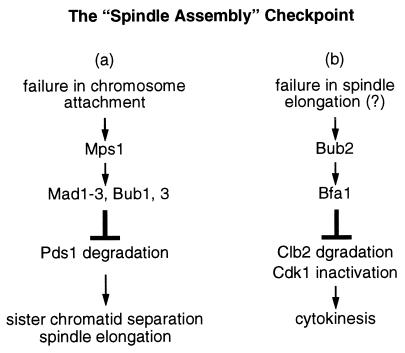
There is strong genetic and biochemical evidence that Mps1, Mad1–3, and Bub1 and 3 function in the same pathway leading to the metaphase arrest in response to kinetochore signals (11, 12, 14, 26, 30). However, there has been a question as to whether Bub2 is part of the same pathway (16). The studies described here strongly suggest that Bub2 and Bfa1, a newly identified mitotic checkpoint protein, function in a genetic pathway separate from those defined by Mps1, Mad1–3, and Bub1 and 3 (Fig. 4). Each pathway is sufficient to delay passage through mitosis, but both are required to achieve a full mitotic arrest when spindle assembly is inhibited.
Figure 4.
A schematic diagram depicting a bifurcation of the mitotic checkpoint pathway. This model hypothesizes that defects in spindle assembly can potentially prevent two mitotic events: chromosome attachment to the spindle (a); and anaphase spindle elongation (b). A defect in a may be monitored at the kinetochores and activates a set of mitotic checkpoint proteins that prevent sister chromatid separation by blocking degradation of chromosome cohesion factors such as Pds1. A defect in b may be monitored at the spindle pole bodies through Bub2 and Bfa1, which prevent cytokinesis by blocking the degradation of mitotic cyclins such as Clb2.
Two of the findings argue against a role for the Bub2/Bfa1-dependent pathway in sensing defects in kinetochore attachment. First, both proteins are found to localize to the spindle poles, and no centromere or nuclear localization was detected. However, it remains possible that Bub2 and Bfa1 monitor kinetochore attachment from a distance, perhaps by sensing the tension exerted on spindle poles. Human Mad1 and Mad2 have also been found to localize to centrosomes during metaphase, in addition to their kinetochore localization during interphase (31). It is not known whether these proteins have different functions when associated with different structures.
Second, overexpression of Bfa1 causes an anaphase arrest without blocking chromosome segregation but prevents degradation of the mitotic cyclin Clb2. Mitotic cyclin degradation and inactivation of Cdk1 have been shown to be required specifically for cytokinesis but not for chromosome segregation and anaphase spindle movement (32). Taken together, the findings described above lead to a hypothesis, as illustrated in Fig. 4, that Bfa1 and Bub2 constitute a bifurcation of the mitotic checkpoint control and function to coordinate anaphase events—chromosome segregation and cytokinesis. By associating with spindle pole bodies, Bfa1 and Bub2 may directly monitor the progression of anaphase B and signal cytokinesis only when chromosomes are successfully segregated to the poles. The direct downstream target of the Bub2/Bfa1 pathway may be Cdh1/Hct1, the anaphase promoting complex activator required for the degradation of mitotic cyclins (33–36), thus preventing cytokinesis and entry into the next cell cycle.
Acknowledgments
I thank A. Murray for providing the mad1 and 2 mutant strains and the Gal-Mps1 plasmid, A. Amon for the anti-Clb2 antibody, D. Drubin for the anti-actin antibody, and P. Silver for the anti-GFP antibody; A. Murray and members of the Li laboratory for helpful discussion and encouragement; and D. Finley, M. Kirschner, F. Mckeon, and D. Winter for comments on the manuscript.
ABBREVIATION
- GFP
green fluorescent protein
References
- 1.Murray A. Curr Opin Cell Biol. 1994;6:872–876. doi: 10.1016/0955-0674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 2.Elledge S J. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 3.Rudner A D, Murray A W. Curr Opin Cell Biol. 1996;8:772–780. [Google Scholar]
- 4.Straight A F. Curr Biol. 1997;7:R613–R616. doi: 10.1016/s0960-9822(06)00315-0. [DOI] [PubMed] [Google Scholar]
- 5.Li R, Murray A W. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 6.Hoyt M A, Totis L, Roberts B T. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 7.Weiss E, Winey M. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen R H, Waters J C, Salmon E D, Murray A W. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Benezra R. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- 10.Taylor S S, McKeon F. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- 11.Chen, R. H., Shevchenko, A., Mann, M. & Murray, A. W. (1998) J. Cell Biol. 283–295. [DOI] [PMC free article] [PubMed]
- 12.Taylor S S, Ha E, McKeon F. J Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basu J, Logarinho E, Herrmann S, Bousbaa H, Li Z, Chan G K T, Yen T J, Sunkel C E, Goldberg M L. Chromosoma. 1998;107:376–385. doi: 10.1007/s004120050321. [DOI] [PubMed] [Google Scholar]
- 14.Hardwick K G, Weiss E, Luca F C, Winey M, Murray A W. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- 15.He X, Patterson T E, Sazer S. Proc Natl Acad Sci USA. 1997;94:7965–7970. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Burke D J. Mol Cell Biol. 1995;15:6838–6844. doi: 10.1128/mcb.15.12.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fankhauser C, Marks J, Reymond A, Simanis V. EMBO J. 1993;12:2697–2704. doi: 10.1002/j.1460-2075.1993.tb05931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song K, Mach K E, Chen C Y, Reynolds T, Albright C F. J Cell Biol. 1996;133:1307–1309. doi: 10.1083/jcb.133.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt S, Sohrmann M, Hofmann K, Woollard A, Simanis V. Genes Dev. 1997;11:1519–1534. doi: 10.1101/gad.11.12.1519. [DOI] [PubMed] [Google Scholar]
- 20.Furge K A, Wong K, Armstrong J, Balasubramanian M, Albright C F. Curr Biol. 1998;8:947–954. doi: 10.1016/s0960-9822(98)70394-x. [DOI] [PubMed] [Google Scholar]
- 21.Sherman F, Fink G, Lawrence C. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1974. [Google Scholar]
- 22.Berben G, Dumont J, Gilliquet V, Bolle P, Hilger F. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- 23.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lippincott J, Li R. J Cell Biol. 1998;140:355–366. doi: 10.1083/jcb.140.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drubin D G, Miller K G, Botstein D. J Cell Biol. 1988;107:2551–2561. doi: 10.1083/jcb.107.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardwick K G, Murray A W. J Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicklas R B. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- 28.Visintin R, Prinz S, Amon A. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 29.Elledge S J. Science. 1998;279:999–1000. doi: 10.1126/science.279.5353.999. [DOI] [PubMed] [Google Scholar]
- 30.Farr K A, Hoyt M A. Mol Cell Biol. 1998;18:2738–2747. doi: 10.1128/mcb.18.5.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin D Y, Spencer F, Jeang K T. Cell. 1998;93:81–91. doi: 10.1016/s0092-8674(00)81148-4. [DOI] [PubMed] [Google Scholar]
- 32.Wheatley S P, Hinchcliffe E H, Glotzer M, Hyman A A, Sluder G, Wang Y l. J Cell Biol. 1997;138:385–393. doi: 10.1083/jcb.138.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zachariae W, Schwab M, Nasmyth K, Seufert W. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- 34.Schwab M, Lutum A S, Seufert W. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 35.Fang G, Yu H, Kirschner M W. Mol Cell. 1998;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- 36.Kramer E R, Gieffers C, Holzl G, Hengstschlager M, Peters J M. Curr Biol. 1998;8:1207–1210. doi: 10.1016/s0960-9822(07)00510-6. [DOI] [PubMed] [Google Scholar]
- 37.Kilmartin J, Wright B, Milstein C. J Cell Biol. 1982;93:576–582. doi: 10.1083/jcb.93.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K. EMBO J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]