Abstract
In fission yeast and multicellular organisms, centromere-proximal regions of chromosomes are heterochromatic, containing proteins that silence gene expression. In contrast, the relationship between heterochromatin proteins and kinetochore function in the budding yeast Saccharomyces cerevisiae remains largely unexplored. Here we report that the yeast heterochromatin protein Sir1 is a component of centromeric chromatin and contributes to mitotic chromosome stability. Sir1 recruitment to centromeres occurred through a novel mechanism independent of its interaction with the origin recognition complex (ORC). Sir1 function at centromeres was distinct from its role in forming heterochromatin, because the Sir2-4 proteins were not associated with centromeric regions. Sir1 bound to Cac1, a subunit of chromatin assembly factor I (CAF-I), and helped to retain Cac1 at centromeric loci. These studies reveal that although budding yeast and mammalian cells use fundamentally different mechanisms of forming heterochromatin, they both use silencing proteins to attract the histone deposition factor CAF-I to centromeric chromatin.
Keywords: Chromatin assembly, centromere, kinetochore, yeast, silencing
Eukaryotic chromosome transmission during cell division depends largely on the formation of stable contacts between the mitotic spindle and chromosomes. Centromeres promote spindle attachment to chromosomes by nucleating assembly of kinetochores, nucleoprotein structures that bind microtubules. In the fission yeast Schizosaccharomyces pombe and in multicellular eukaryotes, heterochromatic domains flank all centromeres and are required for high-fidelity chromosome segregation (Karpen and Allshire 1997). Like other heterochromatic loci, these centromere-flanking domains are distinguished from euchromatin by their ability to silence the transcription of reporter genes in a position-dependent manner. Centromeric heterochromatin helps to position centromeres toward the opposing spindle poles, thereby fostering bipolar microtubule-kinetochore contacts (Bernard et al. 2001).
As in all eukaryotes, epigenetic gene silencing in the budding yeast Saccharomyces cerevisiae is mediated by heterochromatin (Loo and Rine 1995). However, the protein composition of the silencing machinery in the budding yeast S. cerevisiae stands in marked contrast to that of S. pombe. For example, budding yeast lack structural homologs of the Swi6 (HP1) and Clr4[Su(var)3-9] proteins (Moazed 2001). Furthermore, the methylation of Lys 9 on histone H3, a signature of heterochromatin in S. pombe and other eukaryotic organisms, has not been detected in budding yeast (Briggs et al. 2001). Instead, transcriptional silencing of the cryptic mating-type loci HMR and HML in S. cerevisiae requires the Sir1-4 proteins (Loo and Rine 1995).
Two distinct nucleosome assembly factors, CAF-I and the Hir proteins, contribute to the specialized chromatin structures at both silent and centromeric loci in budding yeast (Kaufman et al. 1998; Sharp et al. 2002). Although position-dependent gene silencing has not been described at centromeres in budding yeast, the involvement of CAF-I and Hir proteins at centromeric chromatin led us to test whether Sir1-4 proteins were also present at centromeres. Surprisingly, we found that only the Sir1 protein (and not Sir2, Sir3, or Sir4) was associated with centromeres. Furthermore, recruitment of Sir1 to centromeres and HM loci required distinct protein interactions. Sir1 interacted directly with the large subunit of CAF-I and was required together with Hir1 for normal levels of association of CAF-I with centromeres. Epistasis experiments determined that Sir1 prevented chromosome nondisjunction in a manner that was overlapping with Cac1 and Hir1. Therefore, although Sir1 was previously known to function only in the formation of heterochromatin at HM loci, these data demonstrate an un-suspected role for Sir1 in promoting chromosome stability during mitosis.
Results and Discussion
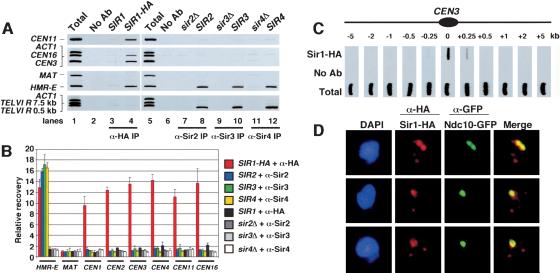
To explore the role of budding yeast silencing proteins in centromere function, chromatin immunoprecipitation (ChIP) was used to test whether the Sir1-4 proteins associated with centromeric regions. Sir1, but not Sir2, Sir3, or Sir4, was detected at both centromeric DNA and at HM loci (Fig. 1A,B). Sir1 was enriched at all centromeres tested, which included CEN1-CEN4, CEN11, and CEN16 (Fig. 1A; data not shown). Control immunoprecipitations confirmed that the enrichment of Sir1-HA at centromeres was dependent on both the presence of anti-HA antibody and the HA epitope on Sir1 (Fig. 1A, lanes 2,3). Furthermore, analysis of four negative control loci revealed that the association of CEN and HM loci with Sir1 was specific. For example, immunoprecipitation of Sir1-HA recovered only background levels of ACT1, subtelomeric regions on Chromosome VI, and the transcriptionally active MAT locus. These data were consistent with previous genetic experiments that demonstrated that the cryptic mating loci HMR and HML, but not telomeres or the MAT locus, are subject to transcriptional control by Sir1 (Loo and Rine 1995). Although Sir2-4 ChIP eluates contained significant amounts of the HMR-E silencer element and telomere-proximal DNA, levels of CEN DNA were quantitatively comparable to MAT and ACT1 negative control loci (Fig. 1A,B). All four Sir proteins were enriched ∼13- to 17-fold at the HMR-E silencer element relative to MAT, but only Sir1 was enriched ∼10- to 14-fold at six centromeres tested (Fig. 1A,B).
Figure 1.
Association of Sir1 with centromeric chromatin. (A) Presence of Sir1, but not Sir2-4, at centromeric loci. Formaldehyde cross-linked chromatin prepared from yeast strains CFY416 (SIR1-HA; lanes 2,4) and PKY346 (SIR1; lane 3) was immunoprecipitated in the presence of monoclonal 12CA5 anti-HA antibody (lanes 3-4) or mock treated (lane 2). Chromatin from yeast strains PKY090 (wild-type SIR2, SIR3, SIR4), PKY3342 (sir2Δ), PKY3343 (sir3Δ), and PKY3344 (sir4Δ) was mock treated (lane 6), or immunoprecipitated with antibodies to Sir2 (lanes 7,8), Sir3 (lanes 9,10), and Sir4(lanes 11,12). PCR was performed to visualize recovery of the core centromeric regions of CEN3, CEN11, CEN16, the HMR-E silencer, MAT, ACT1, and two subtelomeric sequences on the right arm of Chromosome VI (TEL). Total chromatin was titrated to determine the linear range of the PCR (data not shown); a 1:32 dilution that falls within this range is shown in lanes 1 and 5. (B) Quantitation of Sir1-4 chromatin immunoprecipitation experiments. Chromatin immunoprecipitations (n = 3 for each genotype) were performed as described in A. The signal strength of PCR products was measured using Quantity One software (Bio-Rad) and used to calculate the percent recovery of HMR-E, MAT, CEN1-4, CEN11, and CEN16 PCR products. The average percent recovery is expressed as fold enrichment relative to the MAT control locus. (C) Distribution of Sir1 across the CEN3 region. Chromatin was prepared from yeast strain CFY416 (SIR1-HA), and PCR was performed as in A. Anti-HA-precipitated chromatin, mock-precipitated chromatin, and total chromatin was analyzed for recovery of fragments at or flanking the core centromeric region of CEN3 as indicated on the diagram (not to scale). (D) Sir1 colocalizes with centromere protein Ndc10. Yeast strain PKY2648 (SIR1-HA, NDC10-GFP) was prepared for indirect immunofluorescence analysis as described (Loidl et al. 1998). Spread nuclei were stained with anti-HA (red), anti-GFP (green) antibodies, and DAPI (blue). Colocalization of Sir1 and Ndc10 is indicated by the yellow staining in the merged image.
The distribution of individual centromere-associated proteins varies across centromere-proximal chromatin. For example, cohesin subunits (Megee et al. 1999; Tanaka et al. 1999) and Cac1 (Sharp et al. 2002) are distributed across ∼10 kb of centromeric chromatin; in contrast, the centromere-specific histone Cse4 and the CENP-C homolog Mif2 are tightly localized over only the 125-bp core centromeric DNA (Meluh et al. 1998). To determine whether the distribution of Sir1 was similar to either of these patterns, ChIP experiments were used to examine the 10-kb flanking CENIII. Like Cse4, Sir1 was tightly localized over core centromeric DNA (Fig. 1C), suggesting that specific protein-protein interactions at the kinetochore resulted in recruitment of Sir1.
Cytological analysis was used as a second test of Sir1 centromere localization. Budding yeast centromeres are clustered together and generally appear as one or two foci depending on the stage of the cell cycle (Guacci et al. 1997). The Ndc10 protein, an essential component of kinetochores, displays a similar clustered localization pattern. In chromosome spread preparations, we observed that brightly staining Sir1 foci colocalized with Ndc10 (Fig. 1D). Sir1 was also present in multiple weakly staining foci that did not colocalize with Ndc10, demonstrating that the chromatin-associated pool of Sir1 included, but was not limited to, centromeric domains.
Because the function of the kinetochore is to ensure chromosome segregation during cell division, the centromeric localization of Sir1 suggested an undiscovered role for Sir1 in chromosome stability. To test this idea, we measured the rates of chromosome missegregation per cell division (Shero et al. 1991; Sharp et al. 2002) in wild-type and sir1Δ cells. sir1Δ cells displayed an average 21-fold increase in the rate of chromosome loss compared with wild-type cells (wild-type: 2.0 × 10-5 ± 0.44 × 10-5; sir1Δ: 4.1 × 10-4 ± 1.5 × 10-4; n = 3 experiments). In contrast, in sir4Δ cells, the rate of chromosome loss was equivalent to that of wild type, consistent with earlier studies (sir4Δ: 2.2 × 10-5 ± 2.2 × 10-6; n = 3 experiments; Palladino et al. 1993). These data demonstrate that defects in HM silencing per se do not cause chromosome missegregation, and that sir1Δ mutants display a chromosome loss phenotype not observed in another sir mutant. Furthermore, these data are consistent with the localization of Sir1 but not the other Sir proteins to centromeric chromatin (Fig. 1A,B), and support a role for Sir1 at centromeric chromatin that is distinct from its role in recruiting the Sir2-4 proteins to HM silencer elements.
Additional criteria were used to test whether SIR1 interacted genetically with known kinetochore components. First, SIR1 was found to interact with a gene essential for chromosome segregation. Specifically, deletion of SIR1 increased the maximum permissive temperature of cells carrying a mutation in the centromere-specific histone Cse4(Supplementary Fig. 1, cse4-107 allele; Chen et al. 2000), and caused greater resistance of cse4-107 cells to the microtubule-depolymerizing drug benomyl. Second, genetic interactions between a nonessential kinetochore component and SIR1 were also observed. Cells lacking the outer kinetochore protein Mcm19 are sensitive to benomyl, and this sensitivity was partially suppressed by deletion of SIR1 (Supplementary Fig. 1; Ghosh et al. 2001). Third, deletion of the spindle checkpoint gene MAD2 in sir1Δ cells resulted in greatly increased chromosome loss rates. Consistent with earlier studies, mad2Δ cells caused a 10-fold increase in chromosome loss, but chromosome loss rates of sir1Δ mad2Δ cells were increased 270-fold relative to the wild-type control (wild-type: 2.0 × 10-5 ± 0.44 × 10-5; mad2Δ: 2.1 × 10-4 ± 3.5 × 10-5; sir1Δ mad2Δ: 5.4× 10-3 ± 1.3 × 10-3; n = 3 experiments; Li and Murray 1991; Warren et al. 2002). These data indicate that both SIR1 and MAD2 promote chromosome stability in unperturbed mitotic cell divisions. Together, these genetic interaction data demonstrate that Sir1 affects the fidelity of chromosome segregation.
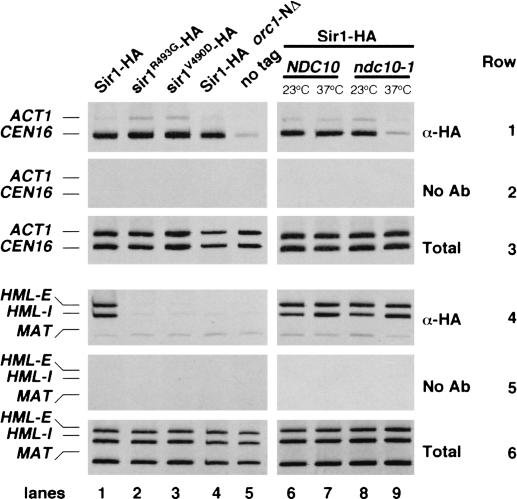
Recruitment of Sir1 to HM silencers occurs through a direct interaction between Sir1 and the N-terminal region of the Orc1 subunit of ORC (Triolo and Sternglanz 1996; Fox et al. 1997; Zhang et al. 2002). Mutant forms of Sir1 (termed “srd”, silencer recognition defective) that fail to interact with Orc1 can still recruit the other Sir proteins for silencing, but must be artificially tethered to an HM silencer to do so (Gardner et al. 1999; Gardner and Fox 2001). To test whether ORC also recruited Sir1 to centromeric regions, we performed ChIP experiments with cells containing HA-tagged wild-type Sir1 or two altered Sir1srd proteins that contain single amino acid changes (Fig. 2, lanes 1-3). We observed that Sir1srd proteins did not associate efficiently with HML silencers, consistent with their previously characterized defects in ORC binding. In contrast, localization of Sir1srd proteins was unperturbed at both CEN16 and CEN3 (Fig. 2; data not shown). Identical results were achieved when the localization of wild-type Sir1 was analyzed in a strain containing an N-terminally truncated Orc1 protein that is unable to bind Sir1 (Fig. 2, lane 4; Triolo and Sternglanz 1996; Gardner et al. 1999). Therefore, the localization of Sir1 to centromeres was independent of ORC, suggesting that a previously undetected set of interactions was required for this localization.
Figure 2.
Sir1 association with centromeric chromatin requires Ndc10, but is independent of the Orc1 N terminus. Chromatin immunoprecipitation was performed as described in Figure 1. Total (rows 3,6), mock immunoprecipitations (rows 2,5), and anti-HA immunoprecipitations (rows 1,4) were then tested for the presence of CEN16, ACT1, HML-E, HML-I, and MAT DNA. Yeast strains were PKY2586 (SIR1-HA; lanes 1,6,7), CFY687 (sir1R493G-HA; lane 2), CFY689 (sir1V490D-HA; lane 3), CFY1392 (SIR1-HA, orc1-NΔ; lane 4), CFY345 (SIR1; lane 5), and PKY2578 (SIR1-HA, ndc10-1; lanes 8,9).
Next, we tested whether the centromeric association of Sir1 depended on intact kinetochore structure. Ndc10 acts at an early step in kinetochore assembly, because ndc10 mutations abolish the centromere localization of every other kinetochore protein tested to date (Biggins and Walczak 2003). Therefore, Sir1 ChIP experiments were performed with strains containing either the temperature-sensitive ndc10-1 mutation or the wild-type NDC10 allele (Fig. 2, lanes 6-9). At the restrictive temperature, centromere association of Sir1 was sharply decreased in ndc10-1 cells, yet recruitment of Sir1 to the HML silencer was unchanged. In contrast, centromere association of Sir1 was independent of the outer kinetochore protein Mcm19 (data not shown). Therefore, the association of Sir1 with centromeric chromatin displayed distinct requirements for the integrity of inner and outer kinetochore structure. Moreover, because Sir1 recruitment responded differently to NDC10 and ORC1 perturbations in a locus-specific manner, the targeting of Sir1 to heterochromatin and centromeric chromatin occurred via independent interactions.
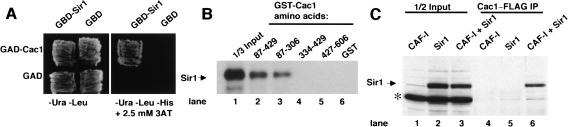
In mammalian cells, a direct link between nucleosome assembly and heterochromatic gene silencing has been established: CAF-I is recruited to pericentric heterochromatin by direct interaction between the large subunit of CAF-I and HP1 (Murzina et al. 1999). The presence of both CAF-I (Sharp et al. 2002) and the silencing protein Sir1 (Figs. 1, 2) at budding yeast centromeres suggested that direct interactions might exist between CAF-I and Sir1 in yeast. Indeed, we detected a physical interaction between Sir1 and the large subunit of CAF-I (Cac1) in multiple assays. First, this interaction was observed in vivo using a yeast two-hybrid assay (Fig. 3A). To determine whether Sir1 and Cac1 interact directly, the interaction was tested in a GST coprecipitation assay. In vitro translated Sir1 coprecipitated with a GST-Cac1 fusion protein (Fig. 3B) and required amino acids 87-306 on GST-Cac1. Additionally, Cac1 and Sir1 interacted when expressed in baculovirus-infected insect cells (Fig. 3C). Coimmunoprecipitation of Sir1 with CAF-I occurred specifically in extracts prepared from cells expressing both Sir1 and CAF-I, and was antibody-dependent. Together, these data demonstrated a direct interaction between Sir1 and the large subunit of CAF-I.
Figure 3.
Biochemical interaction between Sir1 and Cac1. (A) Two-hybrid interaction in yeast. Yeast strain PJ69-4a (James et al. 1996) expressed the indicated activation domain (GAD) or DNA-binding (GBD) fusion proteins. The GAD-Cac1 plasmid encodes amino acids 217-606 of Cac1, and was recovered in a two-hybrid screen using the full-length GBD-Sir1 plasmid. Cells were grown either on media lacking uracil and leucine (-Ura -Leu) to select for the two plasmids or on -Ura -Leu media also lacking histidine and containing 2.5 mM 3-aminotriazole (-His + 3AT) to score for activation of the HIS3 reporter gene, indicating interaction between the fusion proteins. (B) For direct interaction in vitro, 2.5 μg of unfused GST (lane 6) or GST-Cac1 fusion proteins [Krawitz et al. 2002; Cac1 amino acids 87-429 (lane 2), 87-306 (lane 3), 334-429 (lane 4), or 429-606 (lane 5)] prebound to 15 μL of Glutathione Agarose (Sigma) were incubated with 6 μL of 35S-labeled in vitro translated Sir1 (Novagen). Reactions were rotated at 4°C for 1 h prior to three 1-mL washes with Buffer A (25 mM Tris-HCl at pH 7.5, 1 mM EDTA, 0.1% NP-40) plus 250 mM NaCl. Bound proteins were eluted with SDS sample buffer, separated by SDS-PAGE, and detected by autoradiography. Lane 1 contains 2 μL of in vitro translated Sir1 loaded directly onto the gel. (C) Interaction between CAF-I and Sir1 in cell extracts. Overproduction of CAF-I including a Flag-tagged Cac1 subunit and Sir1 in SF9 cells was performed as described (Sharp et al. 2001). Five microliters of nuclear extract containing CAF-I (lane 4), Sir1 (lane 5), or both CAF-I and Sir1 (lane 6) were incubated with 95 μL of Buffer A plus 50 mM NaCl at 4°C for 2 h in the presence of anti-Flag antibodies (Sigma) cross-linked to Protein A-Sepharose. Samples were washed three times with 1 mL of Buffer A plus 500 mM NaCl. Bound proteins were eluted with SDS sample buffer, separated by SDS-PAGE, and analyzed by immunoblotting with a polyclonal anti-Sir1 antibody. Also, 2.5 μL of total nuclear extract from cells expressing CAF-I alone (lane 1), Sir1 alone (lane 2), or both CAF-I and Sir1 (lane 3) were analyzed on the same gel. The asterisk indicates a cross-reacting protein present in the crude cell lysates.
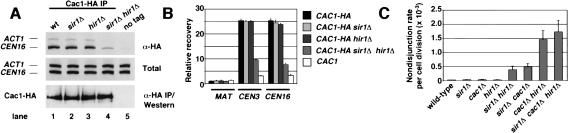
The chromatin-associated pool of CAF-I is enriched at centromeric regions (Fig. 4A; Sharp et al. 2002). The physical interaction between Cac1 and Sir1 suggested that Sir1 might contribute to the centromere association of Cac1 or vice versa. To test this, ChIP experiments were performed in cells lacking the SIR1 gene. Although sir1Δ cells displayed wild-type levels of Cac1 centromere association, a marked reduction was observed in cells containing both sir1Δ and hir1Δ gene deletions (Fig. 4A). In contrast, cac1Δ and hir1Δ mutations alone or in combination had no effect on the association of Sir1 with either CEN16 or HM loci (data not shown). Western blot analysis of Cac1-HA immunoprecipitations showed similar expression levels and immunoprecipitation efficiency from all strains tested (Fig. 4A, bottom panel), indicating that changes observed in the ChIP assay were not attributable to changes in cellular Cac1 protein levels. Quantitation of Cac1-HA ChIP experiments demonstrated an approximately threefold reduction of Cac1 association with CEN3 and CEN16 in sir1Δ hir1Δ cells compared with the wild-type cells (Fig. 4B). These data suggest that the stable association of Cac1 with centromeric loci is mediated by a complex network of protein interactions that includes both Sir1 and Hir1, and possibly other factors yet to be discovered.
Figure 4.
Cac1, Hir1, and Sir1 make overlapping contributions to centromere structure and function. (A) Cac1 association with the core centromeric region requires Sir1 and Hir1. Chromatin immunoprecipitations were performed on yeast strains YB703 (CAC1-HA; lane 1), PKY2615 (CAC1-HA, sir1Δ; lane 2), PKY2617 (CAC1-HA, hir1Δ; lane 3), PKY2619 (CAC1-HA, sir1Δ, hir1Δ; lane 4), and PKY346 (CAC1; lane 5). Total, mock (data not shown), and anti-HA immunoprecipitations were then tested for the presence of CEN16 and ACT1 DNA. A Western blot was performed to compare immunoprecipitation of Cac1-HA from the indicated strains (bottom panel). (B) Quantitation of Cac1-HA chromatin immunoprecipitation experiments. Chromatin immunoprecipitations (n = 3 for each genotype) were analyzed as described in Figure 1B for the MAT, CEN3, and CEN16 loci. (C) SIR1, HIR1, and CAC1 together prevent chromosomal nondisjunction during mitotic cell divisions. Nondisjunction rates per cell division were quantified in haploid yeast strains containing a nonessential chromosome fragment by the half-sector colony color assay as described (Shero et al. 1991; Sharp et al. 2002). Yeast strains are PKY847 (wild-type), PKY2609 (sir1Δ), PKY2216 (cac1Δ), PKY865 (hir1Δ), PKY2655 (sir1Δ hir1Δ), PKY 2610 (sir1Δ cac1Δ), PKY 2217 (cac1Δ hir1Δ), and PKY2656 (sir1Δ cac1Δ hir1Δ). Three experiments for each genotype (except PKY2656, n = 4) were performed; the average chromosome nondisjunction rate per cell division and standard deviation (error bars) are shown.
The CAF-I and Hir nucleosome assembly proteins have overlapping roles in maintaining high-fidelity chromosome segregation: cells simultaneously lacking CAC and HIR genes display greatly increased rates of chromosomal nondisjunction (Fig. 4C; Sharp et al. 2002). We hypothesized that chromosome stability defects caused by loss of Sir1 would also be exacerbated upon deletion of CAC or HIR genes. To test this, chromosome missegregation rates were compared in cells with all possible combinations of sir1Δ, hir1Δ, and cac1Δ gene deletions (Fig. 4C). Consistent with previous data, cac1Δ hir1Δ cells displayed a 75-fold increase in chromosomal nondisjunction rates. Furthermore, synergistic increases in nondisjunction rates were observed in sir1Δ hir1Δ and sir1Δ cac1Δ cells (20-fold and 24-fold, respectively) compared with either wild-type or single mutant cells. Additionally, chromosome nondisjunction rates in sir1Δ cac1Δ hir1Δ triple mutants were statistically indistinguishable from nondisjunction rates of cac1Δ hir1Δ double-mutant cells. Together, these data demonstrate that Sir1 functions in a partially overlapping manner with CAF-I and the Hir proteins to maintain chromosome stability.
In this report, we have demonstrated a novel role for the Sir1 protein in promoting centromeric chromatin structure and function. Sir1 localizes to budding yeast kinetochores and helps maintain mitotic chromosome stability by a mechanism that is independent of interactions known to be critical for forming silent heterochromatin at HM loci. These data are consistent with the finding that budding yeast centromeres do not exert position-dependent silencing on neighboring genes (K. Bloom, pers. comm.), and support the argument that the role of Sir1 in promoting formation of specialized chromatin structures is not limited to the recruitment of Sir2-4 proteins.
Instead, we suggest that the activity of Sir1 is regulated by chromosomal context. In this view, protein partners specific to the kinetochore-associated pool of Sir1 form a specialized chromatin structure distinct from that found at heterochromatin. For example, Sir1 copurifies with the kinetochore protein Mcm19 (N. Krogan and J. Greenblatt, pers. comm.), suggesting that Sir1 may be a peripheral subunit of the outer kinetochore. This concept of chromatin context-dependent function is reinforced by several examples in the literature. First, a budding yeast origin of replication acts as a silencer element only when juxtaposed closely in sequence space to Rap1- and Abf1-binding sites (McNally and Rine 1991; Loo and Rine 1995). Second, the methylation of histone H3 Lys 4 is a modification closely associated with active gene transcription, yet is also required for the efficient silencing of RNA polymerase II transcription within rDNA sequences (Briggs et al. 2001; Bryk et al. 2002; Santos-Rosa et al. 2002). Third, the heterochromatin protein HP1 also contributes to inducible gene expression at some euchromatic loci (Piacentini et al. 2003). Identification of Sir1-containing complexes unique to the kinetochore will be an important step in understanding the centromeric function of Sir1.
Previously, we demonstrated that CAF-I and Hir proteins are critical for maintaining the integrity of specialized chromatin structures found at budding yeast kinetochores (Sharp et al. 2002). Here, our data demonstrate a direct physical interaction between the Sir1 protein and the large subunit of Chromatin Assembly Factor-I. Importantly, we found that Sir1 and Hir1 acted together to maintain normal levels of CAF-I at centromeric regions. Also, we determined that Sir1, Hir1, and CAF-I all have overlapping roles in preventing chromosome nondisjunction during mitosis. We propose that an important aspect of the biology of Sir1 at centromeric chromatin is to participate in a structure that acts to recruit CAF-I. We note that although budding yeast has no HP1 homolog, we have discovered that this organism uses an alternative heterochromatin protein, Sir1, to help maintain CAF-I at centromeres. Therefore, diverse eukaryotic organisms use silencing proteins to recruit histone deposition factors to centromeres to support kinetochore function.
Materials and methods
See Supplemental Material for Material and Methods.
Acknowledgments
We thank J. Rine, Z. Zhang, and B. Stillman for strains; J. Rine for antibodies; and members of the Kaufman and Rine laboratories for comments on the manuscript. This work was supported by grants from the NIH, NSF, and the DOE to P.D.K. and from the NIH to C.A.F. Correspondence and requests for materials should be addressed to P.D.K. (pdkaufman@lbl.gov).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1131103.
Supplemental material is available at http://www.genesdev.org.
References
- Bernard P., Maura, J.F., Partridge, J.F., Genier, S., Javerzat, J.P., and Allshire, R.C. 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294: 2539-2542. [DOI] [PubMed] [Google Scholar]
- Biggins S. and Walczak, C.E. 2003. Captivating capture: How microtubules attach to kinetochores. Curr. Biol. 13: R449-R460. [DOI] [PubMed] [Google Scholar]
- Briggs S.D., Bryk, M., Strahl, B.D., Cheung, W.L., Davie, J.K., Dent, S.Y., Winston, F., and Allis, C.D. 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes & Dev. 15: 3286-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk M., Briggs, S.D., Strahl, B.D., Curcio, M.J., Allis, C.D., and Winston, F. 2002. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 12: 165-170. [DOI] [PubMed] [Google Scholar]
- Chen Y., Baker, R.E., Keith, K.C., Harris, K., Stoler, S., and Fitzgerald-Hayes, M. 2000. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol. Cell. Biol. 20: 7037-7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox C.A., Ehrenhofer-Murray, A.E., Loo, S., and Rine, J. 1997. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science 276: 1547-1551. [DOI] [PubMed] [Google Scholar]
- Gardner K.A. and Fox, C.A. 2001. The Sir1 protein's association with a silenced chromosome domain. Genes & Dev. 15: 147-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner K.A., Rine, J., and Fox, C.A. 1999. A region of the Sir1 protein dedicated to recognition of a silencer and required for interaction with the Orc1 protein in Saccharomyces cerevisiae. Genetics 151: 31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S.K., Poddar, A., Hajra, S., Sanyal, K., and Sinha, P. 2001. The IML3/MCM19 gene of Saccharomyces cerevisiae is required for a kinetochore-related process during chromosome segregation. Mol. Genet. Genomics 265: 249-257. [DOI] [PubMed] [Google Scholar]
- Guacci V., Hogan, E., and Koshland, D. 1997. Centromere position in budding yeast: Evidence for anaphase A. Mol. Biol. Cell 8: 957-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay, J., and Craig, E.A. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen G.H. and Allshire, R.C. 1997. The case for epigenetic effects on centromere identity and function. Trends Genet. 13: 489-496. [DOI] [PubMed] [Google Scholar]
- Kaufman P.D., Cohen, J.L., and Osley, M.A. 1998. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of Chromatin Assembly Factor I. Mol. Cell. Biol. 18: 4793-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawitz D.C., Kama, T., and Kaufman, P.D. 2002. Chromatin Assembly Factor-I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol. Cell. Biol. 22: 614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. and Murray, A.W. 1991. Feedback control of mitosis in budding yeast. Cell 66: 519-531. [DOI] [PubMed] [Google Scholar]
- Loidl J., Klein, F., and Engebrecht, J. 1998. Genetic and morphological approaches for the analysis of meiotic chromosomes in yeast. Methods Cell. Biol. 53: 257-285. [DOI] [PubMed] [Google Scholar]
- Loo S. and Rine, J. 1995. Silencing and heritable domains of gene expression. Ann. Rev. Cell Dev. Biol. 11: 519-548. [DOI] [PubMed] [Google Scholar]
- McNally F.J. and Rine, J. 1991. A synthetic silencer mediates SIR-dependent functions in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 5648-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee P.C., Mistrot, C., Guacci, V., and Koshland, D. 1999. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol. Cell 4: 445-450. [DOI] [PubMed] [Google Scholar]
- Meluh P. B., Yang, P., Glowczewski, L., Koshland, D., and Smith, M.M. 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94: 607-613. [DOI] [PubMed] [Google Scholar]
- Moazed D. 2001. Common themes in mechanisms of gene silencing. Mol. Cell 8: 489-498. [DOI] [PubMed] [Google Scholar]
- Murzina N., Verreault, A., Laue, E., and Stillman, B. 1999. Heterochromatin dynamics in mouse cells: Interaction between chromatin assembly factor 1 and HP1 proteins. Mol. Cell 4: 529-540. [DOI] [PubMed] [Google Scholar]
- Palladino F., Laroche, T., Gilson, E., Axelrod, A., Pillus, L., and Gasser, S.M. 1993. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell 75: 543-555. [DOI] [PubMed] [Google Scholar]
- Piacentini L., Fanti, L., Berloco, M., Perrini, B., and Pimpinelli, S. 2003. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J. Cell Biol. 161: 707-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider, R., Bannister, A.J., Sherriff, J., Bernstein, B.E., Emre, N.C., Schreiber, S.L., Mellor, J., and Kouzarides, T. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419: 407-411. [DOI] [PubMed] [Google Scholar]
- Sharp J.A., Fouts, E.T., Krawitz, D.C., and Kaufman, P.D. 2001. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 11: 463-473. [DOI] [PubMed] [Google Scholar]
- Sharp J.A., Franco, A.A., Osley, M.A., and Kaufman, P.D. 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes & Dev. 16: 85-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shero J.H., Koval, M., Spencer, F., Palmer, R.E., Hieter, P., and Koshland, D. 1991. Analysis of chromosome segregation in Saccharomyces cerevisiae. Methods Enzymol. 194: 749-773. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Cosma, M.P., Wirth, K., and Nasmyth, K. 1999. Identification of cohesin association sites at centromeres and along chromosome arms. Cell 98: 847-858. [DOI] [PubMed] [Google Scholar]
- Triolo T. and Sternglanz, R. 1996. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature 381: 251-253. [DOI] [PubMed] [Google Scholar]
- Warren C.D., Brady, D.M., Johnston, R.C., Hanna, J.S., Hardwick, K.G., and Spencer, F.A. 2002. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol. Biol. Cell 13: 3029-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Hayashi, M.K., Merkel, O., Stillman, B., and Xu, R.M. 2002. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J. 21: 4600-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]