Abstract
RyhB is a small antisense regulatory RNA that is repressed by the Fur repressor and negatively regulates at least six mRNAs encoding Fe-binding or Fe-storage proteins in Escherichia coli. When Fe is limiting, RyhB levels rise, and target mRNAs are rapidly degraded. RyhB is very stable when measured after treatment of cells with the transcription inhibitor rifampicin, but is unstable when overall mRNA transcription continues. We propose that RyhB turnover is coupled to and dependent on pairing with the target mRNAs. Degradation of both mRNA targets and RyhB is dependent on RNase E and is slowed in degradosome mutants. RyhB requires the RNA chaperone Hfq. In the absence of Hfq, RyhB is unstable, even when general transcription is inhibited; degradation is dependent upon RNase E. Hfq and RNase E bind similar sites on the RNA; pairing may allow loss of Hfq and access by RNase E. Two other Hfq-dependent small RNAs, DsrA and OxyS, are also stable when overall transcription is off, and unstable when it is not, suggesting that they, too, are degraded when their target mRNAs are available for pairing. Thus, this large class of regulatory RNAs share an unexpected intrinsic mechanism for shutting off their action.
Keywords: RyhB, Fur, RNase E, RNase III, Hfq, DsrA
RyhB is a small regulatory RNA that modulates multiple mRNAs involved in iron metabolism in Escherichia coli (Massé and Gottesman 2002). Fe is an important nutrient involved in several cellular processes, including respiration, the citric acid cycle, DNA synthesis, elimination of H2O2 and O2-, photosynthesis and others (for review, see Andrews 1998). Accessible iron is very scarce in many environments, particularly for bacteria in mammalian hosts (Ratledge and Dover 2000). Under these conditions, the cell needs ways for accessing both extracellular and intracellular stores of Fe. However, it also needs ways of avoiding too much Fe. Excess Fe, in the presence of oxygen, generates toxic products via the Fenton reaction. Normal aerobic metabolism will generate mildly toxic products such as O2- and H2O2. However, the presence of Fe will catalyze the production of the highly reactive and damaging hydroxyl radical (OH•). To circumvent this problem, a tight regulation of Fe concentration is essential for the cell.
RyhB provides part of the answer to this conflicting need for storing Fe when abundant and using it efficiently when it is scarce. In conditions of Fe abundance, the activated Fur repressor binds to the promoter region of RyhB and shuts down its transcription. In these conditions, the RyhB-targeted messenger RNAs are expressed. These include the genes for tricarboxylic acid cycle enzymes sdhCDAB, fumA, and acnA, the superoxide dismutase gene, sodB, all encoding Fe-S proteins, and two genes encoding Fe storage proteins, ftnA and bfr. In particular, the iron-storage proteins ferritin and bacterioferritin (encoded by ftnA and bfr, respectively) help the cell keep Fe in a soluble, less toxic, and more accessible form, providing the metal when it is needed (Andrews et al. 1992). Ferritins can be found in archeabacteria, eubacteria, and eukaryotes, implying a fundamental role in Fe metabolism. When Fe is depleted, Fur repression is inactivated and RyhB increases rapidly. It pairs with its target mRNAs and causes their disappearance. By reducing the synthesis of these Fe-binding and storage proteins, limiting Fe is available for essential cellular processes. Thus, switching on and switching off Fe entry and storage mechanisms in response to changes in iron levels is critical for cell growth.
RyhB is a member of a major family of small regulatory RNAs in bacteria, those that bind to the RNA chaperone Hfq and require Hfq for activity. Members of this family that have been studied all act by pairing to target messenger RNAs, perturbing their stability and/or translation. More than 20 small RNAs of this class exist in E. coli (Zhang et al. 2003). We report here on the mechanism of RyhB action in helping reorient intracellular Fe use when Fe is limiting, and the reversal of this effect when Fe is abundant. Our results suggest that target mRNAs are rapidly degraded in a RyhB-dependent manner and, more surprisingly, that RyhB itself is also degraded in the process; all of these steps require RNase E. The stages in small RNA action described here apply, at least in part, to other small RNAs of this family as well.
Results
RyhB stimulates degradation of a full-length target mRNA
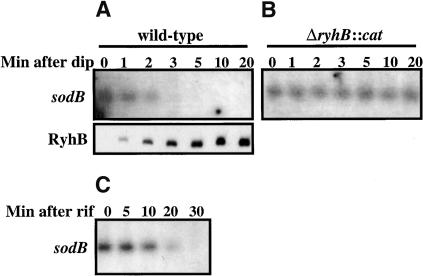
We demonstrated previously that expression of the small regulatory RNA RyhB specifically down-regulates the steady-state level of acnA, fumA, sdhCDAB, bfr, ftn, and sodB target mRNAs (Massé and Gottesman 2002). The ryhB gene is normally repressed by the Fur protein in the presence of Fe2+. It is derepressed in a fur- background or upon addition of the Fe chelator, 2,2′-dipyridyl; the resulting strong expression of RyhB correlates with a rapid decrease in the level of target mRNAs. These experiments did not address whether RyhB was blocking transcription initiation or elongation, or stimulating degradation of the mRNA during or after transcription. To monitor the kinetics of this process, we measured the small RNA RyhB and sodB mRNA levels, by Northern blot, from total RNA isolated from cells as a function of time after the addition of 2,2′-dipyridyl. In Figure 1A, the lower panel shows the rapidly increasing level of RyhB over time in a wild-type strain. The upper panel shows that the sodB steady-state level drops rapidly as RyhB increases. Most of the sodB mRNA has disappeared after 3 min of RyhB expression. In Figure 1B, the same experiment in a ΔryhB::cat background shows that the sodB mRNA level remains unchanged even 20 min after 2,2′-dipyridyl addition. It is clear from these results that, when RyhB is expressed at high levels, only 3 min are sufficient to completely eliminate the target sodB message. The half-life of the sodB message (in the absence of high levels of RyhB) was determined by treating cells growing in LB (in the absence of 2,2′-dipyridyl) with rifampicin; this is an unusually stable mRNA, with a half-life of ∼6 min (Fig. 1C). Because the half-life of the message in the absence of RyhB is long, we can conclude that the total disappearance of all sodB mRNA within 3 min after induction of RyhB means that the small regulatory RNA leads to destruction of full-length mRNA, and, therefore, does not act at the level of transcription initiation or elongation. This is consistent with previous work from Dubrac and Touati (2000), who also found a long half-life for sodB mRNA in wild-type, and a shorter half-life for the message in a fur- mutant, in which RyhB is expressed at high levels.
Figure 1.
Degradation of full-length sodB mRNA. (A) Cells (EM1055, wild-type) were grown at 37°C to an O.D.600 of 0.4. 2,2′-dipyridyl (dip) was added at 250 μM and samples taken as indicated and RNA extracted as described in Materials and Methods. Northern blots for sodB and RyhB were done as described previously (Massé and Gottesman 2002). (B) ryhB- cells (EM1238) were grown and processed as in A. (C) Half-life of sodB message. EM1055 cells were grown as in A, but without the addition of 2,2′-dipyridyl (dip). Rifampicin was added at 250 μg/mL at time 0, and samples taken as indicated.
RNase dependence of RyhB-mediated mRNA degradation
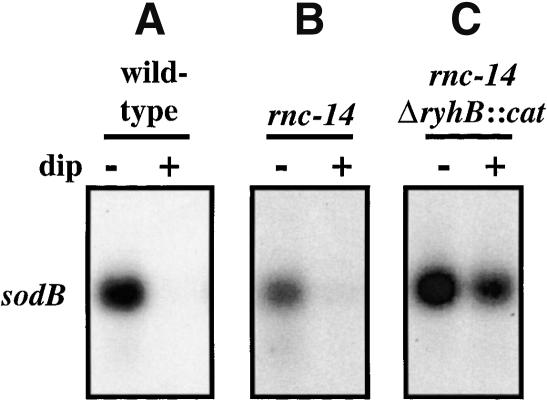
Dubrac and Touati (2002) demonstrated that the regulation of sodB-lacZ fusions by Fur was dependent upon both ribonucleases III and E. Our results with sodB mRNA strongly suggested the involvement of a ribonuclease in the RyhB-mediated loss of specific mRNAs. To test this, we first tested the effect of inactivating RNase III (encoded by rnc). This endoribonuclease specifically cleaves double-stranded RNA and is involved in the stability of mRNAs in E. coli (for review, see Grunberg-Manago 1999). We introduced the rnc-14::Tn10-null mutation into the isogenic wild-type and ΔryhB::cat mutant strains. The resulting strains were grown to exponential phase, RyhB was induced with 2,2′ dipyridyl, and the level of the sodB mRNA determined (Fig. 2). Disappearance of the message was complete by 15 min in both the wild-type and rnc-14::Tn10 (RNase III-) mutant (Fig. 2, cf. A and B), indicating that RNase III is not required for RyhB-mediated sodB mRNA degradation. This experiment does not rule out an effect of RNase III on the kinetics of sodB decay.
Figure 2.
RNase III is not required for sodB degradation. Cells were grown in LB at 37°C. At an O.D.600 of 0.3, the culture was split and 250 μM 2,2′-dipyridyl was added to one culture and growth was continued for 15 min. The total RNA was then extracted as described in Materials and Methods and sodB mRNA detected by Northern blot. (A) EM1055 (wild-type). (B) EM1321 (rnc-14::Tn10). (C) EM1322 (rnc-14::Tn10ΔryhB::cat).
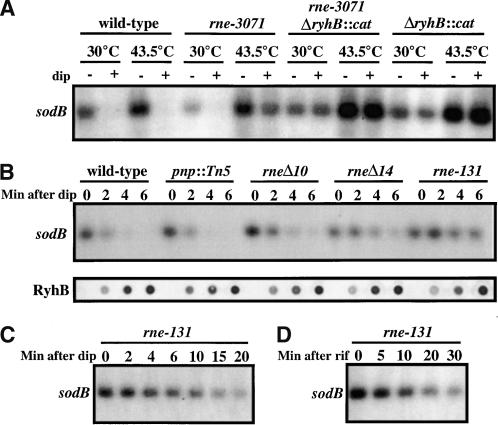
RNase E (encoded by rne) is another endoribonuclease, also involved in mRNA decay as well as processing of tRNAs and ribosomal RNAs. Because RNase E is essential, we used a temperature-sensitive allele of rne, rne-3071. The results (Fig. 3A) show that sodB is resistant to RyhB-mediated degradation when RNase E is inactivated by a 10-min treatment of the rne-3071 strain at 43.5°C. Similar experiments were carried out in which the sdh message was measured, with similar results (data not shown). Therefore, RNase E is required for the degradation of RyhB targets. Interestingly, this enzyme is a single-strand-specific endoribonuclease (see below), suggesting that the paired region of RyhB-sodB interaction is not the site of the initial cleavage. Parallel experiments with another temperature-sensitive rne mutant, rne-50, gave similar results (data not shown).
Figure 3.
RNase E is necessary for degradation of RyhB targets. (A) Strains were grown at 30°C to an O.D.600 of 0.4, a portion of the culture was shifted to 43.5°C for 10 min, and 250 μM 2,2′-dipyridyl was added to a portion of the 30°C and a portion of the 43.5°C cultures. After 10 min, samples were harvested, and RNA extracted and analyzed as described in Materials and Methods. The strains used were EM1279 (wild-type), EM1277 (rne-3071), EM1280 (rne-3071 ΔryhB::cat), and EM1282 (ΔryhB::cat). (B) Strains were grown at 37°C to an O.D.600 of 0.4, induced for 10 min with 2,2′-dipyridyl, and samples removed and processed as in A. Strains used were EM1055 (wild-type), EM1368 (pnp::tn5), EM1375 (rneΔ10), EM1376 (rneΔ14), and EM1377 (rne-131). RyhB levels were determined by dot blot (Materials and Methods). (C) Experiment as in B. The strain used is EM1377 (rne-131), but samples removed up to 20 min after 2,2′-dipyridyl addition. (D) Cells (EM1377, rne-131) grown and treated with rifampicin as in Figure 1C.
Involvement of the degradosome
The N-terminal domain of RNase E, which contains the active site for endonuclease activity, is essential for the processing of tRNA and for cell viability. The C terminus acts as a scaffold, necessary for protein-protein interactions with polynucleotide phosphorylase (PNPase), an RNA helicase and enolase, a complex that has been called the degradosome (Carpousis et al. 1994, 1999; Py et al. 1994, 1996; Miczak et al. 1996). Deletions of portions or all of the C terminus are viable, process ribosomal RNA and tRNAs normally, but are slowed for degradation of some messages (Lopez et al. 1999; Ow et al. 2000; see below). We tested strains deleted for degradosome domains for the degradation of sodB mRNA after RyhB induction (Fig. 3B,C). The rneΔ10 and rneΔ14 mutants are deleted for the interaction site with PNPase and RNA helicase, respectively; rne-131 has a deletion of the complete C terminus of RNase E (Leroy et al. 2002). The three mutants tested, rneΔ10, rneΔ14, and rne-131, all show slower loss of sodB message, with the largest deletion, rne-131, showing the slowest disappearance of the message (Fig. 3B,C). In addition, we tested a strain mutant for one of the proteins that associate with RNase E in the degradosome, polynucleotide phosphorylase (pnp::Tn5). No detectable effect of the pnp mutant on sodB disappearance was detected. In all of these mutants, expression of the small RNA, determined by dot blot, was normal (Fig. 3B).
The degradosome mutant rne-131 was also tested for the degradation of sodB in the absence of high RyhB expression. In this experiment, rifampicin addition was used to stop new sodB synthesis, so the results are not strictly comparable with those in which sodB synthesis is not stopped but message is destroyed by RyhB as soon as it is made (Figs. 1A, 3B). In Figure 1C, we showed that the sodB message had a half-life of ∼6 min. In Figure 3D, the same experiment, done in an rne-131 mutant, gives a half-life estimate of 18 min. We conclude that there is an effect of the degradosome mutant on sodB mRNA turnover. The disappearance of sodB mRNA when RyhB is expressed at high levels occurs in less than 3 min in a rne+ strain (Figs. 1A, 3B) and takes more than 20 min (with an estimated half-life of 10 min) in an rne-131 strain (Fig. 3C). The degradosome thus participates in both RyhB-independent and RyhB-mediated sodB decay. It is not yet clear what component of the degradosome is important.
Turnover of RyhB
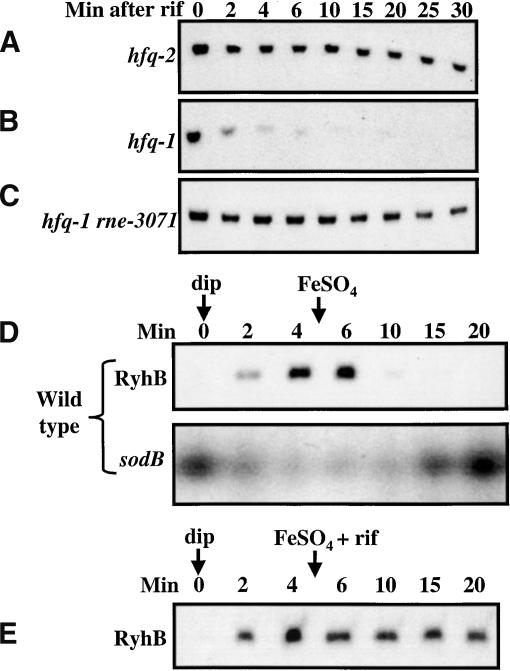
Many small regulatory RNAs, including DsrA, OxyS, and Spot 42, have been shown to have relatively long half-lives after rifampicin treatment (Altuvia et al. 1997; Majdalani et al. 2001; Repoila and Gottesman 2001; Møller et al. 2002a). It has also been reported that the Hfq protein is essential for the stability of some of these small RNAs (Sledjeski et al. 2001; Møller et al. 2002a). We measured the stability of RyhB in the presence and absence of Hfq. The cells were treated for 10 min with 2,2′-dipyridyl to induce RyhB, before rifampicin was added, and total RNA was then extracted. Aliquots of cells were taken at different intervals after addition of rifampicin (Fig. 4A). In the Hfq+ (hfq-2) strain, RyhB had a half-life of >30 min (Fig. 4A); in the Hfq- (hfq-1) background (Fig. 4B), the stability of RyhB is dramatically decreased. This is consistent with what has been observed for DsrA (Sledjeski et al. 2001), and suggests that Hfq is necessary for the stability of RyhB. The degradation of RyhB in the hfq mutant is dependent upon RNase E (Fig. 4C), whereas an rnc mutation had no effect on RyhB stability in the hfq mutant (data not shown).
Figure 4.
Northern blot analysis of RyhB stability. (A) Strains were grown at 37°C to an O.D.600 of 0.4., and 250 μM 2,2′-dipyridyl was added. After 10 min of incubation, rifampicin was added at 250 μg/mL, incubation continued, and samples were removed and RNA was extracted as decribed in Materials and Methods. The strain used is EM1266 (hfq-2; Hfq+ phenotype). (B) Experiment as in A. The strain used is EM1265 (hfq-1:Hfq- phenotype). (C) Strains were grown at 30°C to an O.D.600 of 0.4, then the culture was shifted to 43.5°C for 10 min, and 250 μM 2,2′-dipyridyl was added. After 10 min, samples were harvested and RNA extracted. The strain used is EM1342 (hfq-1rne-3071). (D) Strains were grown at 37°C to an O.D.600 of 0.4., and 250 μM 2,2′-dipyridyl was added. After 5 min of incubation, FeSO4 was added at 100 μM. Samples were removed at the time indicated, and RNA was extracted as decribed in Materials and Methods. The strain used is EM1055 (wild-type). Northern blot analysis for sodB was performed as described (Massé and Gottesman 2002). (E) As in D, except that rifampicin (250 μg/mL) was added simultaneously with FeSO4. The strain used is EM1055 (wild-type).
Interestingly, we noted a significant decrease in the RyhB level between the 0 min time point and 2 min that is not observable for time points taken after 2 min (Fig. 4A). It seemed possible that this decrease is due to mRNA targets of RyhB that are present at the start of the rifampicin treatment. If so, within a few minutes, any mRNA target would be degraded (Fig. 1) and no new ones can be transcribed. This suggested that it would be useful to study the turnover of RyhB under conditions in which the target mRNAs are still transcribed. The reversible and tight repression of RyhB by the Fur repressor and Fe provided a simple way to do this. Using a Fur-regulated lac fusion (to gene fhuF), we determined an Fe concentration sufficient to restore repression after the 2,2′-dipyridyl induction of the Fur regulon (data not shown). Cells were grown, RyhB synthesis was induced with chelator, and 5 min later, an excess of FeSO4 was added. Samples were removed at the times shown in the figure and processed to monitor RyhB levels (Fig. 4D). Under these conditions, in contrast to the stability of RyhB in Figure 4A, we find that RyhB disappears rapidly. Within 5 min after addition of FeSO4, it is barely detectable.
Although we interpret this result to mean that RyhB is unstable when its target mRNAs are available (being transcribed, and presumably pairing with RyhB), Fe could possibly have an effect on the stability of RyhB itself either directly or indirectly. We tested this possibility in two ways. In the first experiment, FeSO4 and rifampicin were added simultaneously; the rifampicin will stop the transcription of both RyhB and its targets. The results clearly show that RyhB becomes stable when FeSO4 and rifampicin are added simultaneously (Fig. 4E). Therefore, transcription is required for RyhB disappearance; FeSO4 is not sufficient.
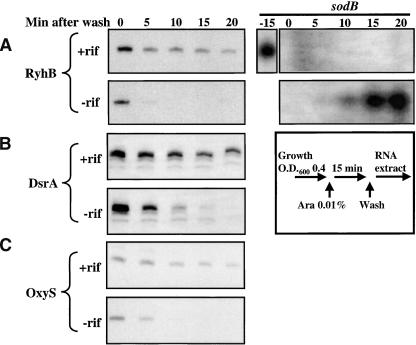
Nor is Fe, per se, necessary. In the second test, RyhB was expressed from a pBAD promoter, regulated by arabinose. RyhB was expressed by addition of arabinose to the culture, and cultures were then washed to remove arabinose and shut off new synthesis, and samples taken, either in the presence or absence of rifampicin (Fig. 5A). As for the results in Figure 4D and E, RyhB was very unstable when arabinose was removed, but became significantly more stable in the presence of rifampicin (Fig. 5A). In the rifampicin-treated cells, it is very apparent that there is an initial drop in RyhB levels (between 0 and 5 min). It seems likely that RyhB made from the pBAD promoter is at a lower level than that for the natural, fully induced promoter. As a result, RyhB levels are closer to the levels of mRNA targets in this case, and, if degradation requires pairing with the mRNA target, ∼50% of the RyhB is degraded. In cells expressing RyhB from the pBAD promoter, the sodB message is not detectable after 15 min of RyhB expression (Fig. 5A, right). Thus, even in cells in which the Fe levels are not changed and the Fur system is not perturbed, RyhB can act effectively. As RyhB disappears, the target sodB messenger reaccumulates (Figs. 4D, bottom, 5A, right). Within 15 min after addition of FeSO4, sodB mRNA levels have returned to the same level as before addition of 2,2′-dipyridyl. Thus, within 3 min, RyhB induction can target all sodB mRNAs for degradation (Fig. 1A); within 15 min after RyhB induction ceases, sodB mRNA levels are fully recovered. This provides a highly effective, reversible switch to regulate cellular Fe use.
Figure 5.
Alternative promoters for small RNA expression. (A) EM1059 + pBAD-ryhB. Cells were grown in LB with ampicillin (50 μg/mL) to an O.D.600 of 0.4, and arabinose added (0.01%) for 15 min. The culture was centrifuged briefly (5 min) to wash out the arabinose, and resuspended in an equal volume of LB with glucose (0.2%), either with or without rifampicin (250 μg/mL). Samples were removed at the intervals shown and RNA extracted and probed for RyhB. The -15 (min) panel shows sodB expression right before addition of arabinose. (B) Same as A but with pBAD-dsrA. The strain used is EM1063 + pBAD-dsrA. (C) Same as A but with pBAD-oxyS. The strain used is EM1059 + pBAD-oxyS.
RyhB decay is mediated by RNase E
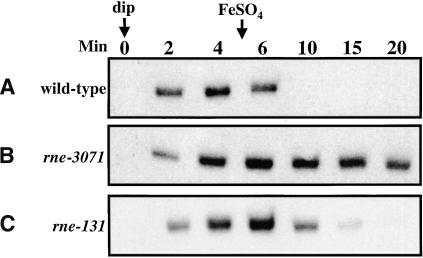
We showed above that RNase E is involved in the degradation of target mRNA after RyhB induction (Fig. 3); RNase E is also responsible for the degradation of RyhB in the absence of Hfq (Fig. 4C). We tested the rne mutant for the fate of RyhB in an experiment like that in Figure 4D. The rne-3071 strain was shifted to 43.5°C for 10 min, induced with 2,2′-dipyridyl for 5 min, and then treated with FeSO4 to repress RyhB expression. Aparallel experiment was done at 37°C with the rne-131 degradosome mutant. The results (Fig. 6) show that RyhB is quite stable after FeSO4 addition in the rne-3071 mutant (Fig. 6B), and degradation is slowed in the degradosome mutant (Fig. 6C). Degradation was unaffected in the rnc (RNase III) mutant (data not shown). Thus, degradation of RyhB is similar to that of the sodB mRNA in its dependence on RNase E and the degradosome.
Figure 6.
RNase dependence of RyhB degradation. (A) Strains were grown at 30°C to an O.D.600 of 0.4, then the culture was shifted to 43.5°C for 10 min, and 250 μM 2,2′-dipyridyl was added. After 5 min of incubation, FeSO4 was added at 100 μM. Samples were removed at time indicated and RNA was extracted as decribed in Materials and Methods. The strain used is EM1279 (wild-type). (B) Same as in A. The strain used is EM1277 (rne-3071). (C) Strains were grown at 37°C to an O.D.600 of 0.4., and 250 μM 2,2′-dipyridyl was added. After 5 min of incubation, FeSO4 was added at 100 μM. Samples were removed at times indicated and RNA was extracted as decribed in Materials and Methods. The strain used is EM1377 (rne-131).
Decay of other small RNAs also depends on transcription
As mentioned above, a large number of other small RNAs that act by pairing with target mRNAs are also dependent upon Hfq, both for stability and activity. Some, like DsrA, act to positively stimulate translation, by pairing with an inhibitory sequence upstream of the translation start site of rpoS (Majdalani et al. 1998). DsrA also acts negatively on hns translation and message stability (Lease and Belfort 2000). OxyS acts negatively on the translation of the fhlA and rpoS regulatory genes (Altuvia et al. 1998; Zhang et al. 1998). Both of these small RNAs are relatively stable (half-lives: OxyS, 15-30 min; Altuvia et al. 1997; Zhang et al. 2002); DsrA, 20 min (Repoila and Gottesman 2001; Sledjeski et al. 2001). We expressed each of them under pBAD control and monitored their disappearance when arabinose was removed, both in the presence and absence of rifampicin. As with RyhB, DsrA and OxyS were cleared from the cell significantly more rapidly in the absence of rifampicin than in its presence (Fig. 5B,C), suggesting that these small RNAs are also degraded as a consequence of pairing to their targets.
Discussion
The data presented here demonstrate that RyhB causes the rapid degradation of target mRNAs in a process that is dependent upon RNase E. In addition, RyhB itself is unstable under conditions of normal transcription—when its transcripts are being made (and rapidly degraded); this degradation is also RNase E dependent. These results, plus others discussed below, lead us to propose the model shown in Figure 7, in which RyhB (and probably all other pairing small RNAs) act stoichiometrically and not catalytically. In this model, RyhB degradation is a consequence of pairing with its target mRNAs, and is occurring in a 1:1 manner with the target mRNAs. As a result of this mode of action, regulation by the small RNA is tightly coupled to its continued induction; when induction ceases, the small RNA will quickly be consumed.
Figure 7.
Model for self-limited regulation by a small pairing RNA. Normal growth: Only small amounts of the small RNA (sRNA) may be expressed; these will be used quickly, clearing them from the cell. Hfq may bind to some portion of the target mRNAs. Step 1: In response to a stress or other regulatory signal (Fe limitation for RyhB), synthesis of the sRNA rapidly increases and is bound by Hfq (yellow ring). Step 2: The small RNA pairs with its target. Pairing is frequently but not always near the start of translation; in some cases, two or more short (7-9 nucleotides) regions of complementarity are used (for review, see Gottesman 2002). Both small RNA and target mRNA are degraded rapidly in an RNase E-dependent process (scissors). As long as the small RNA continues to be synthesized at a high rate, it will accumulate, and target messages will be destroyed. Step 3: After the stress signal is lost, synthesis of the small RNA will drop to basal levels. The sRNA that has accumulated will be degraded as it is used, allowing reaccumulation of the target mRNAs (Normal growth).
In the first step, stress signals cause induction of the appropriate small RNA (RyhB in this work; Fig. 7, step1). Hfq binds to the RyhB small RNA efficiently, protecting it from degradation (Fig. 4B) and presenting it to its targets. RyhB is efficiently immunoprecipitated with Hfq (Wassarman et al. 2001; Zhang et al. 2003), and given the stabilizing effect of Hfq on RyhB, it seems likely that essentially all of the cellular RyhB is bound to Hfq. Hfq probably also binds to the target mRNAs. This has been observed for the target mRNAs for other Hfq-dependent small RNAs (Møller et al. 2002a; Zhang et al. 2002, 2003), and recently for sodB mRNA as well (T.A. Geissmann and D. Touati, pers. comm.). In vitro, Hfq promotes annealing between the small RNA and its target (Møller et al. 2002a; Zhang et al. 2002); we predict the same is occurring here. It is not clear whether Hfq leaves this complex, but both RyhB and its target mRNA (sodB in the work presented here) are rapidly degraded (Fig. 7, step 2); degradation is dependent upon RNase E. When the stress signal is no longer sensed, no new synthesis of RyhB takes place, and pre-existing RyhB is degraded as it is used, eventually depleting RyhB to the basal level (Fig. 7, step 3).
This model for RyhB action is independent of Fe levels or the Fur protein. Whereas relief of Fur repression is normally necessary for expression of RyhB, we can see the same dependence of RyhB degradation on ongoing transcription when RyhB synthesis is regulated from a foreign (pBAD) promoter (Fig. 5A). Thus, Fe sensing for this small RNA is solely through regulation of RyhB synthesis.
RNase E has the same sequence recognition as Hfq
RNase E is necessary for degradation of RyhB in the absence of Hfq (Fig. 4C) and for the degradation of RyhB when it is being consumed during use (Fig. 6B). Thus, it seems possible that Hfq binding directly blocks RNase E access to RyhB. It is striking, therefore, that the recognition sites of Hfq and RNase E are essentially identical. Hfq binding to RNA occurs particularly at AU-rich single-stranded regions (Møller et al. 2002a; Schumacher et al. 2002; Zhang et al. 2002; Brescia et al. 2003). The recognition sequence for RNase E cleavage is also single-stranded AU rich sequences (Mackie 1991; Mackie and Genereaux 1993; McDowall et al. 1994).
What happens after pairing to make RyhB now sensitive to RNase E? We propose that an exchange of Hfq for RNase E may occur when mRNA pairs with the small RNA, leading to the degradation of both. This could occur in a number of ways: (1) by loss of Hfq from both RNAs upon pairing, leaving them open to RNase E cleavage; (2) by inhibition of sodB translation by annealing of RyhB, enhancing degradation by RNase E; possibly the presence of RNase E on the sodB message allows it to also degrade the nearby RyhB RNA; (3) by direct recruitment of RNase E by some aspect of the paired RNAs (even by Hfq itself?).
A relationship between RNase E and Hfq targets has been noted in a number of other instances. The stability of OmpA mRNA is regulated in a growth rate-dependent manner that is also dependent upon Hfq; degradation of ompA message is due to RNase E (Vytvytska et al. 1998). In this case, Hfq binding is believed to stimulate degradation by blocking ribosome binding (Vytvytska et al. 2000), although the participation of an Hfq-dependent small RNA has not been ruled out. The region in ompA defined as sufficient for Hfq binding overlaps with an RNase E cleavage site (Vytvytska et al. 2000). Hfq was found recently to bind to certain processing precursors for the argX-proM tRNA operon; binding appeared to slow RNase E cleavage at the same sites (Zhang et al. 2003).
RNase E has been implicated in the degradation of a number of other small RNAs. The regulatory MicF antisense RNA is a substrate for RNase E (Schmidt and Delihas 1995), and we have demonstrated recently that MicF also binds Hfq, suggesting that MicF pairing may also depend upon Hfq (Zhang et al. 2003). The well-studied RNAI antisense RNA, which controls the replication of plasmid ColE1, has been shown to be regulated by RNase E (Lin-Chao and Cohen 1991). The FinP antisense RNA regulating the FinO mRNA is also an RNase E substrate (Jerome et al. 1999). The CopA regulatory RNA that controls the replication of plasmid R1 is cleaved by RNase E (Soderbom and Wagner 1998). Finally, the Sok antisense RNA is initially cleaved by RNase E before being rapidly degraded by PNPase (Dam Mikkelsen and Gerdes 1997). Whether these plasmid-encoded regulatory RNAs also trade Hfq, or another chaperone, for RNase E, remains to be determined.
Degradosome involvement
RNase E has been shown to act as the organizer of a complex of proteins called the degradosome. The C-terminal domain of RNase E, not essential for its catalytic activity, acts as a scaffold for binding of polynucleotide phosphorylase (PNPase), RhlB helicase, and enolase (Carpousis et al. 1994, 1999; Py et al. 1994, 1996; Miczak et al. 1996). Deletions of this C-terminal domain, although they have little or no effect on the processing of rRNA or tRNAs, significantly slow the degradation of some test substrates (untranslated mRNAs, in particular; Lopez et al. 1999; Ow et al. 2000; Leroy et al. 2002; Ow and Kushner 2002). In our experiments, we find that cells in which the degradosome scaffold of RNase E is deleted slow normal sodB degradation and the RyhB-dependent accelerated degradation of the sodB message (Fig. 3B,C), and also slow the degradation of RyhB itself (Fig. 6C). Thus, we believe that components of the degradosome may be important either in recruiting RNase E to the paired mRNA:RyhB complex or in allowing accessibility of RNase E to the paired complex (displacing Hfq?).
Mechanism of action may be conserved for other small RNAs
The model presented here for RyhB action can probably be extended to many, if not all, pairing small RNAs. Many of these small RNAs have been reported to have long half-lives when measured in the presence of rifampicin to stop new transcription, as in Figure 4A. We show here that at least two of them become unstable when the half-life is measured while transcription is taking place. DsrA, an activator of rpoS translation, has a half-life of >20 min in the presence of rifampicin (Repoila and Gottesman 2001; Sledjeski et al. 2001; Fig. 5B); it is significantly more unstable in the absence of rifampicin (Fig. 5B). OxyS, induced after oxidative stress, has a half-life of 15-30 min (Altuvia et al. 1997; Zhang et al. 2002); again, it is more unstable in the absence of rifampicin (Fig. 5C). We predict that the same will be true of the many other small RNAs of this Hfq-binding class, and that all of these small RNAs are self-limiting in their action, acting stoichiometrically on their target messages and being degraded as they act. Furthermore, we predict that RNase E will be an essential component of the degradation pathway in all of these cases as well. It is less clear how this mechanism will apply to small RNAs that do not cause destruction of their target mRNAs, such as Spot 42 RNA (Møller et al. 2002b). Spot 42 down-regulates the translation of the galK gene without causing message degradation; in cells treated with rifampicin, it has a half-life of >30 min (Møller et al. 2002b). We would argue that it is likely to be degraded during this process, even if the message is not; otherwise it is hard to imagine how it acts as a regulatory molecule or how the down-regulation would be terminated.
How similar is this mechanism of action to the regulatory small RNAs in eukaryotic cells? Like RNAi, RyhB causes rapid destruction of the target messages (for review, see Zamore et al. 2000). However, unlike RNAi or microRNAs, it does not need to be processed from a longer transcript to act. Once made, RNAi is used in ways that require that it be self-perpetuating rather than self-limited, and therefore, whereas degradation in the context of use may well occur, it is not required for proper use. MicroRNAs, on the other hand, are regulatory molecules akin to RyhB and the other Hfq-binding pairing RNAs (for review, see Ambros 2001). Whether or not the target mRNA is degraded as well as being translationally regulated, we would predict that the micro-RNAs are used stoichiometrically, and destroyed while they are used.
This study demonstrates a critical aspect of small RNA action—a mechanism of shutting off signaling once an environmental signal has been removed. RyhB is made when Fe is limiting, and under those conditions, it helps the cell reprogram iron use, by down-regulating synthesis of nonessential iron-binding proteins. In addition to the targets we have described previously, recent work suggests a large number of other iron-binding proteins are also down-regulated when iron is limiting or the Fur repressor is absent, most likely by RyhB action (McHugh et al. 2003; E. Massé, C. Rosenow, and S. Gottesman, unpubl.). When iron is abundant, however, it is critical that RyhB stop functioning, so that ferritins and other iron-binding proteins can help to sequester iron and protect the cell from Fe damage. Cells unable to make ferritins are sensitive to H2O2, which activates Fe damage (Abdul-Tehrani et al. 1999), so are cells that overproduce RyhB (E. Massé, unpubl.). Thus, the finding that RyhB is rapidly consumed during use provides a mechanism for the rapid recovery from Fe starvation, and provides a clear demonstration of the use of a small RNA as a reversible regulatory switch.
Materials and methods
Strains and plasmids
Strains used in this work are described in Table 1. All strains are derivatives of EM1055 (DJ480), made by Pl transduction as indicated, selecting for the appropriate antibiotic-resistant marker. Iron depletion was induced by addition of 250 μM 2,2′-dipyridyl (dip) in the medium. In the Fe repletion experiments (Fig. 4D,E), FeSO4 at 100 μM was added 5 min after the 2,2′-dipyridyl treatment. Half-life determination of RNA was made by addition of 250 μg/mL rifampicin in the culture and RNA extraction from samples at indicated time.
Table 1.
Strains and plasmids
| Strain name | Relevant markers | Reference/source |
|---|---|---|
| AC21 | MC1061 zce-726::Tn10 | (Carpousis et al. 1994) |
| AC22 | MC1061 rne-3071 zce-726::Tn10 | (Carpousis et al. 1994) |
| AC24 | rneΔ10 zce-726::Tn10 | (Leroy et al. 2002) |
| AC27 | rne-131 zce-726::Tn10 | (Leroy et al. 2002) |
| AC28 | rneΔ14 zce-726::Tn10 | (Leroy et al. 2002) |
| JC3560 | pnp::Tn5 | (Portier et al. 1981) |
| CH1828 | MC1061 rne-50 zce-726::Tn10 | (Mudd et al. 1990) |
| TX2758 | hfq-2::Ω(kan;Kpn1) | (Tsui et al. 1994) |
| TX2808 | hfq-1::Ω(kan;Bcl1) | (Tsui et al. 1994) |
| HT115 | rnc-14::Tn10 | (Takiff et al. 1989) |
| CAG12124 | zce-3113::Tn10kan | (Singer et al. 1989) |
| LMG194 | Δara 714 leu:: Tn10 | (Guzman et al. 1995) |
| DDS719 | ΔdsrA5 zed-3069::Tn10 | (Sledjeski et al. 1996) |
| EM1055 | DJ480 [MG1655 Δlac X174] | (Massé and Gottesman 2002) |
| EM1059 | EM1055 Δara 714 leu::Tn10 | EM1055 + P1 (LMG194) |
| EM1062 | EM1055 Δara 714 leu+ | EM1059 + P1 (EM1055) |
| EM1063 | EM1055 Δara 714 ΔdsrA5 zed-3069::Tn10 | EM1062 + P1 (DDS719) |
| EM1238 | EM1055 ΔryhB::cat | (Massé and Gottesman 2002) |
| EM1265 | EM1055 hfq-1::Ω(kan;Bcl1) | EM1055 + P1 (TX2808) |
| EM1266 | EM1055 hfq-2::Ω(kan;Kpn1) | EM1055 + P1 (TX2758) |
| EM1277 | EM1055 rne-3071 zce-726::Tn10 | EM1055 + P1 (AC22) |
| EM1279 | EM1055 zce-726::Tn10 | EM1055 + P1 (AC21) |
| EM1280 | EM1055 rne-3071 zce-726::Tn10 ΔryhB::cat | EM1277 + P1 (EM1238) |
| EM1282 | EM1055 zce-726::Tn10 ΔryhB::cat | EM1279 + P1 (EM1238) |
| EM1321 | EM1055 rnc-14::Tn10 | EM1055 + P1 (HT115) |
| EM1322 | EM1055 rnc-14::Tn10 ΔryhB::cat | EM1238 + P1 (HT115) |
| EM1342 | EM1055 rne-3071 zce-726::Tn10 hfq-1::Ω(kan;Bcl1) | EM1277 + P1 (TX2808) |
| EM1368 | EM1055 pnp::Tn5 | EM1055 + P1 (JC3560) |
| EM1371 | EM1055 rne-50 zce-726::Tn10 | EM1055 + P1 (CH1828) |
| EM1374 | EM1055 rne-50 zce-3113::Tn10kan | EM1371 + P1 (CAG12124) |
| EM1375 | EM1055 rneΔ10 zce-726::Tn10 | EM1374 + P1 (AC24) |
| EM1376 | EM1055 rneΔ14 zce-726::Tn10 | EM1374 + P1 (AC28) |
| EM1377 | EM1055 rne-131 zce-726::Tn10 | EM1374 + P1 (AC27) |
| Plasmids | Description | |
| pNM12 | pBAD24 derivative | (Majdalani et al. 1998) |
| pNM13 | pBAD-dsrA | (Majdalani et al. 1998) |
| pEM1396 | pBAD-ryhB | From pNM12 |
| pAZ#7 | pBAD18-oxyS | A. Zhang (unpubl.) |
To construct the rne deletion strains, the zce-3113::Tn10kan from CAG12124 (Singer et al. 1989) was introduced by P1 transduction into temperature-sensitive EM1371 to replace the zce-726::Tn10. Kanamycin resistant transductants were selected at 30°C and then screened for tetracycline sensitivity and thermo-sensitivity at 42°C (EM1374). The rneΔ10, rneΔ14, and rne-131 allele from AC24, AC28, and AC27, respectively, were transduced into the EM1374 strain with P1, and tetR transductants were selected on LB tetracycline medium incubated at 42°C. The rne alleles were confirmed by Western blot with an anti-RNase E antibody [a kind gift from M. Dreyfus (Laboratoire de Genétiqué Moléculaire, CNRS, École Normale Supérieure, Paris, France)]. The resulting strains are EM1375 (rneΔ10), EM1376 (rneΔ14), and EM1377 (rne-131).
To construct plasmid pBAD-ryhB, the promoterless ryhB gene was amplified by PCR using Pfu DNA polymerase (Stratagene) with the following oligonucleotides: EM34 (5′-CCATGC GATCAGGAAGACCCTCG-3′) and EM35 (5′-GACTGAGA ATTCGGATAAATTGAGAACGAAAGATC-3′) designed with an EcoRI site. The resulting 134-nucleotide DNA fragment was digested by EcoRI and then inserted into pNM12 (Majdalani et al. 1998) digested with MscI and EcoRI. The resulting plasmid was confirmed by sequencing.
RNA extraction and Northern blot analysis
RNA was extracted by using a modified version of the hot phenol techique (Aiba et al. 1981). Briefly, 520 μL of cell culture was removed and mixed with 75 μL of lysis solution (320 mM Na acetate at pH 4.6, 8% SDS, 16 mM EDTA). Lysed cells were mixed for 5 min with an equal volume of 65°C water-buffered phenol. After centrifugation, the supernatant was extracted by phenol-chloroform twice and precipitated with 3 vol of 100% ethanol. The resulting RNA pellet was resuspended in H2O treated with diethyl pyrocarbonate (DEPC) and kept at -70°C. RNA concentration was measured with a spectrophotometer at 260 nm. For RNA analyzed by the Dot-Blot method, 1 μg of total RNA was loaded on a Zeta-probe membrane (Bio-Rad) on a Bio-Rad Bio-dot apparatus as described by the manufacturer. For analysis of RNA on PAGE or agarose gel, the techniques used are described in Massé and Gottesman (2002). Oligonucleotides used as probe for Northern blot of 5S rRNA: 5′-GTTTCACTTC TGAGTTCGGCATGGGGTCAGGTGGG-3′. Probe for OxyS; 5′-GAGATCCGCAAAAGTTCACGTTGGCTTTAGTTATTC GAGT-3′. These probes are biotinylated at their 5′ end and detected by the Brightstar Biodetect detection kit (Ambion). The probes for RyhB and sodB were described earlier in Massé and Gottesman (2002). The probe for DsrA (DsrA SL-II) is described in Majdalani et al. (1998).
Quantification of RNA bands from Northern blots
Films were scanned and intensity of RNA bands was measured with the Stratagene Eagle Sight software. The intensities were plotted and RNA half-life was calculated using the slope from each plot.
Acknowledgments
We are grateful to N. Majdalani, Y.N. Zhou, M. Maurizi, M. Cashel, G. Storz, A. Zhang, A.J. Carpousis, and M. Dreyfus for comments and stimulating discussions; D. Fitzgerald and S. Labbé for comments on the manuscript; A. Zhang, N. Majdalani, D. Court, C Portier, and A. Carpousis for plasmids and bacterial strains; D. Touati for discussions, comments, and communicating results prior to publication; and M. Dreyfus for antibodies and strains. E.M. was supported by a post-doctoral fellowship of the Canadian Institutes for Health Research (CIHR). F.E.E. was supported by the NIH Undergraduate Scholarship Program.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1127103.
References
- Abdul-Tehrani H., Hudson, A.J., Chang, Y.-S., Timms, A.R., Hawkins, C., Williams, J.M., Harrison, P.M., Guest, J.R., and Andrews, S.C. 1999. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J. Bacteriol. 181: 1415-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiba H., Adhya, S., and de Crombrugghe, B. 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 256: 11905-11910. [PubMed] [Google Scholar]
- Altuvia S., Weinstein-Fischer, D., Zhang, A., Postow, L., and Storz, G. 1997. A small stable RNA induced by oxidative stress: Role as a pleiotropic regulator and antimutator. Cell 90: 43-53. [DOI] [PubMed] [Google Scholar]
- Altuvia S., Zhang, A., Argaman, L., Tiwari, A., and Storz, G. 1998. The Escherichia coli oxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J. 17: 6069-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. 2001. microRNAs: Tiny regulators with great potential. Cell 107: 823-826. [DOI] [PubMed] [Google Scholar]
- Andrews S.C. 1998. Iron storage in bacteria. Adv. Microb. Physiol. 40: 281-351. [DOI] [PubMed] [Google Scholar]
- Andrews S.C., Arosio, P., Bottke, W., Briat, J.F., von Darl, M., Harrison, P.M., Laulhere, J.P., Levi, S., Lobreaux, S., and Yewdall, S.J. 1992. Structure, function, and evolution of ferritins. J. Inorg. Biochem. 47: 161-174. [DOI] [PubMed] [Google Scholar]
- Brescia C.C., Mikulecky, P.J., Feig, A.L., and Sledjeski, D.D. 2003. Identification of the Hfq-binding site on DsrA RNA:Hfq binds without altering DsrA secondary structure. RNA 9: 33-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis A.J., van Houwe, G., Ehretsmann, C., and Krisch, H.M. 1994. Copurification of E. coli RNAse E and PNPase: Evidence for a specific association between two enzymes important in RNA processing and degradation. Cell 76: 889-900. [DOI] [PubMed] [Google Scholar]
- Carpousis A.J., Vanzo, N.F., and Raynal, L.C. 1999. mRNA degradation. Atale of poly(A) and multiprotein machines. Trends Genet. 15: 24-28. [DOI] [PubMed] [Google Scholar]
- Dam Mikkelsen N. and Gerdes, K. 1997. Sok antisense RNA from plasmid R1 is functionally inactivated by RNase E and polyadenylated by poly(A) polymerase I. Mol. Microbiol. 26: 311-320. [DOI] [PubMed] [Google Scholar]
- Dubrac S. and Touati, D. 2000. Fur positive regulation of iron superoxide dismutase in Escherichia coli: Functional analysis of the sodB promoter. J. Bacteriol. 182: 3802-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2002. Fur-mediated transcriptional and post-transcriptional regulation of FeSOD expression in Escherichia coli. Microbiology 148: 147-156. [DOI] [PubMed] [Google Scholar]
- Gottesman S. 2002. Stealth regulation: Biological circuits with small RNA switches. Genes & Dev. 16: 2829-2842. [DOI] [PubMed] [Google Scholar]
- Grunberg-Manago M. 1999. Messenger RNA stability and its role in control of gene expression in bacteria and phage. Annu. Rev. Genet. 33: 193-227. [DOI] [PubMed] [Google Scholar]
- Guzman L.-M., Belin, D., Carson, M.J., and Beckwith, J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177: 4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerome L.J., van Biesen, T., and Frost, L.S. 1999. Degradation of FinP antisense RNA from F-like plasmids: The RNA-binding protein, FinO, protects FinP from ribonuclease E. J. Mol. Biol. 285: 1457-1473. [DOI] [PubMed] [Google Scholar]
- Lease R.A. and Belfort, M. 2000. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proc. Natl. Acad. Sci. 97: 9919-9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy A., Vanzo, N.F., Sousa, S., Dreyfus, M., and Carpousis, A.J. 2002. Function in Escherichia coli of the non-catalytic part of RNase E: Role in the degradation of ribosome-free mRNA. Mol. Microbiol. 45: 1231-1243. [DOI] [PubMed] [Google Scholar]
- Lin-Chao S. and Cohen, S.N. 1991. The rate of processing and degradation of antisense RNAI regulates the replication of ColE1-type plasmids in vivo. Cell 65: 1233-1242. [DOI] [PubMed] [Google Scholar]
- Lopez P.J., Marchand, I., Joyce, S.A., and Dreyfus, M. 1999. The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol. Microbiol. 33: 188-199. [DOI] [PubMed] [Google Scholar]
- Mackie G.A. 1991. Specific endonucleolytic cleavage of the mRNA for ribosomal protein S20 of Escherichia coli requires the product of the ams gene in vivo and in vitro. J. Bacteriol. 173: 2488-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie G.A. and Genereaux, J.L. 1993. The role of RNA structure in determining RNase E-dependent cleavage sites in the mRNA for ribosomal protein S20 in vitro. J. Mol. Biol. 234: 998-1012. [DOI] [PubMed] [Google Scholar]
- Majdalani N., Cunning, C., Sledjeski, D., Elliott, T., and Gottesman, S. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. 95: 12462-12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N., Chen, S., Murrow, J., St. John, K., and Gottesman, S. 2001. Regulation of RpoS by a novel small RNA: The characterization of RprA. Mol. Microbiol. 39: 1382-1394. [DOI] [PubMed] [Google Scholar]
- Massé E. and Gottesman, S. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. 99: 4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowall K.J., Lin-Chao, S., and Cohen, S.N. 1994. A + U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J. Biol. Chem. 269: 10790-10796. [PubMed] [Google Scholar]
- McHugh J.P., Rodriguez-Quinones, F., Abdul-Tehrani, H., Svistunenko, D.A., Poole, R.K., Cooper, C.E., and Andrews, S.C. 2003. Global Iron-dependent gene regulation in Escherichia coli: A new mechanism for iron homeostasis. J. Biol. Chem. 278: 29478-29486. [DOI] [PubMed] [Google Scholar]
- Miczak A., Kaberdin, V.R., Wei, C.L., and Lin-Chao, S. 1996. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc. Natl. Acad. Sci. 93: 3865-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller T., Franch, T., Hojrup, P., Keene, D.R., Bachinger, H.P., Brennan, R., and Valentin-Hansen, P. 2002a. Hfq: A bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell 9: 23-30. [DOI] [PubMed] [Google Scholar]
- Møller T., Franch, T., Udesen, C., Gerdes, K., and Valentin-Hansen, P. 2002b. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes & Dev. 16: 1696-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd E.A., Krisch, H.M., and Higgins, C.F. 1990. RNase E, an endoribonuclease, has a general role in the chemical decay of Escherichia coli mRNA: Evidence that rne and ams are the same genetic locus. Mol. Microbiol. 4: 2127-2135. [DOI] [PubMed] [Google Scholar]
- Ow M.C. and Kushner, S.R. 2002. Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli. Genes & Dev. 16: 1102-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow M.C., Liu, Q., and Kushner, S.R. 2000. Analysis of mRNA decay and rRNA processing in Escherichia coli in the absence of RNAse E-based degradosome assembly. Mol. Microbiol. 38: 854-866. [DOI] [PubMed] [Google Scholar]
- Portier C., Migot, C., and Grunberg-Manago, M. 1981. Cloning of E. coli pnp gene from an episome. Mol. Gen. Genet. 183: 298-305. [DOI] [PubMed] [Google Scholar]
- Py B., Causton, H., Mudd, E.A., and Higgins, C.F. 1994. A protein complex mediating mRNA degradation in Escherichia coli. Mol. Microbiol. 14: 717-729. [DOI] [PubMed] [Google Scholar]
- Py B., Higgins, C.F., Krisch, H.M., and Carpousis, A.J. 1996. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381: 169-172. [DOI] [PubMed] [Google Scholar]
- Ratledge C. and Dover, L.G. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54: 881-941. [DOI] [PubMed] [Google Scholar]
- Repoila F. and Gottesman, S. 2001. Signal transduction cascade for regulation of RpoS: Temperature regulation of DsrA. J. Bacteriol. 183: 4012-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M. and Delihas, N. 1995. micF RNA is a substrate for RNase E. FEMS Microbiol. Lett. 133: 209-213. [DOI] [PubMed] [Google Scholar]
- Schumacher M.A., Pearson, R.F., Møller, T., Valentin-Hansen, P., and Brennan, R.G. 2002. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: A bacterial Sm-like protein. EMBO J. 21: 3546-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M., Baker, T.A., Schnitzler, G., Deischel, S.M., Goel, M., Dove, W., Jaacks, K.J., Grossman, A.D., Erickson, J., and Gross, C.A. 1989. A collection of strains containing genetically linked alternating resistance elements for genetic mapping. Microbiol. Rev. 53: 1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledjeski D.D., Gupta, A., and Gottesman, S. 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 15: 3993-4000. [PMC free article] [PubMed] [Google Scholar]
- Sledjeski D.D., Whitman, C., and Zhang, A. 2001. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 183: 1997-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderbom F. and Wagner, E.G. 1998. Degradation pathway of CopA, the antisense RNA that controls replication of plasmid R1. Microbiology 144: 1907-1917. [DOI] [PubMed] [Google Scholar]
- Takiff H.E., Chen, S.M., and Court, D.L. 1989. Genetic analysis of the rnc operon of Escherichia coli. J. Bacteriol. 171: 2581-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui H.-C.T., Leung, H.-C.E., and Winkler, M.E. 1994. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 13: 35-49. [DOI] [PubMed] [Google Scholar]
- Vytvytska O., Jakobsen, J., Balcunaite, G., Andersen, J., Baccarini, M., and von Gabain, A. 1998. Host Factor I, Hfq, binds to Escherichia coli ompA mRNA in a growth-rate dependent fashion and regulates its stability. Proc. Natl. Acad. Sci. 95: 14118-14123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vytvytska O., Moll, I., Kaberdin, V.R., von Gabain, A., and Blasi, U. 2000. Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Genes & Dev. 14: 1109-1118. [PMC free article] [PubMed] [Google Scholar]
- Wassarman K.M., Repoila, F., Rosenow, C., Storz, G., and Gottesman, S. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes & Dev. 15: 1637-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. 2000. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25-33. [DOI] [PubMed] [Google Scholar]
- Zhang A., Altuvia, S., Tiwari, A., Argaman, L., Hengge-Aronis, R., and Storz, G. 1998. The oxyS regulatory RNA represses rpoS translation by binding Hfq (HF-1) protein. EMBO J. 17: 6061-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Wassarman, K.M., Ortega, J., Steven, A.C., and Storz, G. 2002. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 9: 11-22. [DOI] [PubMed] [Google Scholar]
- Zhang A., Wassarman, K.M., Rosenow, C., Tjaden, B.C., Storz, G., and Gottesman, S. 2003. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. (in press) [DOI] [PubMed]