Abstract
Segregation of the replicated genome during cell division requires kinetochores, mechanochemical organelles that assemble on mitotic chromosomes to connect them to spindle microtubules. CENP-A, a histone H3 variant, and CENP-C, a conserved structural protein, form the DNA-proximal foundation for kinetochore assembly. Using RNA interference-based genomics in Caenorhabditis elegans, we identified KNL-1, a novel kinetochore protein whose depletion, like that of CeCENP-A or CeCENP-C, leads to a “kinetochore-null” phenotype. KNL-1 is downstream of CeCENP-A and CeCENP-C in a linear assembly hierarchy. In embryonic extracts, KNL-1 exhibits substoichiometric interactions with CeCENP-C and forms a near-stoichiometric complex with CeNDC-80 and HIM-10, the C. elegans homologs of Ndc80p/HEC1p and Nuf2p—two widely conserved outer kinetochore components. However, CeNDC-80 and HIM-10 are not functionally equivalent to KNL-1 because their inhibition, although preventing formation of a mechanically stable kinetochore-microtubule interface and causing chromosome missegregation, does not result in a kinetochore-null phenotype. The greater functional importance of KNL-1 may be due to its requirement for targeting multiple components of the outer kinetochore, including CeNDC-80 and HIM-10. Thus, KNL-1 plays a central role in translating the initiation of kinetochore assembly by CeCENP-A and CeCENP-C into the formation of a functional microtubule-binding interface.
Keywords: Centromere, mitosis, tubulin, CENP, chromosome, spindle
Eukaryotic chromosome segregation requires kinetochores, organelles that form on condensing chromosomes to provide a dynamic attachment site for spindle microtubules (for review, see Cleveland et al. 2003). Microtubules are polymers of tubulin, one of the most conserved proteins in the eukaryotic kingdom. Thus, despite extensive variation in the DNA sequences upon which kinetochores assemble (Sullivan et al. 2001), fundamental similarities were anticipated in their structure and protein composition. Consistent with this expectation, conserved components have been identified at all levels of kinetochore structure, from its specialized chromatin foundation to the outer domains that interact with spindle microtubules (Cleveland et al. 2003). This molecular similarity extends from organisms such as budding yeast, which assemble point kinetochores at small defined chromosomal loci (for review, see Kitagawa and Hieter 2001), to organisms such as Caenorhabditis elegans that have holocentric chromosomes with diffuse kinetochores that form along their entire length (Howe et al. 2001; Moore and Roth 2001; Oegema et al. 2001).
A major breakthrough in the molecular dissection of kinetochores came with the discovery of CENP-A, a histone H3 variant that is a component of the specialized chromatin that acts as the structural foundation for kinetochore assembly (Sullivan 2001). Centromere-specific histone H3 molecules have now been found at the kinetochores of all eukaryotes examined (Henikoff et al. 2001; Sullivan et al. 2001). CENP-C, a second widely conserved protein discovered using human autoimmune sera (Saitoh et al. 1992), localizes to the same chromatin-proximal region of the kinetochore as CENP-A. In addition to CENP-A and CENP-C, numerous other kinetochore components have been identified, particularly in fungi (Goshima et al. 1999, 2003; He et al. 2001; Kitagawa and Hieter 2001; Cheeseman et al. 2002). The most widely conserved of these are the mitotic checkpoint proteins that transiently localize to kinetochores and translate the state of kinetochore attachment into a signal that regulates the metaphase-anaphase transition (for review, see Cleveland et al. 2003). Also conserved are Ndc80p/HEC1 and Nuf2p, a pair of interacting proteins that localize to the outer kinetochore and are required for proper kinetochore-microtubule interactions (Chen et al. 1997; Wigge et al. 1998; He et al. 2001; Howe et al. 2001; Janke et al. 2001; Nabetani et al. 2001; Wigge and Kilmartin 2001; DeLuca et al. 2002; Martin-Lluesma et al. 2002; McCleland et al. 2003).
Given the essential role of the kinetochore in cell division, function-based identification of its components has been largely limited to genetic organisms with the potential for conditional mutant screens. Recently, the C. elegans embryo has emerged as a powerful genomic system to identify essential genes required for cell division (Gönczy et al. 2000; Zipperlen et al. 2001). In C. elegans, RNA-mediated interference (RNAi) can be used to analyze the first mitotic division of a fertilized embryo following near-complete depletion of any target protein. Large-scale genomic analysis based on RNAi is defining the complete set of nonredundant genes required for embryo viability (Kamath et al. 2003), a subset of which are required for cell division. To use this technical paradigm for studying kinetochores, we developed high-resolution assays to monitor kinetochore assembly and function during the first mitotic division of the C. elegans embryo (Oegema et al. 2001). Previously, we showed that depletion of either the C. elegans CENP-A or CENP-C homolog results in a distinctive “kinetochore-null” phenotype (Oegema et al. 2001). Using the same assays to rescreen gene products implicated in chromosome segregation by RNAi-based functional genomics, we have identified a novel kinetochore component, KNL-1, whose depletion results in a kinetochore-null phenotype. KNL-1 makes a central contribution to kinetochore function by translating the initiation of kinetochore assembly into the formation of a mechanical interface that interacts with spindle microtubules.
Results
Depletion of KNL-1 results in a kinetochore-null phenotype
During an RNAi-based functional genomic screen of C. elegans Chromosome III, we identified six genes required for mitotic chromosome segregation (Gönczy et al. 2000). Because this screen was performed using differential interference contrast microscopy, by which chromosomes are not visible in the C. elegans embryo, defects in chromosome segregation were inferred from the presence of nuclei trapped in the cleavage furrow and/or extra nuclei. Five of the genes identified in this manner (smc-4, hcp-3, cls-2, klp-19, and F54C8.2) were homologous to known proteins. The remaining gene, C02F5.1, which encodes a novel protein, was named knl-1 for reasons described below.
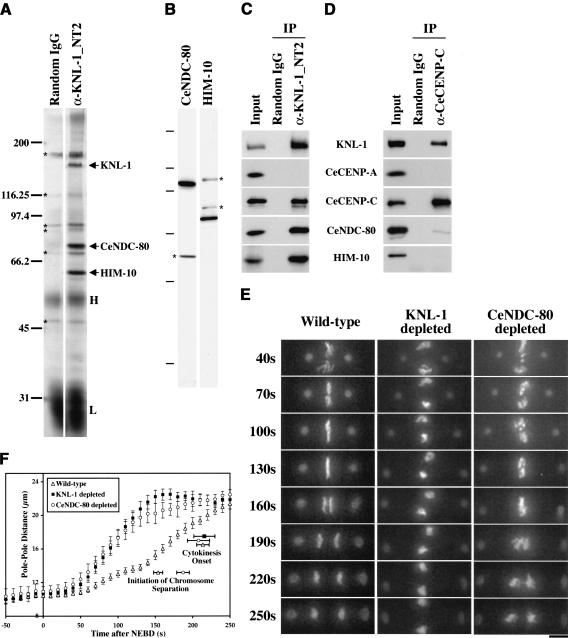
To examine the chromosome segregation defect resulting from depletion of KNL-1, we performed 3D time-lapse analysis of embryos expressing both GFP-histone H2B (to label DNA) and GFP-γ-tubulin (to label spindle poles). KNL-1 is >95% depleted under our RNAi conditions (see Fig. 5C, below). In KNL-1-depleted embryos, chromosomes derived from each of the pronuclei (sperm or oocyte) clumped together, resulting in two DNA masses positioned midway between the two spindle poles (Fig. 1A, 120/130-sec panels). The spindle poles rapidly and prematurely separated, and no anaphase chromosome segregation was observed (Fig. 1A, 200/210-sec panels). Spinning disk confocal microscopy of GFP-β-tubulin-expressing embryos indicated that KNL-1-depleted embryos fail to form a robust spindle and confirmed premature separation of the spindle poles (Fig. 1B).
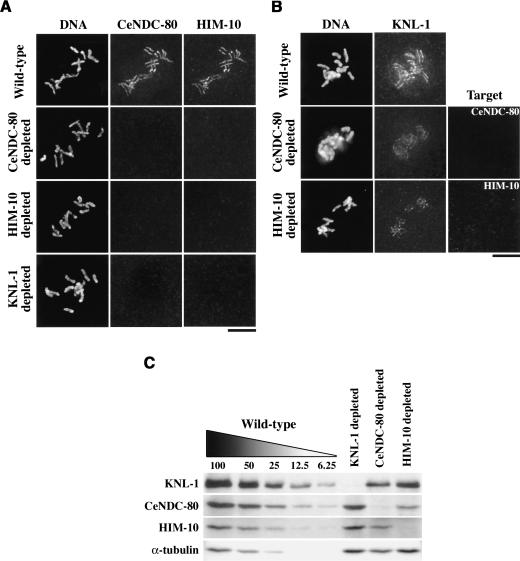
Figure 5.
KNL-1, CeNDC-80, and HIM-10 exhibit asymmetric dependencies during kinetochore assembly. (A) Wild-type, CeNDC-80-depleted (n = 33), HIM-10-depleted (n = 36), and KNL-1-depleted (n = 67) embryos were stained for DNA (left panels), CeNDC-80 (middle panels), and HIM-10 (right panels). Chromosomes from prometaphase-stage one-cell embryos are shown. (B) Wild-type, CeNDC-80-depleted (n = 37), and HIM-10-depleted (n = 40) embryos were stained for DNA (left panels), KNL-1 (middle panels), and the depletion target (right panels). (C) Western blot of extracts prepared from wild-type, KNL-1-depleted, CeNDC-80-depleted, and HIM-10-depleted worms. Serial dilutions of wild-type extract were loaded to quantify depletion levels.
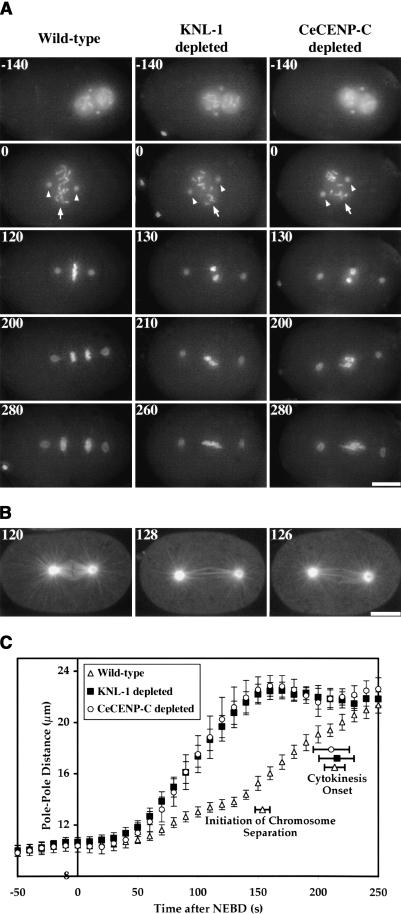
Figure 1.
KNL-1 depletion results in a “kinetochorenull” phenotype. Selected panels from time-lapse sequences of wild-type (left column), KNL-1 depleted (middle column), and CeCENP-C depleted (right column) embryos are shown. Time after NEBD (in seconds) is indicated in the upper left corner of each panel. (A) Chromosome segregation was monitored in embryos coexpressing GFP-histone H2B and GFP-γ-tubulin. The 3D widefield data sets were acquired every 10 sec for wild-type (n = 27), KNL-1-depleted (n = 24), and CeCENP-C-depleted (n = 18) embryos. Time-aligned projections of representative 3D movies are shown (see also Supplementary Videos 1-3). Chromosomes (arrows in 0-sec panels) and centrosomes (arrowheads in 0-sec panels) are easily distinguished in these images. (B) Spindle morphology was monitored by single-section spinning disk confocal imaging of embryos expressing GFP-β-tubulin. Images were acquired every 6-8 sec for wild-type (n = 27), KNL-1-depleted (n = 32), and CeCENP-C-depleted (n = 16) embryos (see also Supplementary Videos 4-6). A single image, concurrent with metaphase in wild type, is shown for each type of embryo. (C) The distance between spindle poles was tracked for 14 wild-type, 15 KNL-1-depleted, and 12 CeCENP-C-depleted embryos filmed as in A. The plot shows average pole-to-pole distance versus time after NEBD. Error bars represent the S.E.M. with a confidence interval of 0.95. The average times for the onset of cytokinesis (for all three conditions) and the initiation of chromosome separation (in wild type) are also indicated. Bars: A,B, 10 μm.
The phenotypic consequences of depleting KNL-1 were remarkably similar to those resulting from depletion of CeCENP-C (product of the hcp-4 gene; Fig. 1A,B; see also Supplementary Videos 1-6) or CeCENP-A (product of the hcp-3 gene; Oegema et al. 2001). Significantly, not only were the phenotypes of KNL-1- and CeCENPC-depleted embryos qualitatively similar, but also the kinetics of premature spindle elongation were quantitatively identical (Fig. 1C). Despite the dramatic chromosome-segregation defect and premature spindle elongation in KNL-1- or CeCENP-C depleted embryos, there was no effect on cell cycle progression as assessed by measurements of the time interval between nuclear envelope breakdown (NEBD) and onset of cytokinesis (Fig. 1C). Because depletion of CeCENP-C (or CeCENP-A) in C. elegans results in failure of kinetochore assembly and a phenotype consistent with complete loss of kinetochore function (Oegema et al. 2001), we named the predicted C02F5.1 gene knl-1 (kinetochore-null).
KNL-1 localizes to kinetochores throughout mitosis
The predicted primary sequence of KNL-1 was confirmed by sequencing two independent cDNAs (yk491a8 and yk444c6). KNL-1 is a 113-kD acidic protein with weak repeats in the N terminus and a predicted coiled coil in the C terminus (Fig. 2A). We identified a homolog of KNL-1 with ∼30% identity (45% similarity) in the related nematode Caenorhabditis briggsae, but sequence analysis failed to reveal strong homologs outside of nematodes. To analyze the subcellular localization of KNL-1, we generated affinity-purified antibodies to two overlapping N-terminal fragments. Both antibodies recognize a single 159-kD band in C. elegans embryo extracts, 46 kD larger than the predicted molecular mass of KNL-1 (Fig. 2B). The in vitro translation product of the predicted KNL-1 primary sequence comigrated with the 159-kD band in embryo extracts (data not shown), indicating that KNL-1 migrates as an anomalously large protein on denaturing protein gels.
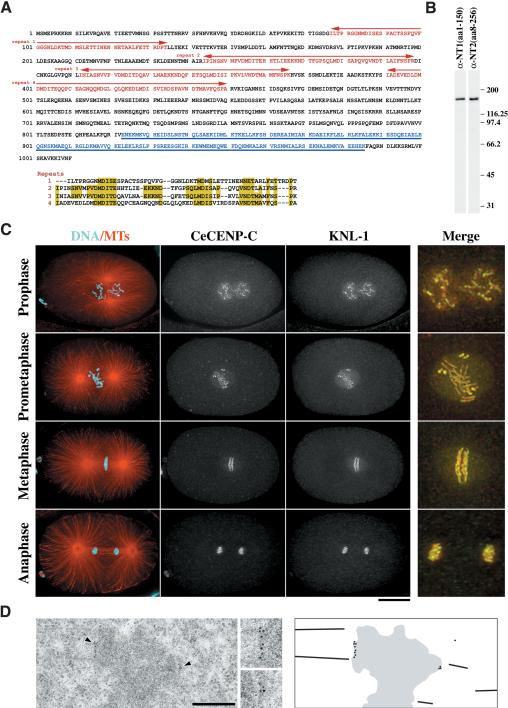
Figure 2.
KNL-1 localizes to kinetochores throughout mitosis. (A) KNL-1 is a 1010-amino-acid novel protein with a predicted molecular mass of 113 kD. The N terminus contains four repeats of a loosely conserved sequence motif (indicated in red and aligned beneath the sequence). The C terminus of KNL-1 has a region predicted to form a coiled coil (underlined and lettered in blue). (B) Western blots of embryo extracts probed using affinity-purified antibodies raised against two overlapping regions of KNL-1 (amino acids 1-150 and amino acids 8-256). (C) Projected 3D data sets of fixed embryos at different cell cycle stages stained for microtubules and DNA (red and cyan; left column), the kinetochore marker CeCENP-C (second column), and KNL-1 (third column). The merged images on the far right show CeCENP-C (red) and KNL-1 (green) at 2.5× higher magnification. (D) Immuno-EM of KNL-1 in multicellular embryos prepared by high-pressure freezing/freeze substitution. Insets were magnified 2×. The schematic to the right illustrates the positions of the gold particles in the field of view. Bars: C, 10 μm; D, 500 nm.
KNL-1 colocalized with CeCENP-C, a bona fide kinetochore marker, throughout mitosis (Fig. 2C). KNL-1 is detected on chromosomes from early prophase until late anaphase. At metaphase, KNL-1 colocalizes with CeCENP-C on the poleward face of the mitotic chromosomes. Between prometaphase and early anaphase, KNL-1 also localizes to the spindle region but does not coalign with spindle microtubules (Fig. 2C). A similar “matrix”-like staining pattern was previously observed for CeBUB-1, the C. elegans homolog of the Bub1 checkpoint kinase (Oegema et al. 2001). Because both KNL-1 antibodies gave identical staining patterns, only the KNL1_NT1(aa1-150) antibody was used in subsequent localization experiments.
To characterize KNL-1 localization at an ultrastructural level, we performed immunoelectron microscopy on high-pressure frozen/freeze-substituted worms. Clusters of secondary antibody-conjugated gold particles were clearly visible close to the surface of the condensed chromatin of prometaphase/metaphase-stage cells in multicellular embryos (Fig. 2D). Cumulatively, the phenotypic and localization analysis indicate that KNL-1 is present at kinetochores throughout mitosis and is essential for mitotic chromosome segregation.
KNL-1 is downstream of CeCENP-A and CeCENP-C in a linear assembly hierarchy
Depletion of KNL-1 results in a kinetochore-null phenotype very similar to that resulting from depletion of CeCENP-A or CeCENP-C. Prior work placed CeCENP-A above CeCENP-C at the top of the kinetochore assembly hierarchy (Moore and Roth 2001; Oegema et al. 2001). Therefore, three possible explanations for the KNL-1 depletion phenotype are: (1) KNL-1 is required for CeCENP-A to form the specialized chromatin upon which kinetochores assemble; (2) KNL-1 is required to localize CeCENP-C in a CeCENP-A-dependent manner; (3) KNL-1 is required downstream of CeCENP-A and CeCENP-C to generate a functional microtubule-binding interface. To distinguish among these possibilities, we performed pairwise depletion and localization analysis. Normal levels of CeCENP-C (Fig. 3A) and CeCENP-A (Fig. 3C) were present at kinetochores in KNL-1-depleted embryos. In contrast, depletion of CeCENP-C (Fig. 3A) or CeCENP-A (data not shown) prevented localization of KNL-1 to chromosomes. On immunoblots of depleted worms, RNAi of their respective genes reduced CeCENP-A and CeCENP-C below the detection level (Fig. 3B). In contrast, KNL-1 levels were unaffected by depletion of either protein, indicating that the observed failure in KNL-1 targeting is not an indirect consequence of its destabilization but is caused by absence of its chromosomal binding site.
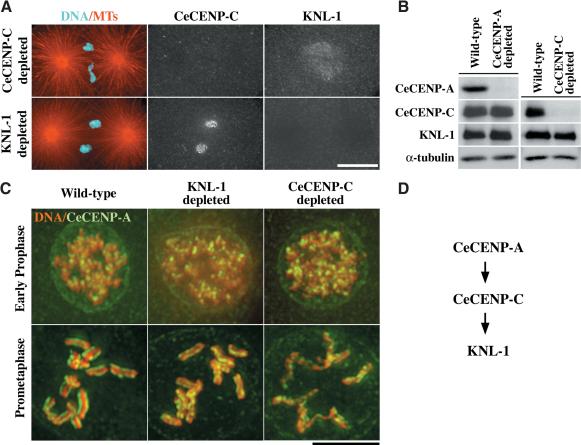
Figure 3.
KNL-1 is downstream of CeCENP-A and CeCENP-C in the kinetochore assembly hierarchy. (A) Embryos were fixed and stained for microtubules and DNA (red and cyan; left panels), CeCENP-C (middle panels), and KNL-1 (right panels). In CeCENP-C-depleted embryos (n = 39), KNL-1 did not localize to chromosomes, although the weaker “matrix” staining in the region of the spindle was still observed. In KNL-1-depleted embryos, CeCENP-A (n = 45; not shown) and CeCENP-C (n = 31) localized normally to kinetochores. Both CeCENP-C (Oegema et al. 2001) and KNL-1 (n = 31 embryos; not shown) failed to target to chromosomes in embryos depleted of CeCENP-A. (B) Western blots of wild-type, CeCENP-A-depleted, and CeCENP-C-depleted worms. α-Tubulin was used as a loading control. (C) Wild-type (left panels), KNL-1-depleted (middle panels), and CeCENP-C-depleted (right panels) embryos were fixed and stained for DNA (red) and CeCENP-A (green). Representative prophase (top row) and prometaphase (bottom row) embryos are shown. (D) Schematic illustrating the linear assembly hierarchy for CeCENP-A, CeCENP-C, and KNL-1. The relationship between CeCENP-A and CeCENP-C was established previously (Moore and Roth 2001; Oegema et al. 2001). Bars: A, 10 μm; C, 5 μm.
CeCENP-C is required for the formation of twinned kinetochore plates in late prophase (Moore and Roth 2001). This failure of sister kinetochore resolution, detected by monitoring CeCENP-A staining, is possibly caused by the perturbation of chromosome structure visible in CeCENP-C-depleted embryos (Fig. 3C). Perturbation of chromosome structure is also evident in CeCENP-A-depleted embryos (data not shown). Consistent with KNL-1 being downstream of CeCENP-A and CeCENP-C in the kinetochore assembly hierarchy, well-condensed chromosomes with two resolved “lines” of CeCENP-A staining were present in KNL-1-depleted embryos (Fig. 3C). However, the spacing between the CeCENP-A lines was reduced in KNL-1-depleted embryos, likely because of a lack of microtubule-mediated forces.
Cumulatively, these results place the three known C. elegans proteins whose depletion leads to a kinetochore-null phenotype in a linear assembly hierarchy (Fig. 3D). Because the kinetochore-null phenotype arises from an inability of kinetochores to interact with spindle microtubules, these results indicate that KNL-1 is a key functional target of CeCENP-A and CeCENP-C in the mitotic C. elegans kinetochore.
KNL-1 forms a complex with the C. elegans homologs of Ndc80p/HEC1 and Nuf2p
The linear assembly hierarchy and similar loss-of-function phenotypes suggested a close functional relationship among KNL-1, CeCENP-C, and CeCENP-A. To test if these proteins directly interact with each other, we immunoaffinity-purified KNL-1-containing complexes from embryo extracts. KNL-1 immunoprecipitates, obtained using either anti-KNL-1 antibody, reproducibly contained KNL-1 (confirmed by mass spectrometry) and two proteins with apparent molecular masses of 58 and 75 kD that appeared to be present with a stoichiometry similar to KNL-1 on silver-stained (Fig. 4A) or Coomassie-stained gels (data not shown). Mass spectrometry identified the copurifying proteins as HIM-10, the previously identified C. elegans homolog of Nuf2p (Howe et al. 2001), and the product of the predicted W01B6.9 gene, which is homologous to Ndc80p/HEC1. Ndc80p/HEC1 and Nuf2p are widely conserved proteins that associate with each other, localize to the outer kinetochore, and are required for kinetochore-microtubule interactions. Because W01B6.9 encodes the only protein homologous to Ndc80p/HEC1 in the C. elegans genome, we named this gene ndc-80 and refer to its protein product as CeNDC-80.
Figure 4.
KNL-1 forms a complex with two widely conserved outer kinetochore proteins. (A) Silver-stained gel of immunoprecipitates from embryo extracts. The three bands consistently detected in KNL-1 (α-KNL-1_NT2) but not control (Random IgG) immunoprecipitates (indicated by arrows; n = 4 extracts) were identified as KNL-1, CeNDC-80, and HIM-10 by mass spectrometry. The same bands were detected in immunoprecipitates using a second KNL-1 antibody (data not shown). Contaminating antibody heavy (H) and light (L) chains and background bands (*) are also indicated. (B) Western blots of embryo extracts probed with affinity-purified antibodies against CeNDC-80 and HIM-10. Asterisks indicate background bands that were not eliminated by RNAi of the corresponding genes. The molecular mass markers were 200, 97.4, 66.2, 45, 31, and 14.4 kD. (C) Western blots of KNL-1 immunoprecipitates probed with the indicated antibodies. Identical results were obtained in immunoprecipitations performed on four independent extracts. (D) Western blots of CeCENP-C immunoprecipitates probed with the indicated antibodies. Identical results were obtained in immunoprecipitations performed on two independent extracts. CeCENP-C was not detectable by silver staining in either KNL-1 or CeCENP-C immunoprecipitates. (E) 3D widefield data sets were acquired every 10 sec for wild-type (n = 27), KNL-1-depleted (n = 24), CeNDC-80-depleted (n = 21), and HIM-10-depleted (n = 22; not shown; see Supplementary Videos 11, 12) embryos. Time-aligned projections of representative 3D movies are shown (see also Supplementary Videos 7-14). The times after NEBD are indicated on the left in seconds. Bar, 5 μm. (F) The distance between spindle poles was tracked for 11 CeNDC-80-depleted embryos filmed as in E. The plot shows average pole-to-pole distance versus time after NEBD. Wild-type and KNL-1-depleted tracking data are replotted from Figure 1C. Error bars represent the S.E.M. with a confidence interval of 0.95. The average times of the onset of cytokinesis (all three conditions) and the average time when chromosome separation was first visible (wild-type and CeNDC-80-depleted embryos) are also indicated.
We confirmed that the two proteins copurifying at near-stoichiometric levels with KNL-1 were CeNDC-80 and HIM-10 by generating affinity-purified antibodies (Fig. 4B) and using them to analyze KNL-1 immunoprecipitates (Fig. 4C). CeCENP-C was also detected in KNL-1 immunoprecipitates, whereas CeCENP-A was not (Fig. 4C). Because we could not identify a band corresponding to CeCENP-C by silver staining, we validated this interaction by showing that CeCENP-C immunoprecipitates also contain KNL-1 (Fig. 4D). CeNDC-80 and HIM-10 were present at very low levels in CeCENP-C immunoprecipitates, suggesting that KNL-1 can interact with CeCENP-C independently of CeNDC-80 or HIM-10. CeCENP-C immunoprecipitates did not contain CeCENP-A, although soluble CeCENP-A was present in the extracts, suggesting that either these two widely conserved proteins interact exclusively in the context of centromeric chromatin or there are unknown components that mediate their interaction at the kinetochore.
Depletion of CeNDC-80 or HIM-10 results in a less severe chromosome-segregation defect than depletion of KNL-1
Our biochemical analysis demonstrated the existence of a soluble complex containing KNL-1, CeNDC-80, and HIM-10. If this complex functions as a structural unit at kinetochores, then depletion of CeNDC-80 or HIM-10 would be predicted to result in a kinetochore-null phenotype. To test this idea, we used live analysis to characterize the first mitotic division in embryos depleted of CeNDC-80 (Supplementary Videos 9, 10, 13) or HIM-10 (Supplementary Videos 11, 12, 14). Qualitatively, both perturbations generated an identical phenotype, and simultaneous depletion of CeNDC-80 and HIM-10 did not increase the severity of the phenotype (data not shown). Contrary to our expectation, depletion of CeNDC-80/HIM-10 resulted in a significantly different phenotype from depletion of KNL-1 (Fig. 4E; for simplicity, only a CeNDC-80-depleted embryo is shown). Mitotic chromosomes in CeNDC-80/HIM-10-depleted embryos, unlike those in KNL-1-depleted embryos, distribute over the central region of the spindle to form a disorganized metaphase plate (Fig. 4E; 100-sec and 130-sec panels). CeNDC-80-depleted (and HIM-10-depleted) embryos also separate their chromosomes, in contrast to KNL-1-depleted embryos, although this separation occurs later than in wild-type embryos and missegregation is visible (Fig. 4E).
Quantitative tracking revealed that in CeNDC-80-depleted embryos, spindle poles initiate premature separation with identical kinetics to KNL-1-depleted embryos (Fig. 4F), suggesting that CeNDC-80 is required to form a robust kinetochore-microtubule interface in the early stages of mitosis. However, in contrast to the persistent rapid elongation observed in KNL-1-depleted embryos, pole elongation slowed in CeNDC-80-depleted embryos concurrently with anaphase onset in wild type, and 34 sec (on average) later, two equivalently sized but misshapen masses of chromosomes separated along the spindle axis. This phenotype suggests that in CeNDC-80-depleted embryos, kinetochores fail to form a mechanically stable interface but interact sufficiently with microtubules to form a disorganized metaphase plate, to partially resist astral pulling forces and to imperfectly segregate chromosomes. Thus, as expected from prior work, CeNDC-80 and HIM-10 play an important role in formation of proper kinetochore-microtubule interactions and accurate chromosome segregation. However, although they form a tight complex with KNL-1, these two conserved kinetochore proteins are not functionally equivalent to KNL-1 during mitotic chromosome segregation, and their inhibition does not result in a kinetochore-null phenotype.
Asymmetric codependency of KNL-1, CeNDC-80, and HIM-10 during kinetochore assembly
The phenotypic differences between KNL-1 and CeNDC-80/HIM-10 depletions prompted us to examine the relationship between these three proteins during kinetochore assembly. Neither CeNDC-80 nor HIM-10 localized to kinetochores in embryos depleted of CeNDC-80, HIM-10, or KNL-1 (Fig. 5A), indicating that CeNDC-80 and HIM-10 are codependent for kinetochore assembly, and both proteins require KNL-1 for their targeting. In contrast, KNL-1 still localized to kinetochores in embryos depleted of either CeNDC-80 or HIM-10 (Fig. 5B). However, the level of kinetochore-localized KNL-1 was significantly reduced relative to identically processed control embryos (Fig. 5B). These results suggest that, although CeNDC-80 and HIM-10 are not absolutely required for KNL-1 targeting, they are required for KNL-1 to attain wild-type levels at kinetochores.
The observed differences between KNL-1 and CeNDC-80/HIM-10, in depletion phenotypes and in kinetochore-targeting dependencies, could arise from variable penetrance of RNAi-mediated depletion. Immunofluorescence analysis suggested that RNAi was equally effective in depleting all three proteins, but to more rigorously exclude this technical caveat, we performed quantitative immunoblotting of RNAi-depleted worms. The blotting analysis revealed that KNL-1 and CeNDC-80 are both >95% depleted, whereas HIM-10 is at least 90% depleted (Fig. 5C). Thus, the differences between KNL-1 and CeNDC80/HIM-10 depletions are unlikely to arise from variable penetrance of RNAi.
The blotting analysis also revealed that depletion of KNL-1 did not significantly alter levels of CeNDC-80 and HIM-10, indicating that their failure to target in KNL-1-depleted embryos is caused by the absence of their chromosomal binding site. However, depletion of CeNDC-80 significantly affected the stability of HIM-10 and vice versa (reducing levels of the other protein by up to 75%), consistent with their phenotypic similarity and codependency for kinetochore targeting. Finally, KNL-1 was reduced to ∼50% of wild-type levels in depletions of either CeNDC-80 or HIM-10, suggesting that reduction of KNL-1 protein levels may contribute to the observed reduction of kinetochore-localized KNL-1 in embryos depleted of CeNDC-80 or HIM-10 (Fig. 5B).
KNL-1 is required to target multiple outer kinetochore components
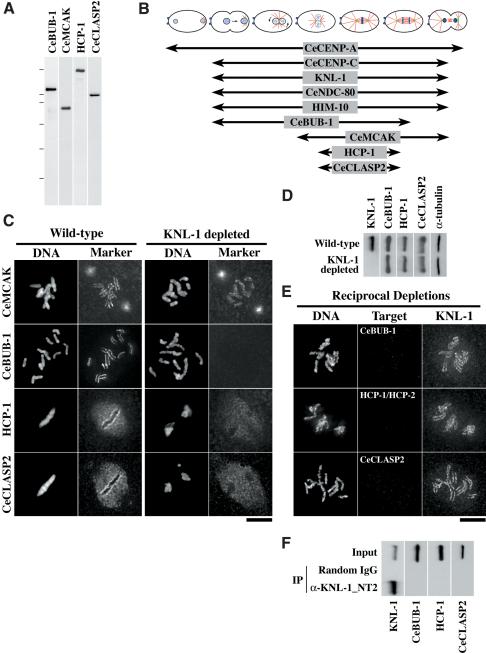
Simultaneous depletion of CeNDC-80 and HIM-10 results in a less severe phenotype than depletion of KNL-1, suggesting that loss of CeNDC-80 and HIM-10 from kinetochores in KNL-1-depleted embryos fails to fully account for KNL-1 function during chromosome segregation. One possible explanation for this finding is that KNL-1 is also required to target other functionally important components of the outer kinetochore. Thus, in CeNDC-80-depleted (or HIM-10-depleted) embryos, reduced levels of kinetochore-localized KNL-1 might suffice to form a partially functional outer kinetochore. To test this idea, we generated affinity-purified antibodies to four other conserved kinetochore components (Fig. 6A) and used them, in conjunction with the antibodies described previously, to define the molecular composition of kinetochores during the first mitotic division of the C. elegans embryo (Fig. 6B; Supplementary Fig. 1). The temporal localization pattern of KNL-1 was essentially identical to that of CeCENP-C, CeNDC-80, and HIM-10, consistent with the biochemical interactions observed for these proteins. CeBUB-1, the C. elegans homolog of the Bub1 protein kinase implicated in mitotic checkpoint function and chromosome segregation, is first observed at kinetochores at the same stage as KNL-1. However, CeBUB-1 levels at kinetochores drop significantly during anaphase (Oegema et al. 2001; Supplementary Fig. 1). CeMCAK, the C. elegans homolog of microtubule-destabilizing Kin I kinesins, localizes to spindle poles and kinetochores. CeMCAK first appears at kinetochores after NEBD and persists throughout anaphase (Oegema et al. 2001; Supplementary Fig. 1). HCP-1, a large coiled-coil-containing protein with weak homology to the mammalian kinetochore protein CENP-F (Moore et al. 1999), and CeCLASP-2, the C. elegans homolog of CLASP-2/Orbit/Mast microtubule-associated proteins (Inoue et al. 2000; Lemos et al. 2000; Akhmanova et al. 2001; Maiato et al. 2002), are only prominent at kinetochores during a relatively narrow time window near metaphase (Fig. 6B,C; Supplementary Fig. 1). In addition to kinetochores, these last two proteins localize strongly to a “matrix” in the region of the spindle (Fig. 6C; Supplementary Fig. 1).
Figure 6.
KNL-1 is required to target multiple outer kinetochore components. (A) Western blots of embryo extracts probed with affinity-purified antibodies against the indicated C. elegans kinetochore components. The molecular mass markers were 200, 116.25, 97.4, 66.2, 45, and 31 kD. (B) Schematic summarizing when during the first embryonic mitosis the nine proteins analyzed in this study localize to kinetochores (Supplementary Fig. 1; Oegema et al. 2001). (C) Wild-type and KNL-1-depleted embryos were stained for DNA (left panels in each pair), KNL-1 (not shown), and one of four kinetochore markers (right panels in each pair) as indicated. KNL-1 was not detectable by immunofluorescence in all cases where it was depleted (data not shown). CeMCAK targets normally to kinetochores in KNL-1-depleted embryos (n = 63), but CeBUB-1 (n = 50), HCP-1 (n = 21), and CeCLASP2 (n = 19) fail to target. (D) Westerns blots of wild-type and KNL-1-depleted worms were probed with antibodies to the three proteins that require KNL-1 for kinetochore targeting. α-Tubulin blotting was used as a loading control. (E) Embryos depleted of CeBUB-1 (n = 55), HCP-1 and HCP-2 (n = 37), or CeCLASP2 (n = 33) were stained for DNA (left panels), the depleted component (middle panels), and KNL-1 (right panels). KNL-1 targeted normally to kinetochores in all cases. (F) Western blots of KNL-1 immunoprecipitates probed with antibodies against the three proteins (CeBUB-1, HCP-1, and CeCLASP2) that require KNL-1 for kinetochore targeting. The control blot panel for KNL-1 is the same as in Figure 4C.
Remarkably, depletion of KNL-1 abolished kinetochore targeting of CeBUB-1, HCP-1, and CeCLASP-2 but not of CeMCAK (Fig. 6C). Immunoblotting of wild-type and KNL-1-depleted embryos excluded the possibility that targeting failures were an indirect consequence of reduction in protein levels (Fig. 6D). To identify potential targeting codependencies, we performed reciprocal depletion experiments. Depletion of CeBUB-1, HCP-1, or CeCLASP-2 had no effect on the kinetochore targeting of KNL-1 (Fig. 6E). Because HCP-1 is functionally redundant with HCP-2 (Moore et al. 1999), the reverse RNAi was performed using a mixture of HCP-1/HCP-2 double-stranded RNAs, and depletion was assessed using a mixture of anti-HCP-1 and anti-HCP-2 antibodies.
The above results indicate that KNL-1 is upstream of CeBUB-1, HCP-1, and CeCLASP-2 in the kinetochore assembly hierarchy. None of these proteins was detected in KNL-1 immunoprecipitates (Fig. 6F), suggesting that KNL-1-dependent targeting of these three components is likely to involve additional intermediates and/or a higher-order structure formed on the chromosome. Cumulatively, these results suggest that the kinetochore-null phenotype of KNL-1-depleted embryos arises from a failure to form the microtubule-binding outer domain of the kinetochore.
Architectural logic of a metazoan kinetochore
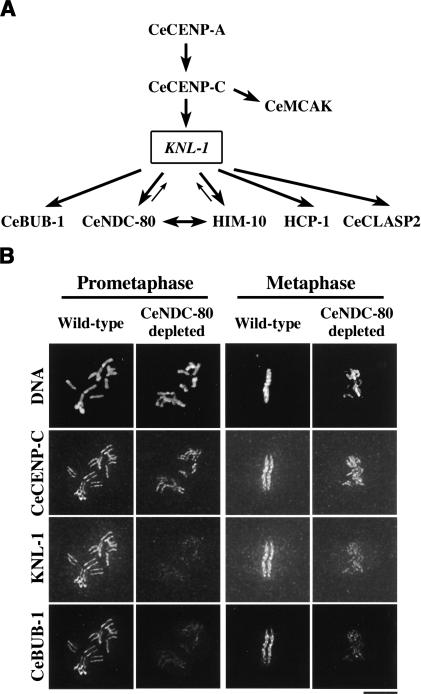
The results described above lead us to propose an architectural hierarchy for the mitotic C. elegans kinetochore (Fig. 7A). In this hierarchy, KNL-1 plays a central role by linking the DNA-proximal elements of the kinetochore to its outer microtubule-binding domain. One interesting prediction of this hierarchy is that depletion of CeNDC-80/HIM-10 should reduce the levels of other outer kinetochore components via a transitive effect through reduction of kinetochore-associated KNL-1. In contrast, components above KNL-1 in the assembly hierarchy, such as CeCENP-C, should not be affected. To test this prediction, we simultaneously analyzed localization of CeCENP-C, KNL-1, and CeBUB-1 in wild-type and CeNDC-80-depleted embryos, using equivalent imaging conditions. Consistent with the prediction, depletion of CeNDC-80 did not significantly reduce the levels of kinetochore-localized CeCENP-C (Fig. 7B). In contrast, both KNL-1 and CeBUB-1 were reduced to similar extents at kinetochores (Fig. 7B), supporting the proposed hierarchy.
Figure 7.
The architectural logic of a mitotic metazoan kinetochore. (A) Schematic of the kinetochore assembly hierarchy in C. elegans based on phenotypic analysis, targeting dependencies, and biochemical data. KNL-1 plays a central role in propagating the initiation of kinetochore assembly by CeCENP-A and CeCENP-C into the formation of a functional microtubule-binding interface. (B) A test of the proposed kinetochore assembly hierarchy. Wild-type and CeNDC-80-depleted embryos were fixed and stained for DNA (top row), CeCENP-C (second row), KNL-1 (third row), and CeBUB-1 (bottom row). Exposures for each kinetochore marker were identical for wild-type and CeNDC-80-depleted embryos. Images were equivalently processed to facilitate comparisons of staining intensities. Similar diminution of KNL-1 and CeBUB-1 staining, but not of CeCENP-C staining, was observed in 16 prophase and 20 prometaphase-early anaphase CeNDC-80-depleted one-cell-stage embryos. Bar, 5 μm.
Discussion
A functional genomic strategy for identification of core kinetochore components
Nearly 40 years ago, the ultrastructure of the kinetochore was defined by electron microscopy. Following chemical fixation, kinetochores in most eukaryotes have a multilayered structure (for review, see Rieder 1982). Pioneering work with human autoimmune sera identified CENP-A and CENP-C, two constituents of the inner plate, the chromatin-proximal structural foundation of the kinetochore. Distal to the chromatin, microtubule plus ends directly embed in the outer plate of the kinetochore, forming a dynamic interface that couples kinetochore motility to changes in the polymerization dynamics of bound microtubules (Rieder and Salmon 1998). Whereas the majority of characterized outer kinetochore proteins are depleted from kinetochores concomitant with microtubule attachment (Hoffman et al. 2001), the outer plate of the kinetochore persists as a morphologically discrete structural entity throughout mitosis, with a stable complement of embedded microtubule ends (Rieder 1982). This discrepancy suggests that important structural components of the kinetochore outer plate remain to be identified.
We have established a function-based genomic approach in the one-cell stage C. elegans embryo to identify core kinetochore components. Using high-resolution fluorescence assays to rescreen gene products identified by functional genomic analysis of Chromosome III, we identified KNL-1, a kinetochore protein that translates the initiation of kinetochore assembly by CeCENP-A and CeCENP-C into the formation of a microtubule-binding interface on mitotic chromosomes. Extension of this approach to the rest of the genome should generate a comprehensive function-based molecular view of a metazoan kinetochore.
KNL-1 is a key target of CeCENP-A and CeCENP-C in the mitotic C. elegans kinetochore
CeCENP-A, CeCENP-C, and KNL-1 are the only known C. elegans proteins whose depletion results in a kinetochore-null phenotype. Depletion of other known kinetochore or chromosomal proteins does not result in this phenotype but is instead associated with less severe chromosome segregation defects (Oegema et al. 2001; Hagstrom et al. 2002; this study). Three pieces of evidence place KNL-1 downstream of CeCENP-A and CeCENP-C in the kinetochore assembly hierarchy. First, depletion of KNL-1 does not affect the chromosomal localization of CeCENP-A or CeCENP-C. Second, depletion of either CeCENP-A or CeCENP-C prevents KNL-1 from localizing to chromosomes. Third, analysis of protein levels excludes indirect effects on targeting caused by changes in KNL-1 stability. Cumulatively, these findings support a linear assembly hierarchy and exclude codependencies during assembly. In agreement with the proposed placement of KNL-1, depletions of CeCENP-A or CeCENP-C result in chromosome structure defects that are absent in KNL-1-depleted embryos. In addition, the presence of well-condensed chromosomes in KNL-1-depleted embryos with two “lines” of CeCENP-A and CeCENP-C staining indicates that KNL-1 is not required to establish the holocentric nature of C. elegans chromosomes.
KNL-1 is required to assemble the microtubule-binding interface of the mitotic C. elegans kinetochore
The kinetochore-null defect observed in KNL-1-depleted embryos is likely explained by its role in targeting multiple components of the outer kinetochore. Five conserved outer kinetochore proteins—CeNDC-80, HIM-10, CeBUB-1, HCP-1, and CeCLASP2—require KNL-1 to target to chromosomes. Of these, CeBUB-1, HCP-1, and CeCLASP2 are not present in KNL-1 immunoprecipitates, suggesting that additional components may mediate their interaction with KNL-1. In contrast, KNL-1 forms a near-stoichiometric complex with CeNDC-80 and HIM-10. Consistent with this interaction, depletion of CeNDC-80/HIM-10 reduces but does not eliminate kinetochore-localized KNL-1. The mechanical integrity of the microtubule-binding interface of the kinetochore is severely impaired in embryos depleted of KNL-1/CeNDC-80/HIM-10, suggesting that this protein complex is required to build a stable kinetochore-microtubule interface. KNL-1 lacks any known microtubule-binding motif and is an acidic protein with an isoelectric point of 4.9, whereas most known microtubule-binding proteins are basic. Although this suggests that KNL-1 does not directly bind microtubules, biochemical reconstitution of the KNL-1 complex or its purification in a native state will be necessary to analyze microtubule-binding activity.
Interestingly, CeMCAK, the C. elegans member of the microtubule-destabilizing Kin I kinesin subfamily, does not require KNL-1 to target to kinetochores, although it does require CeCENP-A and CeCENP-C (Oegema et al. 2001). In contrast to vertebrates, where MCAK exhibits a complex localization pattern (Wordeman and Mitchison 1995), CeMCAK cannot be spatially distinguished on chromosomes from CeCENP-A, CeCENP-C, and KNL-1 by light microscopy. Nevertheless, the independence of CeMCAK targeting from KNL-1 and the relative absence of defects in chromosome-microtubule interactions in embryos depleted of CeMCAK (Grill et al. 2001), both argue for a functional distinction between CeMCAK and outer kinetochore components. The biochemical activity of Kin I kinesins as microtubule destabilizers (Desai et al. 1999) is consistent with this distinction, as it appears likely that microtubule binders and destabilizers are segregated within the fine structure of the kinetochore.
Relationship between KNL-1 and CeNDC-80/HIM-10
Despite its presence in a near-stoichiometric complex with CeNDC-80 and HIM-10, KNL-1 is functionally more important than these two conserved proteins during chromosome segregation. Depletion of KNL-1 results in a “kinetochore-null” phenotype, whereas depletion of CeNDC-80 and/or HIM-10 leads to a less severe chromosome segregation defect. In addition, whereas KNL-1 is absolutely required to target CeNDC-80 and HIM-10 to kinetochores, depletion of CeNDC-80 or HIM-10 reduces but does not eliminate kinetochore-localized KNL-1. The reduction of kinetochore-localized KNL-1 (and CeBUB-1; Fig. 7) in embryos depleted of CeNDC-80 is not due to the action of spindle microtubules as it is clearly evident prior to nuclear envelope breakdown. Cumulatively, the asymmetric targeting dependencies and differences in phenotypic severity suggest that Ndc80p/Nuf2p-associated proteins, such as KNL-1, are of significantly greater functional importance than previously appreciated.
Biochemical analysis of Ndc80p/Nuf2p-containing complexes in budding yeast has revealed the presence of two associated proteins, Spc24p and Spc25p (Janke et al. 2001; Wigge and Kilmartin 2001). Neither of these two proteins has recognizable sequence homologs outside of fungi. A greater severity for the phenotype of spc24 and spc25 mutants relative to ndc80 and nuf2 mutants has been suggested in the context of mitotic checkpoint activation (Janke et al. 2001; Wigge and Kilmartin 2001; McCleland et al. 2003). However, a comparative analysis of kinetochore assembly in different mutants of ndc80, nuf2, spc24, and spc25 has not been performed. It should be noted that the effect we observed, namely, a diminution of outer kinetochore components (Fig. 7B), might be difficult, if not impossible, to establish in the point kinetochores of budding yeast, but may be addressable in vertebrates.
KNL-1 is the first Ndc80p/Nuf2p-associated protein identified outside of fungi. The existence of a functional ortholog of KNL-1 in vertebrates is suggested by the inability to functionally complement Xenopus egg extracts immunodepleted of their Ndc80p or Nuf2p homologs using purified proteins (McCleland et al. 2003). A functional KNL-1 ortholog could also explain the seemingly contradictory effects generated by antibody injection versus RNAi-mediated depletion of vertebrate Ndc80p/Nuf2p homologs (DeLuca et al. 2002; Martin-Lluesma et al. 2002; McCleland et al. 2003). RNAi-mediated depletion of either HEC1 (human Ndc80p) or hNuf2 results in a failure to generate proper kinetochore-microtubule attachments and persistent activation of the mitotic checkpoint (DeLuca et al. 2002; Martin-Lluesma et al. 2002). In contrast, inhibition of Xenopus Ndc80p or Nuf2p by antibody injection results in a more severe phenotype—premature exit from mitosis without any detectable chromosome congression or anaphase movements (McCleland et al. 2003). Our analysis of KNL-1 suggests that coinhibition of an Ndc80p/Nuf2p-associated protein, likely to occur by antibody injection but not by RNAi-mediated depletion, may account for this discrepancy.
Variation in kinetochore structure—it's not just the DNA
The kinetochore offers a particularly fascinating puzzle for students of essential cellular protein machines. The lack of a common theme to the DNA sequences upon which kinetochores assemble has been appreciated for quite some time. Some similarity in their protein composition was expected given that kinetochores interact with microtubules, polymers formed from one of the most conserved proteins in eukaryotes. However, the accumulating evidence for commonalities in kinetochore composition must be tempered by the very low sequence similarity of “conserved” kinetochore components between different species and the presence of many components apparently unique to closely related species. Our analysis of KNL-1 suggests that the junction between the DNA-proximal foundation for kinetochore assembly, established by CENP-A and CENP-C, and the outer kinetochore is formed by proteins that have diverged significantly during evolution. This finding suggests that variation in kinetochore structure is not just restricted to the interface between the DNA and centromere-specific histone variants (Malik and Henikoff 2002) but may extend further down the assembly hierarchy. Such variation may be linked to the postulated role of centromeres/kinetochores in the establishment and/or maintenance of species barriers (Henikoff and Malik 2002).
Materials and methods
All fixed images and movies, as well as all reported n values, are exclusively of mitotic one-cell-stage embryos.
dsRNA and antibody production
Oligonucleotides used for dsRNA and antigen production are listed in Supplementary Table 1. DNA templates for dsRNA synthesis were amplified from N2 genomic DNA. For antibody production, fragments of the coding region from either genespecific or first-strand worm cDNA templates were cloned into pGEX6P-1 (Amersham). Purified GST fusions were outsourced for injection into rabbits. For affinity purification, GST fusions were cleaved using Prescission protease, and antigen fragments were covalently coupled to HiTrap-NHS columns (Amersham). For two of the antibodies [anti-KNL1_NT2(aa8-256), anti-CeCLASP2], rabbit sera were depleted of anti-GST antibodies by four passages over a GST alone-coupled column and then affinity-purified on columns with covalently coupled uncleaved antigens. Purified antibodies were directly labeled with Cy3 and Cy5 fluorescent dyes as described (Francis-Lang et al. 1999).
RNAi and blotting of dsRNA-injected worms
L4 hermaphrodites were injected with dsRNA and incubated either at 16°C for 45-48 h or at 20°C for 36 h prior to analysis. For immunoblotting following RNAi, 60 injected worms were picked into 0.5 mL of M9, gently pelleted, and washed three times with 500 μL of M9. All but 15 μL of M9 was removed, an equal volume of 2× SDS-PAGE sample buffer was added, and the sample was sonicated at 80°C in a bath sonicator for 15 min. Then 10 μL of the sonicated sample was used for Western blotting and analysis of protein levels using ECL Plus (Amersham). Equal numbers of control and injected worms were always processed in parallel, and α-tubulin blotting was used as a loading control.
Live imaging
Mounting for live analysis and spinning disk confocal microscopy of embryos expressing GFP-β-tubulin was performed as described (Oegema et al. 2001). Chromosome segregation was followed in embryos expressing GFP-histone and GFP-γ-tubulin using a DeltaVision deconvolution microscope (Applied Precision) equipped with a CoolSnap CCD camera (Roper Scientific). At 10-sec intervals, six z-sections were acquired at 2-μm steps using a 100×, 1.3 NA Olympus U-Planapo objective with 2 × 2 binning and a 384 × 384-pixel area. Illumination was attenuated using a 10% transmission neutral density filter, and each exposure was 250 msec. Quantitative analysis of spindle pole elongation was performed on movies where the two poles remained in the same or neighboring focal planes. Z stacks were projected and imported into Metamorph (Universal Imaging) and spindle poles were manually tracked. Quantitative estimates suggest there is a <5% distance measurement error from projection of the stacks. Average plots were generated by aligning sequences using NEBD as a time reference. NEBD was defined as the time when GFP fluorescence was equalized between both pronuclei and the cytoplasm. Cytokinesis onset was defined as the time with the first visible sign of cleavage furrow ingression. Tracking data were analyzed and plotted using a custom macro in Microsoft Excel. Error bars represent the S.E.M. with a confidence interval of 0.95 (Grill et al. 2001).
Immunofluorescence
In pairwise depletion-localization assays, ∼60 injected worms were processed in parallel with an equal number of uninjected worms using freeze-cracking followed by -20°C methanol fixation. The time of fixation was chosen to maximize sensitivity for detecting the depleted component. A mixture of directly labeled antibodies to α-tubulin (FITC; Sigma), RNAi target (Cy3/Cy5), and assayed component (Cy5/Cy3) was used after blocking slides with AbDil (PBS + 0.1% Triton X-100 + 4% BSA). Prior to mounting, slides were incubated for 5 min with 1 μg/mL Hoechst 33342 to label the DNA. Four-color 3D stacks at 0.2-μm steps were collected throughout the volume of the embryo using an Olympus 100×, 1.3 NA U-Planapo objective, computationally deconvolved and projected for presentation. Exposure times were chosen to limit maximal intensities in control embryos at ∼70% camera saturation. For the experiment in Figure 7B, a mixture of directly labeled antibodies to CeCENP-C (FITC), KNL-1 (Cy3), and CeBUB-1 (Cy5) was used, and exposure times and subsequent image processing steps were kept constant between control and depleted embryos. Immuno-EM was performed as described (Kirkham et al. 2003).
Immunoprecipitations and mass spectrometry
Synchronous worm cultures were generated using starved L1 larvae. Embryo isolation was performed by alkaline bleach dissolution of adult worms, soon after they initiated embryo production. The volume of the final embryo pellet, estimated from its wet weight, was mixed with an equal volume of H100 buffer (50 mM HEPES at pH 7.5, 100 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10% glycerol, 0.05% NP-40, 1 mM DTT, protease inhibitors) and sonicated for a total of 3 min at 40% amplitude (microtip on model Branson 450) with intermittent cooling on ice. Crude extracts were clarified at 4°C at 20,000 rpm (22,000g) for 10 min followed by 50,000 rpm (135,000g) for 20 min in a TLA100.3 rotor. Twenty-five micrograms of affinity-purified or control rabbit antibodies were covalently linked to Protein A beads, pre-eluted with glycine (Moritz et al. 1998), incubated with 700-1000 μL of fresh extract at 4°C for 1 h, washed five times with HB100 and once with HB100 without NP-40, and eluted by heating at 50°C in 50 μL of 2× SDS-PAGE Sample Buffer without DTT. The eluted supernatant was supplemented with DTT (100 mM final) and boiled for 3 min. Then 5 μL was loaded for silver staining and 25 μL for protein identification. Coomassie-stained bands were in-gel-digested with trypsin (Shevchenko et al. 1996), and proteins were identified by MALDI MS peptide mapping (Havlis et al. 2003). For immunoblots, the elution was diluted 20-fold, and 5-10 μL was loaded per lane. Immunoblots were performed using 0.5-1 μg/mL affinity-purified primary antibody and HRP-linked anti-rabbit secondary antibody and detected using ECL (Amersham).
Acknowledgments
We are grateful to Yuji Kohara (National Institute of Genetics, Mishima, Japan) for genespecific cDNAs, Martin Srayko (Max Planck Institute, Dresden, Germany) for worm cDNA, the Caenorhabditis Genetics Center (University of Minnesota, St. Paul, MN, USA) for strains, Bianca Habermann for help with bioinformatics, and Iain Cheeseman and Paul Maddox for comments on the manuscript. A.D. was funded by EMBO and the American Cancer Society; K.O. was funded by the Helen Hay Whitney Foundation and the Max Planck Institute. A.D. and K.O. are presently supported by the Ludwig Institute for Cancer Research.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1126303.
Corresponding authors.
References
- Akhmanova A., Hoogenraad, C.C., Drabek, K., Stepanova, T., Dortland, B., Verkerk, T., Vermeulen, W., Burgering, B.M., De Zeeuw, C.I., Grosveld, F., et al. 2001. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell 104: 923-935. [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Anderson, S., Jwa, M., Green, E.M., Kang, J., Yates III, J.R., Chan, C.S., Drubin, D.G., and Barnes, G. 2002a. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111: 163-172. [DOI] [PubMed] [Google Scholar]
- Chen Y., Riley, D.J., Chen, P.L., and Lee, W.H. 1997. HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol. Cell. Biol. 17: 6049-6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D.W., Mao, Y., and Sullivan, K.F. 2003. Centromeres and kinetochores: From epigenetics to mitotic checkpoint signaling. Cell 112: 407-421. [DOI] [PubMed] [Google Scholar]
- DeLuca J.G., Moree, B., Hickey, J.M., Kilmartin, J.V., and Salmon, E.D. 2002. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J. Cell Biol. 159: 549-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Verma, S., Mitchison, T.J., and Walczak, C.E. 1999. Kin I kinesins are microtubule-destabilizing enzymes. Cell 96: 69-78. [DOI] [PubMed] [Google Scholar]
- Francis-Lang H., Minden, J., Sullivan, W., and Oegema, K. 1999. Live confocal analysis with fluorescently labeled proteins. Methods Mol. Biol. 122: 223-239. [DOI] [PubMed] [Google Scholar]
- Gönczy P., Echeverri, C., Oegema, K., Coulson, A., Jones, S.J., Copley, R.R., Duperon, J., Oegema, J., Brehm, M., Cassin, E., et al. 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408: 331-336. [DOI] [PubMed] [Google Scholar]
- Goshima G., Saitoh, S., and Yanagida, M. 1999. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes & Dev. 13: 1664-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Kiyomitsu, T., Yoda, K., and Yanagida, M. 2003. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160: 25-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill S.W., Gonczy, P., Stelzer, E.H., and Hyman, A.A. 2001. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature 409: 630-633. [DOI] [PubMed] [Google Scholar]
- Hagstrom K.A., Holmes, V.F., Cozzarelli, N.R., and Meyer, B.J. 2002. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes & Dev. 16: 729-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havlis J., Thomas, H., Sebela, M., and Shevchenko, A. 2003. Fast-response proteomics by accelerated in-gel digestion of proteins. Anal. Chem. 75: 1300-1306. [DOI] [PubMed] [Google Scholar]
- He X., Rines, D.R., Espelin, C.W., and Sorger, P.K. 2001. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell 106: 195-206. [DOI] [PubMed] [Google Scholar]
- Henikoff S. and Malik, H.S. 2002. Centromeres: Selfish drivers. Nature 417: 227. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Ahmad, K., and Malik, H.S. 2001. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293: 1098-1102. [DOI] [PubMed] [Google Scholar]
- Hoffman D.B., Pearson, C.G., Yen, T.J., Howell, B.J., and Salmon, E.D. 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell 12: 1995-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe M., McDonald, K.L., Albertson, D.G., and Meyer, B.J. 2001. HIM-10 is required for kinetochore structure and function on Caenorhabditis elegans holocentric chromosomes. J. Cell Biol. 153: 1227-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y.H., do Carmo Avides, M., Shiraki, M., Deak, P., Yamaguchi, M., Nishimoto, Y., Matsukage, A., and Glover, D.M. 2000. Orbit, a novel microtubule-associated protein essential for mitosis in Drosophila melanogaster. J. Cell Biol. 149: 153-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Ortiz, J., Lechner, J., Shevchenko, A., Magiera, M.M., Schramm, C., and Schiebel, E. 2001. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 20: 777-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R.S., Fraser, A.G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231-237. [DOI] [PubMed] [Google Scholar]
- Kirkham M., Müller-Reichert, T., Oegema, K., Grill, S., and Hyman, A.A. 2003. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell 112: 575-587. [DOI] [PubMed] [Google Scholar]
- Kitagawa K. and Hieter, P. 2001. Evolutionary conservation between budding yeast and human kinetochores. Nat. Rev. Mol. Cell. Biol. 2: 678-687. [DOI] [PubMed] [Google Scholar]
- Lemos C.L., Sampaio, P., Maiato, H., Costa, M., Omel'yanchuk, L.V., Liberal, V., and Sunkel, C.E. 2000. Mast, a conserved microtubule-associated protein required for bipolar mitotic spindle organization. EMBO J. 19: 3668-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Sampaio, P., Lemos, C.L., Findlay, J., Carmena, M., Earnshaw, W.C., and Sunkel, C.E. 2002. MAST/Orbit has a role in microtubule-kinetochore attachment and is essential for chromosome alignment and maintenance of spindle bipolarity. J. Cell Biol. 157: 749-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik H.S. and Henikoff, S. 2002. Conflict begets complexity: The evolution of centromeres. Curr. Opin. Genet. Dev. 12: 711-718. [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma S., Stucke, V.M., and Nigg, E.A. 2002. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297: 2267-2270. [DOI] [PubMed] [Google Scholar]
- McCleland M.L., Gardner, R.D., Kallio, M.J., Daum, J.R., Gorbsky, G.J., Burke, D.J., and Stukenberg, P.T. 2003. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes & Dev. 17: 101-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L.L. and Roth, M.B. 2001. HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J. Cell Biol. 153: 1199-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L.L., Morrison, M., and Roth, M.B. 1999. HCP-1, a protein involved in chromosome segregation, is localized to the centromere of mitotic chromosomes in Caenorhabditis elegans. J. Cell Biol. 147: 471-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M., Zheng, Y., Alberts, B.M., and Oegema, K. 1998. Recruitment of the γ-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J. Cell Biol. 142: 775-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabetani A., Koujin, T., Tsutsumi, C., Haraguchi, T., and Hiraoka, Y. 2001. A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: A link between the kinetochore function and the spindle checkpoint. Chromosoma 110: 322-334. [DOI] [PubMed] [Google Scholar]
- Oegema K., Desai, A., Rybina, S., Kirkham, M., and Hyman, A.A. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153: 1209-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L. 1982. The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int. Rev. Cytol. 79: 1-58. [DOI] [PubMed] [Google Scholar]
- Rieder C.L. and Salmon, E.D. 1998. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 8: 310-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H., Tomkiel, J., Cooke, C.A., Ratrie III, H., Maurer, M., Rothfield, N.F., and Earnshaw, W.C. 1992. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell 70: 115-125. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm, M., Vorm, O., and Mann, M. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68: 850-858. [DOI] [PubMed] [Google Scholar]
- Sullivan B.A., Blower, M.D., and Karpen, G.H. 2001. Determining centromere identity: Cyclical stories and forking paths. Nat. Rev. Genet. 2: 584-596. [DOI] [PubMed] [Google Scholar]
- Sullivan K.F. 2001. A solid foundation: Functional specialization of centromeric chromatin. Curr. Opin. Genet. Dev. 11: 182-188. [DOI] [PubMed] [Google Scholar]
- Wigge P.A. and Kilmartin, J.V. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152: 349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P.A., Jensen, O.N., Holmes, S., Soues, S., Mann, M., and Kilmartin, J.V. 1998. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 141: 967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L. and Mitchison, T.J. 1995. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J. Cell Biol. 128: 95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipperlen P., Fraser, A.G., Kamath, R.S., Martinez-Campos, M., and Ahringer, J. 2001. Roles for 147 embryonic lethal genes on C. elegans chromosome I identified by RNA interference and video microscopy. EMBO J. 20: 3984-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]