Abstract
Fruiting body formation in Myxococcus xanthus depends on ordered changes in cell movements from swarming to aggregation in response to starvation. We show that appropriately starved individual cells change behavior during fruiting body formation. Specifically, from the time of initiation of aggregation, individual wild-type cells began to move with increased gliding speeds, the duration of the mean gliding interval increased, and the stop frequency decreased whereas the duration of the mean stop interval and the reversal frequency remained unchanged. Mutants lacking the cell surface-associated, intercellular C-signal (csgA mutants) failed to aggregate. Likewise, appropriately starved individual csgA cells did not change their behavior during development. In the absence of other cell–cell interactions, the motility defect of individual csgA cells was corrected in a time- and concentration-dependent manner after C-signaling was reestablished by exogenous MalE-CsgA protein. The C-signal-induced stimulation of motility depended on the cytoplasmic Frz signal transduction system. We propose that C-signal instructs cells to move with high speed and low stop and reversal frequencies into aggregation centers during development.
The formation of multicellular fruiting bodies in Myxococcus xanthus depends on temporally and spatially controlled changes in organized cell movements in response to starvation. M. xanthus cells move by gliding, a process whereby bacterial cells move in the direction of their long axes on a solid surface (1). After starvation, swarming behavior of vegetative cells is constrained, and cells begin to congregate in aggregation centers. Aggregation culminates 24 hr after the initiation of starvation, with the formation of haystack-shaped multicellular fruiting bodies, each of which contains ≈105 cells. Within the nascent fruiting bodies, the rod-shaped motile cells differentiate into spherical, nonmotile spores (2). A key question in understanding fruiting body formation concerns how the cells are directed into the aggregation centers.
At least five intercellular signaling systems are important for fruiting body formation (3, 4). Cells with mutations in the csgA gene do not generate the C-signal and are impaired in aggregation and sporulation (5–9). Moreover, expression of genes that are normally induced after 6 hr of starvation is reduced or abolished in csgA mutants (10). The csgA gene codes for a cell surface-associated protein that shows homology to the short-chain alcohol dehydrogenase protein family (11–13). However, it is not yet known whether CsgA acts as an enzyme to produce the C-signal or whether CsgA is the actual C-signal molecule. The developmental defects in csgA mutants are corrected by codevelopment with wild-type (wt) cells (3), by exogenous C-factor protein, which is encoded by csgA (8), or by exogenous MalE-CsgA fusion protein (12).
Several observations have suggested a correlation between C-signaling and the organized cell movements that lead to fruiting body formation. First, cells need to be motile to engage in productive C-signaling, and it has been suggested that motility is a prerequisite to establish specific end-to-end contacts between cells required for C-signal transmission (8, 14–18). Second, the earliest developmental defects in csgA mutants coincide with the first signs of aggregation in wt cells after 6 hr of starvation. Third, restored C-signaling in csgA mutants by exogenous C-factor protein or MalE-CsgA protein results in aggregation, suggesting that C-signal may induce the cell movements that lead to aggregation (8, 12). Finally, C-signal induces chemical modification of the Frz signal transduction system (17), which is involved in controlling directed cell movements (19). Thus, productive C-signaling initially requires cell motility and also may induce the changes in motility that ultimately result in aggregation.
To explore the effect of C-signal on cell behavior, we have developed a single cell motility assay. Using this assay, we provide evidence that C-signal independently of other cell–cell interactions has a direct effect on single cell motility during starvation. Specifically, cells are stimulated to move with higher transient gliding speeds in longer gliding intervals and with reduced stop frequencies. This response depends on the Frz system.
MATERIALS AND METHODS
M. xanthus Strains and Growth.
Strains DK5204 (Tn5 lac Ω4435) (20), DK5253 (Tn5 lac Ω4435, csgA∷Tn5–132 ΩLS205) (10), DK9033 (Δfrz(′CD-F)∷Kanr) (21), and DK9035 (Δfrz(′CD-F)∷Kanr, csgA∷Tn5–132 ΩLS205) (21) were grown in liquid CTT medium (10 g/liter Difco casitone/10 mM Tris·HCl, pH 8.0/1 mM KPO4, pH 7.6/8 mM MgSO4) or were maintained on CTT agar plates (22). Tn5 lac Ω4435 in DK5204 did not interfere with fruiting body formation or the motility response to Mal-CsgA (data not shown).
Purification of MalE-CsgA Protein.
MalE-CsgA protein was purified from Escherichia coli as described (12). The specific activity of the protein was 20 C-factor units (Cfu)/mg protein as determined in the C-signal bioassay; 1 Cfu is defined as the amount of protein that restores wt fruiting body formation and sporulation to 2.5 × 108 csgA cells (8). MalE was purchased from New England Biolabs. Protein concentrations were determined in the Bio-Rad protein assay as described by the manufacturer. Before being used, MalE-CsgA and MalE proteins were dialyzed against 4 liters of A50-starvation buffer (10 mM Mops, pH 7.2/1 mM CaCl2/1 mM MgCl2/50 mM NaCl) (7) for 18 hr at 4°.
Video Microscopy.
Cells were grown at 32°C in CTT medium to a density of 5 × 108 cells/ml and were harvested and resuspended in A50-buffer to a calculated density of 5 × 109 cells/ml. A portion (20 μl) of this suspension was spotted on A50–1.5% agarose and was incubated at 32°C to induce development. At the indicated timepoints, cells were gently harvested from the agarose surface, were resuspended in 50 μl of A50-buffer, and were diluted 100-fold in 400 μl of A50. A portion (5 μl) of this cell suspension was spotted on 1.5-mm-thick A50–1.5% agarose prepared on a sterile standard microscope slide. Unless otherwise indicated, cells were spotted on the A50-agarose slides 5 min after being harvested. Video microscopy of cells was initiated immediately after the droplets had dried (4–5 min). All plates were prepared 24 hr before use and were stored at 22°C. MalE-CsgA or MalE protein was added to cells after they had been diluted 100-fold. Cells were exposed to protein for the indicated periods of time in suspension at 22°C with gentle shaking. Cells on agarose slides were observed by using a Leica IMB/E inverted microscope equipped with a 20× objective using phase contrast. The microscope image was captured with a Sony DXP-935 3CCD video camera connected to a personal computer and by using the image analysis program image pro 3.0 (Media Cybernetics, Silver Spring, MD). Frames were captured every 15 s. Cells were monitored for 900 s. All observations were made at 22°C. Typical microscopic fields contained 8–12 cells. Development occurred with normal timing on A50–1.5% agarose (data not shown).
Computerized Motion Analysis of Cells.
The captured microscopic fields were calibrated by using the image of a slide micrometer. The x, y coordinates of a cell in a given frame were defined as the position of the centroid of the cell. The x, y coordinates of each cell in each frame were determined by the software and were exported to a datafile together with the time, t, of the frame capture. Transient gliding speeds were calculated as follows. The displacement, d, of a cell was determined as the shortest distance between the position of the centroid of that cell in two subsequent frames: di, i − 1 = √[(xi − xi − 1)2 + (yi − yi − 1)2]. The displacement divided by the time interval between the two frames then gives the transient gliding speed, si, in that time interval, si = di, i − 1/(ti − ti − 1). In each experiment, the transient gliding speed for each cell in each 15-s time interval was calculated. The transient gliding speeds in the first recording interval (0–15 s) were not significantly different from the transient gliding speeds in the last recording interval (885–900 s), according to a Kruskal-Wallis test (23). Therefore, an experiment in which 10 cells were followed for 900 s contributed 590 independent transient gliding speed measurements, and distributions of transient gliding speeds in the populations could be prepared. To test for the statistical difference between transient gliding speed distributions, the data sets underlying the distributions were subjected to a Kruskal-Wallis test. Mean gliding speed was calculated by using only transient gliding speeds >0.35 μm/min, the lower detection limit for active movement. Gliding intervals were calculated as the duration between a start and a stop; stop intervals were calculated as the duration between a stop and a start. A stop is a decrease in transient speed to <0.35 μm/min; a start is an increase in transient gliding speed to >0.35 μm/min. Stop frequency was calculated as the number of stops per minute of gliding with transient speeds >0.35 μm/min. Reversal frequency was calculated as the number of reversals per minute of gliding with transient speeds >0.35 μm/min. Motion analyses were performed on a total of 20–30 cells in each experiment.
RESULTS
To analyze whether cells change behavior during fruiting body morphogenesis, a motility assay was developed in which the behavior of single cells was followed. In brief, cells were starved on an agarose medium at the high cell density required for fruiting body formation. Subsequently, cells were harvested and spotted on agarose starvation medium at a low cell density, and single cell movements were recorded by using time-lapse video microscopy (see Materials and Methods). To ensure that cell movements were not influenced by interactions with other cells during the recordings, only cells that were visible in the microscopic field throughout the recording session, and were separated from other cells or slimetrails laid down by other cells by >5 μm, were included in the analyses. So, in this assay, cells are stimulated by cell–cell interactions while at the high cell density, and then their behavior is scored at the low cell density. To distinguish active cell movement from background noise, movements of cells fixed with 1.0% glutaraldehyde were recorded (data not shown). The speed of these cells was 0.20 ± 0.15 μm/min. Thus, the lower detection limit for active cell movement in this assay was 0.35 μm/min, and only gliding speeds >0.35 μm/min were taken to indicate active cell movement.
Motility Behavior of Individual wt Cells Changes During Starvation.
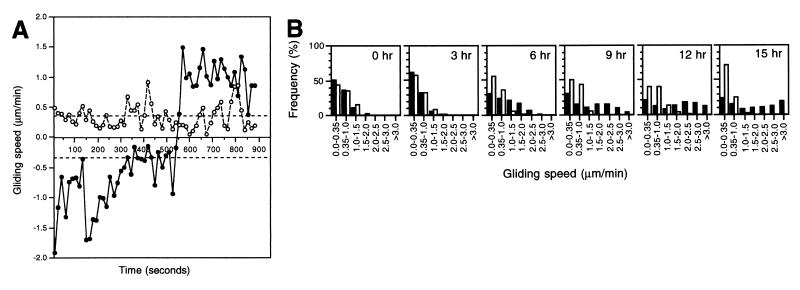
The behavior of individual cells were analyzed in terms of transient gliding speeds, mean gliding speed and duration of mean gliding and stop intervals as well as stop and reversal frequencies (see Materials and Methods for a description of how these parameters were calculated). Images of cells were recorded every 15 s for 900 s, and transient gliding speeds were calculated for each cell for each 15-s interval. Speed profiles of isolated wt cells (DK5204) that had been starved for 6 hr at the high cell density, before being recorded at the low cell density, showed that the cells did not maintain a constant speed but, rather, exhibited a range of values (Fig. 1A) in a manner similar to that of wt cells on rich medium (24). The variation in transient gliding speeds also is reflected in the broad distribution of transient gliding speeds observed for 21 isolated wt cells that had been starved for 6 hr (Fig. 1B, 6-hr panel). Because 30% of the transient gliding speeds in this distribution were <0.35 μm/min, an average cell was gliding 70% of the time under these conditions. All cells experienced periods of no active movement, with the duration of the mean gliding interval being 4.38 ± 0.80 min and the duration of the mean stop interval being 3.53 ± 0.71 min (Table 1). The stop and reversal frequencies were similar, and cells on the average reversed their direction of movement every 7.7 min (Table 1). Thus, the reversal frequency is similar to that previously reported for M. xanthus cells (25, 26).
Figure 1.
Motility parameters of individual wt and csgA cells during development. (A) Gliding speed profiles of typical individual cells that had been starved for 6 hr at a high cell density and had been harvested and spotted at the low cell density. Closed circles, wt cell (DK5204); open circles, csgA cell (DK5253). The dotted lines indicate the lower detection limit for active movement. A reversal is indicated by a change in speed from a positive to a negative value or vice versa. (B) Distributions of transient gliding speeds in individual wt (closed bars) and csgA (open bars) cells that had been starved for the indicated periods of time at a high cell density and had been harvested, diluted, and spotted at the low cell density.
Table 1.
Motility parameters of single wt and csgA cells during starvation
| Hr of starvation | Gliding speed, μm/min*
|
Duration of gliding interval, min†
|
Duration of stop interval, min‡
|
Stop frequency, stop/min§
|
Reversal frequency, reversal/min¶
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| wt | csgA | wt | csgA | wt | csgA | wt | csgA | wt | csgA | |
| 0 | 0.74 ± 0.08 | 0.87 ± 0.07 | 1.69 ± 0.43 | 2.18 ± 0.49 | 3.12 ± 0.33 | 3.23 ± 0.48 | 0.56 ± 0.05 | 0.49 ± 0.11 | 0.09 ± 0.05 | 0.11 ± 0.01 |
| 3 | 0.68 ± 0.02 | 0.54 ± 0.03 | 1.31 ± 0.30 | 1.41 ± 0.21 | 3.38 ± 0.35 | 3.29 ± 0.30 | 0.58 ± 0.12 | 0.59 ± 0.07 | 0.06 ± 0.02 | <0.05 |
| 6 | 1.06 ± 0.02 | 0.65 ± 0.08 | 4.38 ± 0.80 | 1.59 ± 0.32 | 3.53 ± 0.71 | 3.44 ± 0.35 | 0.18 ± 0.04 | 0.55 ± 0.06 | 0.13 ± 0.04 | <0.03 |
| 9 | 1.75 ± 0.26 | 0.67 ± 0.03 | 3.61 ± 0.36 | 1.33 ± 0.07 | 3.06 ± 0.44 | 2.57 ± 0.24 | 0.22 ± 0.03 | 0.69 ± 0.05 | 0.23 ± 0.01 | 0.02 ± 0.01 |
| 12 | 2.07 ± 0.10 | 0.84 ± 0.14 | 4.73 ± 0.36 | 2.00 ± 0.92 | 2.93 ± 1.17 | 2.00 ± 0.25 | 0.14 ± 0.01 | 0.62 ± 0.25 | 0.14 ± 0.01 | 0.05 ± 0.04 |
| 15 | 2.16 ± 0.19 | 0.80 ± 0.15 | 5.10 ± 0.34 | 1.42 ± 0.32 | 3.91 ± 0.87 | 4.52 ± 0.46 | 0.14 ± 0.03 | 0.75 ± 0.17 | 0.15 ± 0.05 | 0.11 ± 0.03 |
The mean values and standard errors of the mean are shown for three independent experiments. Strains used: wt, DK5204; csgA, DK5253.
Mean gliding speed was calculated by using only transient gliding speeds >0.35 μm/min, the lower detection limit for active movement.
A gliding interval was the period of time between a start and a stop.
A stop interval was the period of time between a stop and a start.
Stop frequency was calculated as the number of stops per minute of gliding with transient speeds >0.35 μm/min.
Reversal frequency was calculated as the number of gliding reversals per minute of gliding with transient speeds >0.35 μm/min.
To analyze the motility behavior systematically during development, wt cells were starved between 0 and 15 hr at the high cell density and then were recorded at the low density. The transient gliding speed distributions were broad at all timepoints for these cells (Fig. 1B). However, the distribution changed significantly during development. After 3 hr of starvation, only 40% of the transient gliding speeds were above the detection limit for active cell movement whereas, in cells starved for 6 hr, 70% of the transient gliding speeds were above this limit. After 9, 12, or 15 hr of starvation, 70, 80, and 75%, respectively, of the transient gliding speeds were above the detection limit. Also, more cells were gliding with higher transient gliding speeds the longer the cells had been starved. Consistently, the mean gliding speed increased until after 9 hr of starvation and then remained constant (Table 1). Speed profiles of individual cells showed that, at all timepoints, cells did not maintain a constant speed (Fig. 1A and data not shown). Thus, at all timepoints, the broad distributions of transient gliding speeds reflected that cells were changing speed rather than reflecting the existence of populations of cells moving with different but constant speeds. Between 3 and 6 hr, the duration of the mean gliding interval increased 3- to 4-fold whereas the duration of the mean stop interval remained unchanged throughout starvation (Table 1). Consistently, the stop frequency decreased ≈3-fold from 3 to 6 hr (Table 1). The reversal frequency did not change significantly, and from 6 hr on, the stop and reversal frequencies were similar (Table 1).
Motility Behavior of Individual csgA Cells and wt Cells Is Different During Starvation.
The motility parameters of csgA cells (DK5253) starved at a high cell density for various periods of time and then recorded at the low cell density were determined in the same manner as described for wt cells. At all timepoints, gliding speed profiles showed that individual csgA cells were moving with changing and low gliding speeds and exhibited frequent periods of no active movement (Fig. 1A and data not shown). The motility parameters of csgA cells were similar to those of wt cells after 0 and 3 hr of starvation (Fig. 1B; Table 1). However, after this time, the csgA cells did not manifest the increased motility characteristics of wt cells; that is, the transient gliding speed distributions, the mean gliding speed, the duration of the mean gliding interval, and the stop frequency remained unchanged. The timepoint at which the behavior of wt and csgA cells diverged coincides with the earliest differences observed between wt and csgA cells during fruiting body formation, suggesting that the increased motility in isolated wt cells could be C-signal-dependent.
C-Signal Stimulates Motility.
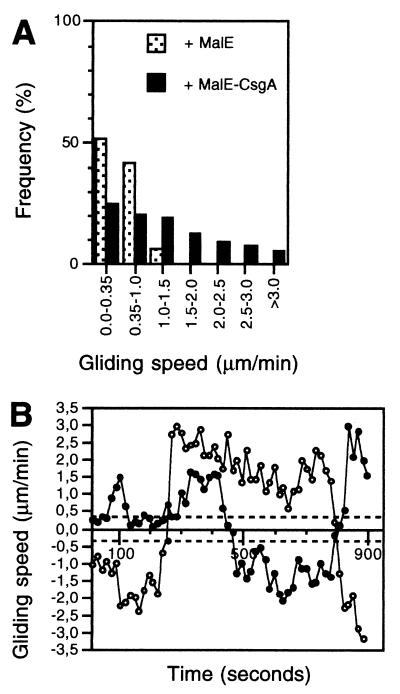
If C-signal stimulates motility in starved cells, then the prediction is that restoring C-signaling in csgA cells should stimulate the motility in these cells. Exogenous MalE-CsgA protein restores development in csgA mutants (12). Therefore, to test this prediction, csgA cells were starved for 6 hr at a high cell density, were harvested, were exposed to 1.0 Cfu of MalE-CsgA protein for 30 min, and then were recorded at the low cell density (see Materials and Methods). In the presence of MalE-CsgA, 75% of the transient gliding speeds were above the detection limit for active movement, compared with only 45% in the absence of MalE-CsgA (Fig. 2A; compare to Fig. 1B). Moreover, MalE-CsgA induced the csgA cells to move with higher transient gliding speeds. Consistently, the mean gliding speed of the csgA cells increased after addition of MalE-CsgA (Table 2). Gliding speed profiles of individual csgA cells showed that the broad transient gliding speed distribution reflected that cells were changing speed (Fig. 2B). In the presence of MalE-CsgA, the duration of the mean gliding interval of the csgA cells increased ≈3-fold whereas the duration of the mean stop interval was unchanged (Table 2). Consistently, MalE-CsgA induced a 2- to 3-fold decrease in the stop frequency (Table 2). Moreover, MalE-CsgA caused an increase in the reversal frequency of csgA cells (Table 2). To ensure that the effect of the MalE-CsgA on the behavior of csgA cells was specific for the CsgA moiety, MalE protein was added in the same concentration as the MalE-CsgA to csgA cells starved for 6 hr. csgA cells exposed to MalE behaved in a manner similar to nonstimulated csgA cells (Fig. 2A and Table 2; compare to Fig. 1B and Table 1).
Figure 2.
Effect of exogenous MalE-CsgA protein on motility pattern of csgA cells. (A) Transient gliding speed distributions of individual csgA cells (DK5253) exposed to 1.0 Cfu of MalE-CsgA protein (closed bars) or an equivalent amount of MalE protein (dotted bars) for 30 min and then spotted at the low cell density. Cells had been starved for 6 hr at the high density before being exposed to protein. (B) Gliding speed profile of two typical individual csgA cells (DK5253) stimulated with 1.0 Cfu of MalE-CsgA protein. The dotted lines indicate the lower detection limit for active movement. A reversal is indicated by a change in speed from a positive to a negative value or vice versa.
Table 2.
Effect of MalE-CsgA on motility parameters of single wt and csgA cells
| Protein added | Gliding speed, μm/min*
|
Duration of gliding interval, min*
|
Duration of stop interval, min*
|
Stop frequency, stop/min*
|
Reversal frequency, reversal/min*
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| wt | csgA | wt | csgA | wt | csgA | wt | csgA | wt | csgA | |
| MalE-CsgA† | 1.99 ± 0.23 | 1.82 ± 0.09 | 4.05 ± 0.77 | 4.12 ± 0.40 | 2.91 ± 0.69 | 2.30 ± 0.52 | 0.17 ± 0.03 | 0.18 ± 0.05 | 0.14 ± 0.02 | 0.17 ± 0.03 |
| MalE† | n.d. | 0.73 ± 0.11 | n.d. | 1.54 ± 0.46 | n.d. | 3.13 ± 0.23 | n.d. | 0.58 ± 0.06 | n.d. | <0.07 |
The mean values and standard errors of the mean are shown for three independent experiments. Strains used: wt, DK5204; csgA, DK5253. n.d., not determined.
See Table 1.
1.0 Cfu of MalE-CsgA protein was added to the cells. MalE was added to a final concentration equivalent to that of MalE-CsgA protein in μg/ml.
Because wt cells were observed to increase their mean gliding speed between 6 and 9 hr of starvation (Table 1), an analysis as to whether this increase was caused by C-signal was performed. Accordingly, wt cells that had been starved for 6 hr were exposed to 1.0 Cfu of MalE-CsgA and were observed at the low cell density. The stimulated wt cells had a transient gliding speed distribution similar to that of wt cells that had been starved for 9 hr (data not shown). Likewise, the speed profiles of the stimulated wt cells were similar to those of wt cells that had been starved for 9 hr (data not shown). Moreover, the mean gliding speed of the 6-hr cells was increased to a level similar to that of the 9-hr cells (Table 2; compare to Table 1). The remaining motility parameters in the 6-hr wt cells were unaffected by the addition of MalE-CsgA (Table 2; compare to Table 1).
C-Signal Stimulation of Motility Is Concentration- and Time-Dependent.
To analyze whether C-signal stimulation of motility depended on the concentration of MalE-CsgA, csgA cells starved for 6 hr at a high cell density were stimulated with 0.25 and 4.0 Cfu of MalE-CsgA. The addition of 0.25 Cfu of MalE-CsgA did not evoke any changes in the motility pattern of single csgA cells. However, csgA cells stimulated with 4.0 Cfu of MalE-CsgA behaved similarly to csgA cells stimulated with 1.0 Cfu of MalE-CsgA (data not shown).
The stimulation of motility of csgA cells described in the previous section was observed after cells had been exposed to MalE-CsgA for 30 min. To analyze the timing of the C-signal-dependent stimulation of motility and to analyze whether cells adapted to the stimulation, csgA cells that had been starved for 6 hr at a high cell density were exposed to 1.0 Cfu of MalE-CsgA for different periods of time and then were recorded at the low cell density. Exposure of csgA cells to MalE-CsgA for 10 min was insufficient to significantly change cell behavior. However, cells exposed to MalE-CsgA for 30, 50, or 70 min were stimulated and displayed similar behavior (data not shown).
C-Signal Stimulation of Motility Depends on the Frz Signal Transduction System.
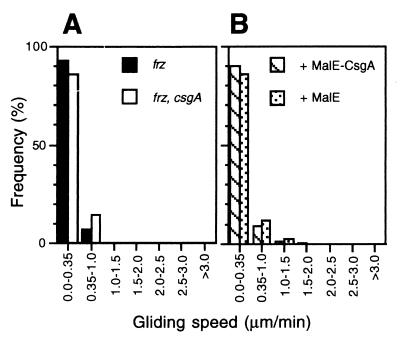
The proteins in the Frz system are components in the motility branch of the C-signal transduction pathway (17, 21). To test whether C-signal stimulation of motility depended on the Frz system, two experiments were carried out. First, the motility parameters of frz cells (DK9033) and frz, csgA cells (DK9035) starved for 6 hr were determined as described above. The two cell types displayed similar behavior and only exhibited limited movements (Fig. 3A; Table 3). Both cell types displayed a narrower spectrum of transient gliding speeds than csgA cells starved for 6 hr, and only 10–15% of the transient gliding speeds were above the detection limit for active movement. Likewise, the duration of the mean gliding interval was shorter than in csgA cells starved for 6 hr, and the duration of the mean stop interval was longer than that in wt and csgA cells. Consistently, the stop frequencies were higher than that of csgA cells. Second, we tested whether 1.0 Cfu of MalE-CsgA could stimulate the motility of csgA, frz cells that had been starved for 6 hr at the high cell density. MalE-CsgA had the same effect as MalE and did not cause significant changes in the motility parameters (Fig. 3B; Table 3). These observations strongly suggest that the Frz signal transduction system is required for the C-signal-dependent stimulation of motility.
Figure 3.
Effect of frz mutation on MalE-CsgA-induced motility stimulation. (A) Transient gliding speed distribution of individual frz (DK9033) (closed bars) and frz, csgA (DK9035) (open bars) cells that had been starved at a high cell density for 6 hr and had been harvested, diluted, and spotted at the low cell density. (B) Effect of exogenous MalE-CsgA on transient gliding speed distribution of individual frz, csgA cells (DK9035). frz, csgA cells were starved at a high cell density for 6 hr and were harvested, diluted, and exposed to 1.0 Cfu of MalE-CsgA protein (cross-hatched bars) or an equivalent amount of MalE protein (dotted bars) for 30 min and then spotted at the low cell density.
Table 3.
Motility parameters of single frz and frz, csgA cells during starvation
| Protein added | Gliding speed, μm/min*
|
Duration of gliding interval, min*
|
Duration of stop interval, min*
|
Stop frequency, stop/min*
|
Reversal frequency, reversal/min*
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| frz | frz, csgA | frz | frz, csgA | frz | frz,csgA | frz | frz, csgA | frz | frz,csgA | |
| None | 0.62 ± 0.11 | 0.59 ± 0.09 | 0.72 ± 0.18 | 0.83 ± 0.30 | 10.99 ± 0.97 | 8.06 ± 0.79 | 1.54 ± 0.31 | 1.26 ± 0.20 | <0.10 | <0.09 |
| MalE-CsgA* | n.d. | 0.51 ± 0.05 | n.d. | 0.87 ± 0.21 | n.d. | 7.64 ± 1.85 | n.d. | 1.25 ± 0.28 | n.d. | <0.07 |
| MalE* | n.d. | 0.53 ± 0.03 | n.d. | 0.79 ± 0.25 | n.d. | 8.51 ± 1.05 | n.d. | 1.41 ± 0.19 | n.d. | <0.10 |
The mean values and standard errors of the mean are shown for three independent experiments. Strains used: frz, DK9033; frz, csgA, DK9035. n.d., not determined.
See Table 2.
DISCUSSION
A central question related to M. xanthus is how cell behavior is changed during fruiting body formation. We have addressed this question by using a motility assay in which movements of single cells are quantified. In this assay, cells proceed correctly through the different stages of fruiting body formation at a high cell density. At specific timepoints during fruiting body formation, the motility characteristics of cells are scored by removing them from the high cell density context. The cells then are dispersed on a solid surface at a density that does not allow cell–cell interactions, followed by time-lapse video microscopy. The advantage of this system is that it allows a systematic, quantitative analysis of motility parameters during development.
The experiments demonstrate that C-signal directly stimulates the motility of starved single cells in the absence of other cell–cell interactions. Specifically, C-signal caused an increase in transient gliding speeds in the duration of the mean gliding interval and a decrease in the stop frequency (an orthokinetic response) whereas the duration of the mean stop interval remained unaffected by C-signal. Several lines of evidence combine to support these conclusions. First, single wt and csgA cells behaved similarly until 3 hr of starvation and only displayed limited movements. From then on, wt cells began to move with higher transient gliding speeds for longer periods of time and with reduced stop frequencies whereas csgA cells continued to display limited movements. The time of divergence of behavior in wt and csgA cells coincides with the time at which C-signal becomes essential for aggregation. Second, the motility defects in csgA cells starved for 6 hr were corrected by exogenous MalE-CsgA protein. Moreover, the effect of MalE-CsgA specifically depended on the CsgA moiety of the protein. Third, wt cells starved for 9–15 hr moved with higher speeds than wt cells starved for 6 hr. The level of C-signaling increases during development (27, 28), suggesting that the higher speeds of cells starved for 9 to 15 hr were caused by the increased level of C-signaling. This hypothesis was supported by the observation that the difference in behavior between wt cells starved for 6 hr and wt cells starved for 9–15 hr was eliminated by addition of MalE-CsgA to the 6-hr cells. Fourth, restored fruiting body formation in csgA mutants occurs in a narrow concentration interval of C-signal (7, 12, 27). Stimulation of motility in csgA cells by MalE-CsgA was observed in the same narrow concentration interval. Thus, the orthokinetic response to C-signal occurs at physiological concentrations. Finally, cells were exposed to MalE-CsgA under conditions that eliminate direct cell–cell interactions, arguing that stimulation is direct and independent of other cell–cell interactions.
Normally, C-signal transmission occurs by a contact-dependent mechanism at a high cell density. The MalE-CsgA protein restores fruiting body formation in csgA cells (12). Here, we have shown that MalE-CsgA stimulates the behavior of single csgA cells in such a way that their motility pattern is indistinguishable from that of single wt cells stimulated by cell–cell interactions while at the high cell density. Therefore, MalE-CsgA can substitute for C-signal produced by wt cells. The net effect of the C-signal-dependent changes in cell behavior described here is an increase in the distance traveled by a cell per minute. This is the combined result of cells gliding with higher speeds, in longer gliding intervals, with reduced stop frequencies and unchanged duration of stop intervals. C-signal may have additional effects on cell behavior that depend on other cell–cell interactions and that are not scored in the assay used here (see also below). So, to fully understand the C-signal-dependent modulation of cell behavior, the next step will be to analyze the effect of C-signal in the context of other cell–cell interactions.
Two genetic systems govern two behavioral patterns in M. xanthus (29). The S-motility system controls gliding of groups of cells, and the A-motility system controls gliding of single cells and allows single cells to move. Here, C-signal stimulates the motility of single cells. This type of motility is regarded as reflecting the A-motility system. However, the properties of the A- and S-motility systems during development are unexplored, and, therefore, additional experiments are required to distinguish whether C-signal stimulation of motility depends on or stimulates the A- or S-motility systems.
The stimulatory effect of MalE-CsgA on cell motility in csgA cells was detected 30 min after cells had been exposed to protein, and then the behavior of cells remained unchanged over the next 55 min. Thus, cells did not show adaptation with respect to gliding speed, gliding interval, or stop frequency. The response time of 30 min is long compared with the instantaneous increase in swimming speed observed in bacteria such as Rhodobacter sphaeroides in response to certain chemicals (30). The swimming speed in this species is several orders of magnitude higher than the gliding speed of M. xanthus cells. Clearly, a fast-moving organism has a need for a faster motility response than a slow moving organism. Thus, the system design of motility pathways may be related to the speed of the organism.
The Frz proteins are important for directed cell movements, and control the reversal frequency in M. xanthus (19). We observed that, after 6 hr of starvation, frz mutants displayed very limited motility. Moreover, the C-signal-dependent stimulation of motility was observed to depend on the Frz system. In vegetative cells, the Frz system is involved in an orthokinetic response to an unknown substance produced by swarming cells (31). Taken together, these observations strongly suggest that the Frz proteins may not only regulate the reversal frequency but also may be important in the regulation of other motility parameters.
C-signal, independently of other cell–cell interactions, excites the Frz system as measured by an increase in the level of methylation of FrzCD, a cytoplasmic methyl-accepting chemotaxis protein homolog, (17). By analogy to the methylation of methyl-accepting chemotaxis proteins in enteric bacteria, FrzCD methylation is likely to be part of an adaptation response to excitation (32–34). Therefore, C-signal is predicted to cause a transient decrease in the reversal frequency (a klinokinetic response with adaptation) (17, 32, 35). Intriguingly, MalE-CsgA did not induce cells to move with low reversal frequencies in our assay. In fact, stimulated csgA cells had a higher reversal frequency than unstimulated cells. Moreover, wt cells starved for 6 hr and then exposed to MalE-CsgA did not experience a decrease in the reversal frequency. In vegetative cells, compounds that increase the level of FrzCD methylation do not suppress reversals. In the presence of such compounds, cells do not begin to move with a low reversal frequency until conditions that allow cell–cell interactions are attained (32). By analogy, we propose that other cell–cell interactions are a prerequisite for C-signal to manifest its effect on the reversal frequency. In this model, C-signaling directly results in two chemokinetic responses: an orthokinetic response without adaptation in which gliding speed, gliding interval, and stop frequency are changed independently of other cell–cell interactions and a klinokinetic response with adaptation in which reversal frequency is transiently decreased contingent on auxiliary cell–cell interactions.
According to current models, orthokinetic responses without adaptation result in dispersal of cells whereas klinokinetic responses with adaptation result in accumulation of cells if reversals are suppressed when cells move up a gradient of the chemical (36). In the case of the C-signal, both types of responses are predicted to be induced at high cell densities. We, therefore, suggest that C-signal is a cell density-dependent aggregation signal during fruiting body formation and that cells are stimulated directly by C-signal to glide with high speeds and low stop and reversal frequencies toward aggregation centers. The aggregation-inducing activity of C-signal is spatially constrained to areas with a high cell density by two mechanisms: C-signal transmission requires direct contact between cells, and auxiliary cell–cell interactions are a prerequisite for C-signal to manifest its effect on the reversal frequency. In our model for C-signal-induced aggregation (17), cells are recruited, by end-to-end contacts with C-signaling, into chains in which cells are moving with a low reversal frequency toward aggregation centers. From the results reported here, the model can now be modified to include that cells recruited to the chains are not only moving with a low reversal frequency but also with high speeds and a low stop frequency. In the chains, cells are predicted to be exposed to increasing levels of C-signal because of two signal amplification loops. In one loop, C-signaling is required for full C-signal production; thus, C-signaling leads to more C-signal production (27, 28). In the second loop, C-signaling results in aggregation and, therefore, a higher cell density. Because C-signaling requires direct cell–cell contact, the prediction is that aggregation per se results in increased C-signaling levels. The increasing levels of C-signaling may ensure that cells not only experience a transient decrease in reversal frequency in response to excitation of the Frz system and adaptation when recruited to a chain but continue to move with a low reversal frequency as long as they are in a chain.
Acknowledgments
We thank M. Ward and D. Kaiser for many helpful suggestions and comments on the manuscript. This work was supported by the Danish Natural Science Research Council and the Carlsberg Foundation.
ABBREVIATIONS
- Cfu
C-factor unit
- wt
wild-type
References
- 1.Henrichsen J. Bacteriol Rev. 1972;36:478–503. doi: 10.1128/br.36.4.478-503.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dworkin M. Microbiol Rev. 1996;60:70–102. doi: 10.1128/mr.60.1.70-102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagen D C, Bretscher A P, Kaiser D. Dev Biol. 1978;64:284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- 4.Downard J, Ramaswamy S V, Kil K S. J Bacteriol. 1993;175:7762–7770. doi: 10.1128/jb.175.24.7762-7770.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimkets L J, Gill R E, Kaiser D. Proc Natl Acad Sci USA. 1983;80:1406–1410. doi: 10.1073/pnas.80.5.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimkets L J, Rafiee H. J Bacteriol. 1990;172:5299–5306. doi: 10.1128/jb.172.9.5299-5306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S K, Kaiser D. Proc Natl Acad Sci USA. 1990;87:3635–3639. doi: 10.1073/pnas.87.10.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim S K, Kaiser D. Cell. 1990;61:19–26. doi: 10.1016/0092-8674(90)90211-v. [DOI] [PubMed] [Google Scholar]
- 9.Hagen T J, Shimkets L J. J Bacteriol. 1990;172:15–23. doi: 10.1128/jb.172.1.15-23.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kroos L, Kaiser D. Genes Dev. 1987;1:840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- 11.Lee K, Shimkets L J. J Bacteriol. 1996;178:977–984. doi: 10.1128/jb.178.4.977-984.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee B-U, Lee K, Mendez J, Shimkets L J. Genes Dev. 1995;9:2964–2973. doi: 10.1101/gad.9.23.2964. [DOI] [PubMed] [Google Scholar]
- 13.Baker M E. Biochem J. 1994;301:311–312. doi: 10.1042/bj3010311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S K, Kaiser D. Science. 1990;249:926–928. doi: 10.1126/science.2118274. [DOI] [PubMed] [Google Scholar]
- 15.Kroos L, Hartzell P, Stephens K, Kaiser D. Genes Dev. 1988;2:1677–1685. doi: 10.1101/gad.2.12a.1677. [DOI] [PubMed] [Google Scholar]
- 16.Kim S K, Kaiser D. Genes Dev. 1990;4:896–904. doi: 10.1101/gad.4.6.896. [DOI] [PubMed] [Google Scholar]
- 17.Søgaard-Andersen L, Kaiser D. Proc Natl Acad Sci USA. 1996;93:2675–2679. doi: 10.1073/pnas.93.7.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sager B, Kaiser D. Genes Dev. 1994;8:2793–2804. doi: 10.1101/gad.8.23.2793. [DOI] [PubMed] [Google Scholar]
- 19.Ward M J, Zusman D R. Mol Microbiol. 1997;24:885–893. doi: 10.1046/j.1365-2958.1997.4261783.x. [DOI] [PubMed] [Google Scholar]
- 20.Kroos L, Kuspa A, Kaiser D. Dev Biol. 1986;117:252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- 21.Søgaard-Andersen L, Slack F, Kimsey H, Kaiser D. Genes Dev. 1996;10:740–754. doi: 10.1101/gad.10.6.740. [DOI] [PubMed] [Google Scholar]
- 22.Hodgkin J, Kaiser D. Proc Natl Acad Sci USA. 1977;74:2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zar J H. Biostatistical Analysis. Englewood Cliffs, NJ: Prentice–Hall; 1996. [Google Scholar]
- 24.Spormann A M, Kaiser A D. J Bacteriol. 1995;177:5846–5852. doi: 10.1128/jb.177.20.5846-5852.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kearns D B, Shimkets L J. Proc Natl Acad Sci USA. 1998;95:11957–11962. doi: 10.1073/pnas.95.20.11957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blackhart B D, Zusman D R. Proc Natl Acad Sci USA. 1985;82:8767–8770. doi: 10.1073/pnas.82.24.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S K, Kaiser D. J Bacteriol. 1991;173:1722–1728. doi: 10.1128/jb.173.5.1722-1728.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li S, Lee B U, Shimkets L J. Genes Dev. 1992;6:401–410. doi: 10.1101/gad.6.3.401. [DOI] [PubMed] [Google Scholar]
- 29.Hodgkin J, Kaiser D. Mol Gen Genet. 1979;171:177–191. [Google Scholar]
- 30.Armitage J P, Schmitt R. Microbiology. 1997;143:3671–3682. doi: 10.1099/00221287-143-12-3671. [DOI] [PubMed] [Google Scholar]
- 31.Ward M J, Mok K C, Zusman D R. J Bacteriol. 1998;180:440–443. doi: 10.1128/jb.180.2.440-443.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi W, Köhler T, Zusman D R. Mol Microbiol. 1993;9:601–611. doi: 10.1111/j.1365-2958.1993.tb01720.x. [DOI] [PubMed] [Google Scholar]
- 33.Shi W, Zusman D R. J Bacteriol. 1994;176:1517–1520. doi: 10.1128/jb.176.5.1517-1520.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blair D F. Annu Rev Microbiol. 1995;49:489–522. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- 35.McBride M J, Köhler T, Zusman D R. J Bacteriol. 1992;174:4246–4257. doi: 10.1128/jb.174.13.4246-4257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schnitzer M J, Block S M, Berg H, Purcell E M. In: Strategies for Chemotaxis. Armitage J P, Lackie J M, editors. Cambridge, U.K.: Cambridge Univ. Press; 1990. pp. 15–34. [Google Scholar]