Abstract
Effective use of conditional Cre recombinase-loxP gene modification requires Cre-expressing mouse strains with defined patterns of expression. To assess the in vivo functionality of Cre-expressing mice, we have engineered an improved reporter strain for monitoring Cre-mediated excisions. The β-galactosidase-neomycin phosphotransferase fusion gene (βgeo)-trapped ROSA26 locus was modified by gene targeting such that βgeo is expressed only after Cre-mediated excision of loxP-flanked DNA sequences. βgeo from the excised ROSA26 allele is expressed ubiquitously in embryos and adult mice. By mating the reporter strain with Cre-expressing transgenic mice, we have shown that the loxP-flanked ROSA26 allele is accessible to Cre during early embryogenesis, as well as in a specific hematopoietic lineage (T lymphocytes). This improved reporter strain should facilitate monitoring in vivo Cre-mediated excision events in a variety of experimental contexts.
Gene targeting in mouse embryonic stem (ES) cells and mice has been used widely to study in vivo functions of genes during development and in adult life (1). Though standard gene knockouts have been highly informative, early embryonic lethality or complex phenotypes often obscure the roles of subject genes at later stages of development or in specific tissues. Conditional gene targeting provides a means to circumvent certain limitations of conventional gene targeting (2). In one approach, the site-specific recombinase Cre is used in vivo to excise critical gene segments flanked by loxP recognition sequences (floxed). Implementation of this strategy requires the creation of a conditional floxed strain and mice expressing Cre in a defined spatial or temporal pattern (3–6). Depending on the pattern of Cre expression, the gene of interest can be modified in the germ line or in somatic tissues in a cell-type-restricted and/or inducible manner. Gain-of-function as well as loss-of-function mutations can be engineered with the Cre-loxP system (7).
The success of Cre-mediated conditional gene targeting depends critically on stringent regulation of Cre-expressing strains with which floxed mice are to be mated (8). If carried in a transgenic construct, Cre may be expressed in a mosaic or leaky fashion, rendering excision incomplete or in too broad a pattern, respectively. If Cre sequences are introduced into a previously characterized locus, hitherto unknown sites of expression may complicate conditional targeting. Many of these complexities can be addressed by characterizing Cre-expressing strains with reporter mice that permit in vivo monitoring of Cre-mediated excision events in diverse tissues and at different developmental stages. For example, a reporter strain carrying a floxed stopper sequence preceding the Escherichia coli β-galactosidase (β-gal) driven by the chicken β-actin promoter has been used to score excision events in selected tissues, such as the nervous system, as well as heart and skeletal muscle (9). A clear need exists, however, for improved reporter strains that permit monitoring excision events more completely both in temporal and spatial dimensions during development and in adult life.
To engineer a suitable reporter strain, we chose to modify a ubiquitously expressed gene locus. An excellent candidate for an appropriate locus is that defined by a gene trap mouse line, ROSA26 (10). In this line, retroviral sequences and a β-gal-neomycin resistance fusion gene (βgeo) were integrated into the locus ROSA26. The wild-type ROSA26 locus expresses two transcripts with no significant ORFs and a third in the antisense orientation that may encode a protein. Transcription of the proviral βgeo gene is initiated from exon-1 of the ROSA26 allele and is ubiquitous during embryonic development and also in adult tissues (11). As cells of the proviral ROSA26 line contain abundant β-gal activity, these mice have been used widely as a marker line in chimera experiments.
To engineer a reporter strain, we modified the gene-trapped proviral ROSA26 locus in ES cells by insertion of a floxed stopper fragment. On excision of these sequences by Cre recombinase, expression of βgeo is restored. The pattern of expression of the excised ROSA26 locus mirrors that of the unmodified locus during embryogenesis and in adult tissues. Interbreeding with different Cre transgenic lines that lead to germ-line- and T cell-specific excision has been used to validate the reporter mice.
MATERIALS AND METHODS
Construction of Targeting Vector.
A 1.3-kb BglII–EcoRI “stopper” fragment from plasmid cAct-XstopXnZ contains transcriptional/translational terminating signals with a loxP site at the 3′ end (9). The stopper fragment was placed 5′ to a HindIII fragment of pGT1.8Iresβgeo vector containing βgeo sequences. A selection cassette, containing phosphoglycerate kinase-1 (PGK)-PuroR and PGK-cytosine deaminase (CD) gene flanked by two loxP sites, then was inserted in front of the stopper fragment. The proviral region of the 3′ long terminal repeat (LTR) and SA was amplified by PCR from ROSA26 genomic DNA by using an intron primer 5′-GGGGAGTGTTGCAATACCTTT-3′ and a lacZ-specific primer 5′-CACGACGTTGTAAAACGACGG-3′. XbaI-digested PCR DNA was ligated with an EcoRI–XbaI fragment of wild-type ROSA26 to constitute the 5′ homology region for targeting (see Fig. 1). A herpes simplex viral thymidine kinase cassette was used as a negative selection marker. Details of plasmid construction are available on request.
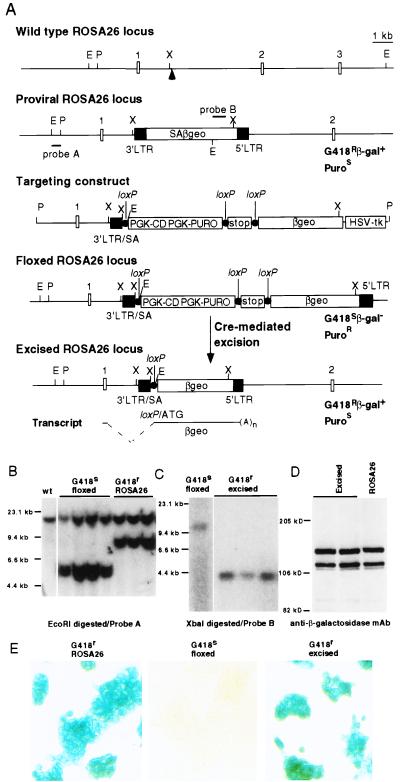
Figure 1.
Modification of the proviral ROSA26 locus. (A) Structure of wild-type ROSA26 locus, proviral ROSA26 locus, targeting vector, floxed ROSA26 locus, and excised ROSA26 locus. The arrowhead indicates the integration site of the provirus. Small open boxes indicate exon regions. Closed boxes denote the 3′ LTR and 5′ LTR. Closed circles indicate loxP sites with the same orientation. E, EcoRI; P, PacI; X, XbaI; Puro, puromycin. Positions for probe A, a EcoRI–PacI genomic fragment, and probe B, a fragment from neor gene are also indicated. (B) Southern blot analysis of genomic DNA from G418S and G418R ES clones. The sizes of wild-type allele, proviral ROSA26 allele, and targeted or floxed allele are 15 kb, 8.7 kb, and 5.4 kb, respectively. (C) Southern blot analysis of G418R clones derived from floxed ROSA26 ES cells. The sizes of floxed allele and excised allele are 10 kb and 4 kb, respectively. (D) Western blot analysis of βgeo expression in ES cells. Whole cell lysates from excised ROSA26 and proviral ROSA26 ES cells were prepared, separated by SDS/PAGE, and blotted with anti-β-gal mAb (Boehringer Mannheim). (E) X-Gal (5-bromo-4-chloro-3-indolyl β-d-galactoside) staining of proviral, floxed, and excised ROSA26 ES cells.
ES Cell Targeting.
PacI-linearized targeting vector was ligated with a PacI-compatible hairpin-shaped oligonucleotide, 5′-TCCGGTACATGATCGAGGGGACTGACAAGACGGCCAGTCCTCGATCATGTACCGGAAT-3′ (12), and electroporated into ROSA26 ES cells. ES cell clones were selected in Puro (1.5 μg/ml) and ganciclovir (2 μM) for 9–10 days. ES clones (n = 244) were obtained, and their sensitivity to G418 (300 μg/ml) was tested. Of the ES clones, 15 were G418S. Southern blot analysis was performed by using a PacI–EcoRI probe just upstream of the 5′ homology region (Fig. 1). Two targeted ES clones were injected into C57BL/6 blastocysts to generate chimeric mice, which were then bred with C57BL/6 mice to obtain germ-line transmission of the modified ROSA26 allele.
In Vitro Excision of Floxed ROSA26 in ES Cells.
ES cells from two clones with a modified ROSA26 allele were electroporated with a plasmid containing the cytomegalovirus promoter/enhancer-driven Cre gene. ES cell clones were selected in G418 (300 μg/ml) for 8 days. Sensitivity to Puro (1.5 μg/ml) was tested. Excision of PGK-CD/PGK-Puro and stopper sequences from the targeted ROSA26 allele was checked by Southern blot by using a neo probe. Two ES clones with the excised ROSA26 allele were injected into C57BL/6 blastocysts to generate germ-line-transmitting chimeras. As a control, unmodified (proviral) ROSA26 ES cells were used to generate mice with a germ-line-transmitted proviral ROSA26 allele.
Generation of GATA1-Cre Transgenic Mice.
The CRE gene was inserted at the NotI site of a GATA1 genomic expression vector (13). Purified, vector-free DNA was injected into the pronuclei of fertilized CD-1 oocytes. Transgenic mice were maintained by mating hemizygous founders with Swiss–Webster mice. Mice carrying the transgene were identified by PCR analysis of tail DNA by using the CRE-specific primers 5′-CAAAACAGGTAGTTATTCGG-3′ and 5′-CGTATAGCCGAAATTGCCAG-3′.
Fluorescence-Activated Cell Sorter (FACS)-β-gal Analysis.
Single-cell suspensions were prepared from thymus, spleen, lymph node, and bone marrow. Expression of β-gal was examined by using fluorescein di-β-d-galactopyranoside (FDG; Molecular Probes; ref. 14). Cells loaded with FDG were incubated on ice for 30 min and then labeled with phycoerythrin (PE)-conjugated antibodies (0.5 μg/ml) for 20 min on ice. Thy1.2-PE, CD4-PE, CD8-PE, B220-PE, and Mac1-PE were obtained from PharMingen. Cells were analyzed on a FACScaliber (Becton Dickinson) machine, and FACS data were analyzed with flowjo software (Tree Star, CA).
T Cell Purification and Activation.
T cells from lymph node were purified by negative selection by using sheep anti-mouse IgG-coated Dynabeads (Dynal, Oslo), giving greater than 98% T cells (Thy1+B220−) by FACS analysis. T cells were stimulated with ConA (2.5 μg/ml; Sigma) and IL-2 (10 ng/ml; R & D Systems) at a density of 5 × 105 cells per ml in 12- or 24-well plates. RPMI medium 1640 was supplemented with 10% (vol/vol) FCS, 2-mercaptoethanol (50 μM), glutamine (2 μM), penicillin (50 units/ml), and streptomycin (50 μg/ml).
X-Gal Staining and Histology.
Embryos from timed matings and organs from 2-week-old mice were fixed and stained overnight with X-Gal at 37°C (15). Embryos were embedded in paraffin, and sections were prepared by standard methods.
RESULTS
Targeting of Proviral ROSA26 Locus.
Heterozygous ROSA26 ES cells containing the βgeo gene trapped (proviral) ROSA26 allele are resistant to G418 and express β-gal (Fig. 1A). A targeting vector was designed to insert floxed PGK-CD/PGK-Puro and stopper sequences between SA and βgeo sequences of the proviral allele. Correctly targeted ES cells would be PuroR and G418S and would not express β-gal. Of 244 PuroR/ganciclovirR clones, 15 were G418S. Representative Southern blots performed with a PacI–EcoRI probe lying outside the 5′-homology region are shown in Fig. 1B. Germ-line transmission of the floxed ROSA26 allele was obtained. Heterozygous mice (ROSA26flox/+) and homozygous floxed mice appeared normal.
Expression of the Excised ROSA26 Allele Mirrors That of the Proviral Allele.
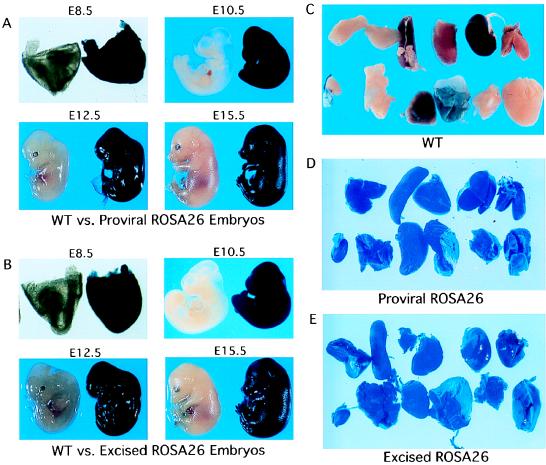
To test the properties of the floxed ROSA26 allele, we first introduced expressible Cre recombinase into the targeted ES cells to excise the floxed sequences. Resulting clones had the expected Southern blot pattern (Fig. 1C) and were G418R and PuroS. The effects of the loxP site and cloning sequences in the excised ROSA26 allele on the expression level and/or pattern of the βgeo gene during development was investigated further. Western blot analysis established that levels of βgeo protein after excision were indistinguishable from those in the proviral ROSA26 ES cells (Fig. 1D). Histochemical staining showed that β-gal activity was absent in cells with the floxed locus but robust after Cre-mediated excision (Fig. 1E). To examine expression of βgeo in vivo, excised ROSA26 ES cells were used to generate chimeric mice, which were then bred to C57BL/6 mice to obtain germ-line transmission. Embryonic day (E) 8.5–E15.5 embryos from timed matings were dissected and stained with β-gal substrate X-Gal. As shown in Fig. 2 A and B, β-gal activity of the excised ROSA26 allele in both yolk sac and embryo proper was widespread and mirrored that of proviral ROSA26 embryos. β-gal activity in adult organs of excised ROSA26 strains was ubiquitous and indistinguishable from that of the proviral ROSA26 strain (Fig. 2 C–E). These data show that residual sequences in the excised ROSA26 allele do not adversely influence either the level or pattern of βgeo expression.
Figure 2.
The comparison of the expression patterns of βgeo from proviral (A) and excised (B) ROSA26 loci. Littermate embryos were fixed and stained overnight with X-Gal; genotypes were performed by Southern blot analysis. WT, wild-type. (C–E) Adult organs are displayed (from left to right): thymus, spleen, liver, kidney, and heart (top rows); testis, skin, lung, stomach, muscle, and brain (bottom rows).
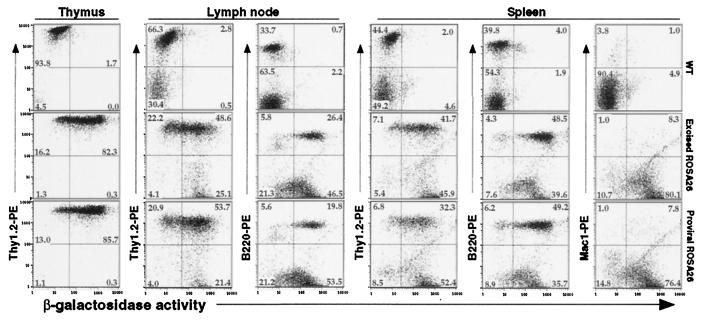
To examine βgeo expression in individual hematopoietic lineages of adult mice, FACS-β-gal analyses were performed by using single-cell suspensions of thymus, spleen, lymph node, and bone marrow. Cells obtained from wild-type excised ROSA26 and original (proviral) ROSA26 mice were loaded with FDG substrate in hypotonic buffer and stained with different antibodies for cell surface molecules. As shown in Fig. 3, thymocytes (Thy1+), peripheral T cells (Thy1+), and B cells (B220+) in lymph node, T cells (Thy1+), B cells (B220+), and myeloid cells (Mac1+) in spleen from excised ROSA26 and proviral ROSA26 mice expressed β-gal activity in a similar pattern. Nucleated bone marrow cells were similarly positive for FDG staining (data not shown).
Figure 3.
Flow cytometric analysis of βgeo expression in the hematopoietic lineages from heterozygous mice containing either proviral or excised ROSA26 locus. Wild-type C57BL/6 was included as a control. Cells from thymus, lymph node, or spleen were analyzed by FACS for expression of β-gal by using loaded FDG substrate, and for cell surface markers by using PE-conjugated antibodies. Thy1.2, T cells; B220, B cells; Mac1, myeloid cells.
Excision of the Floxed ROSA26 Allele in Vivo During Early Embryogenesis.
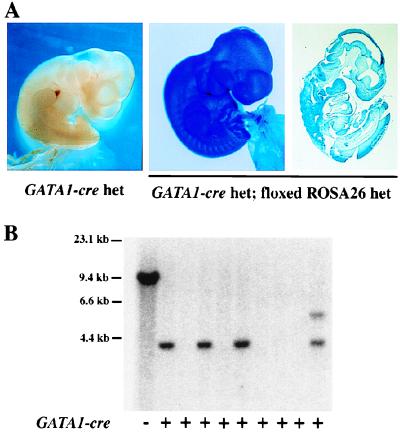
We next examined whether the floxed ROSA26 allele was accessible to Cre-mediated excision in vivo. In other studies, we have generated transgenic mice harboring Cre sequences linked to the regulatory element of the mouse GATA1 gene. Although we had anticipated that such mice would be useful for erythroid- or megakaryocyte-restricted gene targeting, in practice, we have observed highly efficient excision of floxed sequences in tail DNA, internal organs, and germ cells of intercrossed mice (data not shown). This pattern presumably is caused by leaky Cre expression at an early stage of embryogenesis. To test accessibility of the floxed ROSA26 locus for Cre-mediated recombination in early embryogenesis, heterozygous floxed ROSA26 mice were mated with transgenic mice homozygous for the GATA1-Cre transgene (line 2). A total of 10 embryos were analyzed at E10.5 for excision-activated βgeo expression. Of these, all four compound heterozygous embryos showed ubiquitous X-Gal staining in all embryonic and extraembryonic tissues (Fig. 4A, representative samples). In contrast, the embryos containing either GATA1-Cre (Fig. 4A) or floxed ROSA26 allele (data not shown) alone did not show any β-gal activity as expected. Southern blot was used to identify complete (three embryos) or mosaic (one embryo) excision of floxed DNA sequences (Fig. 4B). We conclude that the floxed ROSA26 allele is accessible to Cre-mediated excision during early embryogenesis.
Figure 4.
Ubiquitous excision/activation of βgeo reporter during mouse development. (A) E10.5 littermate embryos generated by interbreeding heterozygous floxed ROSA26 and homozygous GATA1-Cre transgenic mice (line 2) were stained with X-Gal. (Right) A sagittal section of the embryo with complete excision event is shown. (B) Southern blot analysis of Cre-mediated excision in littermate embryos. Genomic DNA from a floxed ROSA26 ES cell was used as a control. DNA samples were digested with XbaI, and the blot was hybridized with probe B as shown in Fig. 1A. Incomplete excision of the floxed allele was observed in one of the DNA samples from compound ROSA26flox/+;GATA1-Cre mice.
Lineage-Restricted Excision of Floxed ROSA26 Sequences by lck-Driven Cre Expression.
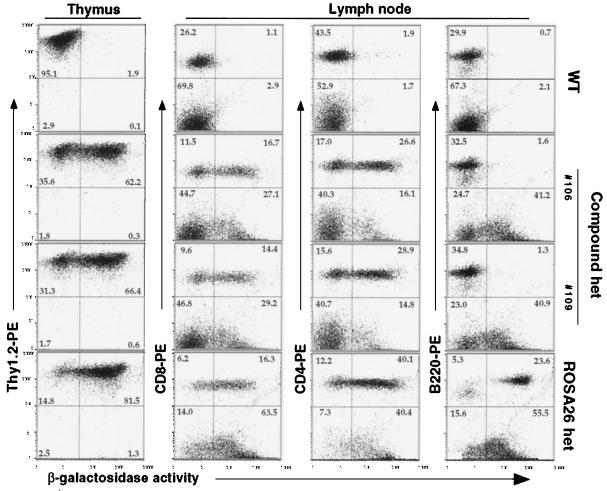
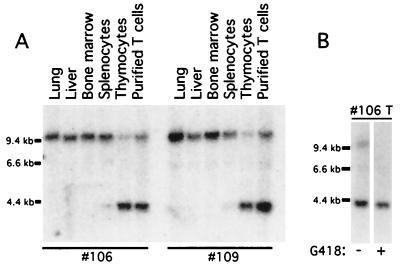
An important application of reporter strains is in monitoring tissue- or lineage-restricted excision events initiated by the Cre transgene. To validate the floxed ROSA26 strain further, ROSA26flox/+ mice were bred to mice carrying a T cell-specific Cre transgene (lck-Cre) (5). Excision at the floxed ROSA26 allele in the compound heterozygous mice was assessed first by FACS-β-gal analysis of hematopoietic cells. As shown in Fig. 5, the majority of thymocytes (Thy1+) cells of ROSA26flox/+; lck-Cre mice were β-gal+, signifying Cre-mediated excision in thymocytes. In the lymph node, most single-positive (CD4+ or CD8+) cells were β-gal+, whereas B220+ B cells remained β-gal−. In the spleen, most single-positive T cells were β-gal+, whereas B220+ B cells and Mac1+ myeloid cells were β-gal− (not shown). These results indicate that activation of βgeo expression occurred in T lymphoid cells. As reflected in the comparison of FACS profiles of cells from ROSAflox/+; lck-Cre and proviral ROSA26 mice, excision did not occur in all cells of compound heterozygous mice. Quantitative Southern blot analysis (Fig. 6A), indicated that 79%/83% of thymocytes and 70%/74% of purified single-positive T cells from lymph node carried the excised ROSA26 allele in the two compound heterozygous mice (106 and 109).
Figure 5.
Tissue-specific in vivo excision of floxed ROSA26 allele by lck-Cre transgene. Expression of the βgeo reporter in thymocytes and peripheral lymphocytes from lymph node was analyzed by FACS by using FDG substrate and PE-conjugated antibody against cell surface markers. One wild-type and two compound heterozygous mice were from the same litter. Age-matched heterozygous proviral ROSA26 mice were used as a positive control.
Figure 6.
Enriched fraction of β-gal+ cells after G418 selection. (A) Southern blot analysis of Cre-mediated excision in genomic DNAs from lung, liver, bone marrow, spleen, thymus, and purified T cells from lymph node. DNA samples were digested with XbaI, and the blot was hybridized with probe B (as shown in Fig. 1A). (B) Purified lymph node T cells were stimulated with ConA and IL-2. Expanded T cells were then cultured in the presence or absence of G418 for 4 days. Excision was examined by Southern blot analysis of T cell DNA from compound heterozygous mice 106. Expression of βgeo also was analyzed by FACS by using FDG substrate (data not shown).
Because the expressed βgeo protein is bifunctional, cells harboring an excised ROSA26 locus should be G418R. We sought to verify this prediction, because this property might prove useful in selecting excised cells in vitro. Purified lymph node T cells were expanded in culture media containing polyclonal lectin Con A and IL-2 and then were incubated in varying concentrations of G418; 375 μg/ml G418 was sufficient to kill wild-type but not proviral ROSA26 T cells. After culture of activated T cells from ROSA26flox; lck-Cre mice in 500 μg/ml G418 for 4 days, Southern blot analysis (shown in Fig. 6B) was performed to quantify the percentage of cells having an excised allele. Although 78% of expanded peripheral T cells (from lymph node of mouse 106) cultured in the absence of G418 had the excised allele, nearly 100% the cells contained the excised allele after G418 selection. Enrichment of T cells with excised ROSA26 allele also was obtained from another compound heterozygous mouse (109). FACS analysis of β-gal activity in these cells confirmed this finding (data not shown). Therefore, the fraction of β-gal+ cells was enriched on G418 selection in vitro.
DISCUSSION
Here, we describe modification of the ROSA26 gene trap locus to engineer a reporter mouse strain suitable for in vivo monitoring of Cre recombinase-mediated excisions. Our data show that βgeo gene expression from the excision-activated ROSA26 locus mimics that of the proviral ROSA26 locus. The reporter line was tested with two transgenic Cre mouse lines to demonstrate the accessibility and functionality of the modified, floxed ROSA26 locus during early embryogenesis and development of a particular hematopoietic lineage.
Two important features of the reporter strain were validated by interbreeding with GATA1-Cre transgenic mice. Although this transgenic line was generated to drive hematopoietic-specific DNA excision, we have observed a high-frequency and nearly complete germ-line excision of floxed sequences in several different conditional mouse strains (data not shown), as reported for CMV-Cre, EIIa-Cre, Zp3-Cre, and PrmCre mice (5, 16–19). Nonetheless, efficient excision within the floxed ROSA26 locus on mating with GATA1-Cre transgenic mice shows accessibility of the reporter allele at a very early stage in mouse embryogenesis. Furthermore, we observed expression of β-gal in all tissues of the embryos (Fig. 4A), illustrating the ubiquitous character of the excision-activated locus. Both aspects are relevant in testing the utility of the reporter line.
T cell-restricted excision of floxed sequences and βgeo expression after mating with lck-Cre transgenic mice provides the complementary validation of the reporter strain. As in other studies that used this lck-Cre transgene, excision, though substantial, is not complete among T cells (3, 5). Very likely this property reflects, in part, some mosaic expression of the multicopy lck-Cre transgene. In our experiments, incomplete excision presumably takes place in the absence of selection for unexcised T cells. Where selection against cells carrying an inactive allele occurs in vivo, incomplete excision may present a serious obstacle to functional analysis. In such instances, specific isolation or purification of cells undergoing complete excision, either in vivo or in vitro, may be required to establish the consequences of gene inactivation. The expression of bifunctional protein βgeo after excision in the floxed ROSA26 line may be particularly useful. We have shown that in vitro culture of T cells from ROSA26flox/+; lck-Cre mice in the presence of G418 enriches for βgal+ T cells (Fig. 6B). This feature of the reporter strain should facilitate in vitro isolation of excised cell populations in various conditional gene-targeting situations.
The reporter strain we have generated has notable advantages compared with prior lines (20–23). Despite the use of promoters that are generally considered to be active in various cell types, previous transgenic reporter genes have not been stably or ubiquitously expressed because of the lack of critical additional cis-regulatory elements, chromosomal positional effects, or the instability of multicopy transgenes. As a consequence, not all lineages sustaining Cre-mediated excision have reporter gene expression. This difficulty often is compounded by variegated expression of Cre transgenes. Given the ubiquitous expression pattern of the excision-activated ROSA26 allele (Figs. 2 and 3), the floxed ROSA26 mice we have engineered provide an improved reporter strain for in vivo assay of the function of Cre, which might be delivered as a transgene, as a virus, or by microinjection. Because the floxed ROSA26 line harbors a single-copy reporter gene, potential problems such as chromosome loss associated with Cre-mediated recombination of multicopy floxed alleles should be avoided (24). It is known that the proviral ROSA26 allele transcribes the βgeo gene as early as the morula/blastocyst stage. Therefore, the excised ROSA26 line is expected to follow the same pattern, although this expectation needs to be confirmed in future studies. The floxed ROSA26 reporter mice also may be useful in the study of cell fate and cell migration during embryogenesis through recombinase-activated tagging. If Cre is specifically expressed in one progenitor cell or a group of progenitor cells, excision-activated βgeo expression will mark progeny both temporally and spatially thereafter. In cases where Cre is expressed only transiently in a group of cells in a developing embryo, progeny could be traced by the constant expression of βgeo, even in the absence of Cre expression at later stages of development. Such approaches have been used successfully in the study of neural progenitors (25, 26). In addition, the reporter mice may serve as an assay system for in vivo gene transfer to somatic cells by using Cre delivered either by viral or by other vectors. Furthermore, our data suggest that the ROSA26 locus may be a particularly favorable site for introduction of sequences to be expressed ubiquitously during embryogenesis and in adult life.
A critical parameter in all in vivo reporter systems is the accessibility of the locus at different stages of development and in different tissues. Although we have established accessibility at a very early stage of mouse development and in the T cell lineage, our data have not addressed the degree of accessibility of the floxed ROSA26 locus as compared with other floxed loci elsewhere in the genome. The extent to which a disparity exists between Cre-mediated excision of reporter and test loci will either overestimate or underestimate the domain in which functional consequences of conditional gene modification might be observed. Nonetheless, the availability of the reporter strain described here should facilitate application of in vivo genetic experiments that use Cre recombinase.
As this work was nearing completion, a reporter strain containing a modified wild-type ROSA26 locus was described (27).
Acknowledgments
We thank Philippe Soriano for providing ROSA26 locus plasmids, Elizabeth Robertson for heterozygous ROSA26 ES cells, Jamey D. Marth for lck-cre transgenic mice, David J. Anderson for cAct-XstopXnZ plasmid, I. Moreno de Alboran for help in FACS analysis, Carol Browne for help in ES cell experiments, and Kerrianne Cunniff and Aimee Chapdelaine for help in mouse blastocyst injection. This research was supported in part by a National Institutes of Health grant to S.H.O., an Investigator of the Howard Hughes Medical Institute. The reporter strain will be deposited for distribution in the collection of The Jackson Laboratory.
ABBREVIATIONS
- SA
splicing acceptor
- β-gal
β-galactosidase
- βgeo
β-gal-neomycin phosphotransferase fusion gene
- CD
cytosine deaminase
- LTR
long terminal repeat
- Puro
puromycin
- X-Gal
5-bromo-4-chloro-3-indolyl β-d-galactoside
- FDG
fluorescein di-β-d-galactopyranoside
- FACS
fluorescence-activated cell sorter
- En
embryonic day n
- ES
embryonic stem
- PGK
phosphoglycerate kinase-1
- PE
phycoerythrin
References
- 1.Thomas K R, Capecchi M R. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 2.Rajewsky K, Gu H, Kuhn R, Betz U A, Muller W, Roes J, Schwenk F. J Clin Invest. 1996;98:600–603. doi: 10.1172/JCI118828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu H, Marth J D, Orban P C, Mossmann H, Rajewsky K. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 4.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 5.Hennet T, Hagen F K, Tabak L A, Marth J D. Proc Natl Acad Sci USA. 1995;92:12070–12074. doi: 10.1073/pnas.92.26.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orban P C, Chui D, Marth J D. Proc Natl Acad Sci USA. 1992;89:6861–6865. doi: 10.1073/pnas.89.15.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lakso M, Sauer B, Mosinger B, Jr, Lee E J, Manning R W, Yu S H, Mulder K L, Westphal H. Proc Natl Acad Sci USA. 1992;89:6232–6236. doi: 10.1073/pnas.89.14.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rossant J, McMahon A. Genes Dev. 1999;13:142–145. doi: 10.1101/gad.13.2.142. [DOI] [PubMed] [Google Scholar]
- 9.Tsien J Z, Chen D F, Gerber D, Tom C, Mercer E H, Anderson D J, Mayford M, Kandel E R, Tonegawa S. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- 10.Friedrich G, Soriano P. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 11.Zambrowicz B P, Imamoto A, Fiering S, Herzenberg L A, Kerr W G, Soriano P. Proc Natl Acad Sci USA. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nehls M, Kyewski B, Messerle M, Waldschutz R, Schuddekopf K, Smith A J, Boehm T. Science. 1996;272:886–889. doi: 10.1126/science.272.5263.886. [DOI] [PubMed] [Google Scholar]
- 13.McDevitt M A, Fujiwara Y, Shivdasani R A, Orkin S H. Proc Natl Acad Sci USA. 1997;94:7976–7981. doi: 10.1073/pnas.94.15.7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nolan G P, Fiering S, Nicolas J F, Herzenberg L A. Proc Natl Acad Sci USA. 1988;85:2603–2607. doi: 10.1073/pnas.85.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanes J R, Rubenstein J L, Nicolas J F. EMBO J. 1986;5:3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwenk F, Baron U, Rajewsky K. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lakso M, Pichel J G, Gorman J R, Sauer B, Okamoto Y, Lee E, Alt F W, Westphal H. Proc Natl Acad Sci USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewandoski M, Wassarman K M, Martin G R. Curr Biol. 1997;7:148–151. doi: 10.1016/s0960-9822(06)00059-5. [DOI] [PubMed] [Google Scholar]
- 19.O’Gorman S, Dagenais N A, Qian M, Marchuk Y. Proc Natl Acad Sci USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Araki K, Araki M, Miyazaki J, Vassalli P. Proc Natl Acad Sci USA. 1995;92:160–164. doi: 10.1073/pnas.92.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akagi K, Sandig V, Vooijs M, Van der Valk M, Giovannini M, Strauss M, Berns A. Nucleic Acids Res. 1997;25:1766–1773. doi: 10.1093/nar/25.9.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danielian P S, Muccino D, Rowitch D H, Michael S K, McMahon A P. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 23.Thorey I S, Muth K, Russ A P, Otte J, Reffelmann A, von Melchner H. Mol Cell Biol. 1998;18:3081–3088. doi: 10.1128/mcb.18.5.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewandoski M, Martin G R. Nat Genet. 1997;17:223–125. doi: 10.1038/ng1097-223. [DOI] [PubMed] [Google Scholar]
- 25.Zinyk D L, Mercer E H, Harris E, Anderson D J, Joyner A L. Curr Biol. 1998;8:665–668. doi: 10.1016/s0960-9822(98)70255-6. [DOI] [PubMed] [Google Scholar]
- 26.Dymecki S M, Tomasiewicz H. Dev Biol. 1998;201:57–65. doi: 10.1006/dbio.1998.8971. [DOI] [PubMed] [Google Scholar]
- 27.Soriano P. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]