Abstract
The embryonic cellular events that set the asymmetry of the genetic control circuit controlling left-right (L-R) axis determination in mammals are poorly understood. New insight into this problem was obtained by analyzing mouse mutants lacking the KIF3A motor subunit of the kinesin-II motor complex. Embryos lacking KIF3A die at 10 days postcoitum, exhibit randomized establishment of L-R asymmetry, and display numerous structural abnormalities. The earliest detectable abnormality in KIF3A mutant embryos is found at day 7.5, where scanning electron microscopy reveals loss of cilia ordinarily present on cells of the wild-type embryonic node, which is thought to play an important role in setting the initial L-R asymmetry. This cellular phenotype is observed before the earliest reported time of asymmetric expression of markers of the L-R signaling pathway. These observations demonstrate that the kinesin-based transport pathway needed for flagellar and ciliary morphogenesis is conserved from Chlamydomonas to mammals and support the view that embryonic cilia play a role in the earliest cellular determinative events establishing L-R asymmetry.
Considerable work has established a genetic control cascade that generates left-right (L-R) asymmetry in mammalian development (1). The cellular events that set the asymmetry of this control circuit remain obscure, although there have been persistent hints and suggestions that ciliary activity or ciliary formation on cells of the early embryo may be crucial to determine L-R asymmetry (2–4). In particular, the consistent association of ciliary morphogenesis defects in mice (2, 3) and humans (4) with randomized L-R asymmetry has led to the view that early embryonic cilia might play an important role in setting the earliest embryonic L-R axis. Whether these cilia are motile has been controversial (2, 5, 6), as has their possible role in establishing the L-R determination pathway.
Recent work in Chlamydomonas, Caenorhabditis, sea urchins, and mice identified a kinesin-II-mediated transport pathway necessary for ciliary formation in diverse contexts (2, 7–11). Considerable biochemical analysis has shown that kinesin-II is often a heterotrimeric complex consisting of two different motor subunits and one nonmotor subunit (12–16). In Chlamydomonas reinhardii, a kinesin-II subunit called FLA-10 is essential for flagellar morphogenesis. Biochemical and morphological studies convincingly established that the FLA-10 kinesin is a motor protein that powers transport of flagellar components from sites of synthesis in the cell body to sites of utilization at the growing flagellar tip (8, 11, 17–20). In Caenorhabditis, a kinesin-II relative called osm-3 is found in the evolutionarily divergent immotile chemosensory cilia and appears to be needed for their proper function and morphogenesis (10, 21). Microinjection experiments in sea urchin established that kinesin-II is needed for the formation of embryonic cilia (7). Finally, work in mouse demonstrated that kinesin-II is built from motor proteins of the KIF3 family of proteins, which has three known members, KIF3A, KIF3B, and KIF3C (15, 22–24). There is considerable evidence that heterodimers between KIF3A and KIF3B or KIF3C are formed, but that homodimers or heterodimers between KIF3B and KIF3C are not common (23, 24). Recently, KIF3B null mutants were reported to have defects in the formation of the earliest cilia of the mouse embryo, namely those on the cells of the embryonic node (2). This ciliary malformation defect preceded the appearance of defects in the L-R determination pathway and resulted in randomized L-R determination in the embryos. Here we describe an analogous phenotype for KIF3A mutants, which establishes that both major motor subunits of kinesin-II are needed for ciliary formation in the nodal cells of the early mouse embryo and strengthens the case that these early cilia are important in the establishment of the embryonic L-R axis.
MATERIALS AND METHODS
Generation of KIF3A Null Animals.
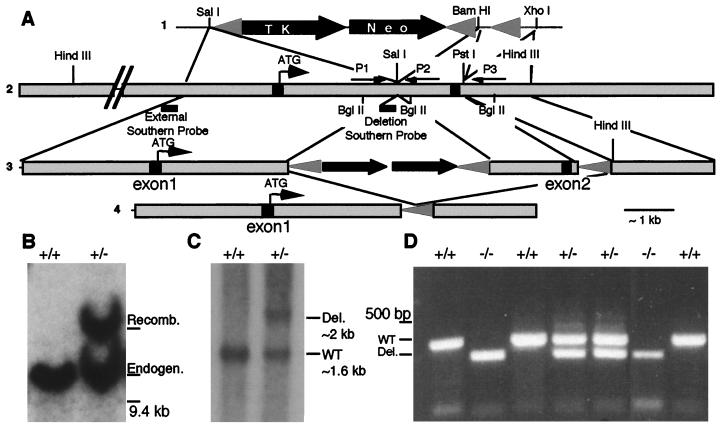
Exon 1 (5′ untranslated and first 5 nt of coding sequence) and exon 2 (nt 6–280) were isolated and identified from a 129/SV/J genomic library (a gift from the Rossant lab) by using the first 350 nt of KIF3A coding sequence. A pflox-KIF3A targeting construct was created (Fig. 1A) and introduced into R1 embryonic stem (ES) by standard methods (25) [pflox vector was a gift from Jamey Marth, ES cells were a gift from Andras Nagy, and recombinant LIF was a gift (26)]. Targeted KIF3A allele was confirmed by Southern blotting of HindIII-cut ES cell DNA, probed with a BamHI/EcoRI genomic DNA fragment external from the targeting construct (Fig. 1A). Genomic DNA flanked by lox P sites was removed by standard methods (25) (pNuKCre was a gift from Jamey Marth). Southern blotting of BglII-cut ES cell DNA probed with an NcoI/EcoRI 160 base pair probed confirmed loss of exon 2 from several clones. Chimeric animals were generated by using standard methods (25). Embryos were genotyped by PCR with extraembryonic DNA by using a common 5′ primer (P1-AGGGCAGACGGAAGGGTGG) and a mixture of primers specific for either the wild-type allele (P2-TCTGTGAGTTTGTGACCAGCC) or the mutant allele (P3-GGTGGGAGCTGCAAGAGGG).
Figure 1.
Generation and molecular analysis of KIF3A mutant mice. (A) Knockout strategy used for generating the KIF3A mutant. (i) Restriction enzyme sites used in pflox vector and (ii) genomic DNA regions that were used to make the KIF3A knockout allele construct. PCR primer locations are indicated by arrows and Southern probe locations by lines. (iii) KIF3A null mutant targeting construct that was introduced into ES cells; proper targeting was confirmed with the external Southern probe; (iv) KIF3A null mutant allele after Cre recombinase treatment that removed exon 2 and the selectable markers. (B) Southern blot of HindIII-digested ES cell DNA to confirm proper targeting of the KIF3A allele; wild-type allele is ≈10 kb and recombinant allele is ≈14 kb. (C) Southern blotting of BglII-digested ES cell DNA after transfection with Cre recombinase confirming the presence of the deletion allele (D) PCR of extraembryonic membrane DNA from a 9.5-days p.c. litter by using the primers described in A.
Western and Northern Analysis.
Western (5 μg of embryo or 30 μg of brain total extract) and Northern (20 μg of total RNA) analysis were performed by using standard methods (27). The KIF3A and KIF3B antibodies were obtained from Babco (Richmond, CA) and were characterized for specificity by Western blotting. These antibodies are directed toward the tail portion of mouse KIF3A and rat KIF3B proteins, respectively. The KIF3A antibody crossreacts equally with an unknown protein of ≈70 Kd in all embryo protein extracts examined. Anti-α-tubulin mAb (DMA1) was a gift from Don Cleveland. KIF3A message was identified with KIF3A cDNA (HindIII nt 1,620 to end) (22).
Scanning Electron Microscopy.
Plugged females were identified in the morning [day 0.5 postcoitum (p.c.)], sacrificed by cervical dislocation at the desired time, and the embryos harvested. Extraembryonic membranes were harvested for genotyping by PCR and the embryos fixed in 4% paraformaldehyde/0.1 M Sorenson’s phosphate for cardiac looping analysis and in situ analysis. For scanning electron microscopy, animals were fixed in 2.5% paraformaldehyde/2.5% gluteraldehyde/0.1 M Sorenson’s phosphate for >24 hr. Preparation of samples for scanning electron microscopy was performed by using standard methods (5, 6).
Whole-Mount in Situ Hybridization Analysis.
In situ hybridization analysis was performed by using standard methods as described (28). Whole-mount in situ hybridization by using the ventricular-specific marker MLC2v (29) identifies cardiac myocytes and indicates that cardiac differentiation occurs in KIF3A mutant mice. The Pitx2 DNA used was as described (30). All of the wild-type animals used in every experiment were littermate controls.
RESULTS AND DISCUSSION
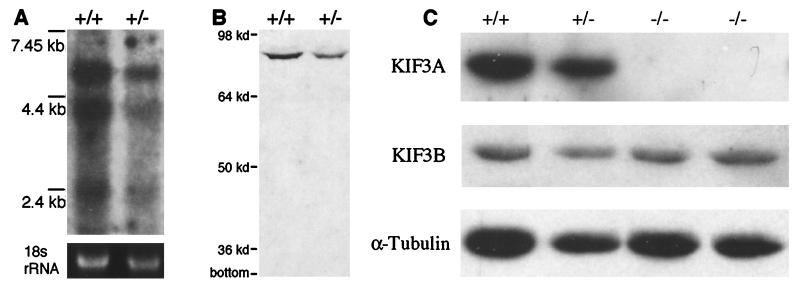
To probe the function of KIF3A in vertebrates, a KIF3A null mutant allele was generated in mice by removing exon 2 (Fig. 1). When exon 1 splices to exon 3, the resulting KIF3A transcript will be translated out of the normal reading frame, thus generating a 62 amino acid nonsense protein that has only the initiation methionine in common with wild-type KIF3A protein. This null mutant allele was introduced into ES cells (Fig. 1A), and then mice were generated by blastocyst injections of the ES cells. Southern blotting and PCR (Fig. 1 B–D) confirmed the molecular identity of the mutants constructed. Northern and Western blotting of heterozygous KIF3A mutants demonstrated that no fragments or aberrant products were made from the mutant gene (Fig. 2 A and B). In addition, examination of embryo extracts from embryos revealed a complete absence of KIF3A protein from mutant embryos, while KIF3B and α-tubulin levels remained effectively unchanged (Fig. 2C).
Figure 2.
KIF3A null mutant allele does not produce any KIF3A protein. (A) Northern blot that shows that a truncated KIF3A message is not produced in KIF3A heterozygous mutant animals. rRNA (18 s) is shown as a control for RNA loading. (B) Immunoblot demonstrating that no truncated KIF3A protein products are generated by the deletion allele. Note that adult brain was used for A and B. (C) Western immunoblot of 9.5-days p.c. embryos indicating an absence of KIF3A protein while the levels of KIF3B and α-tubulin are effectively unchanged.
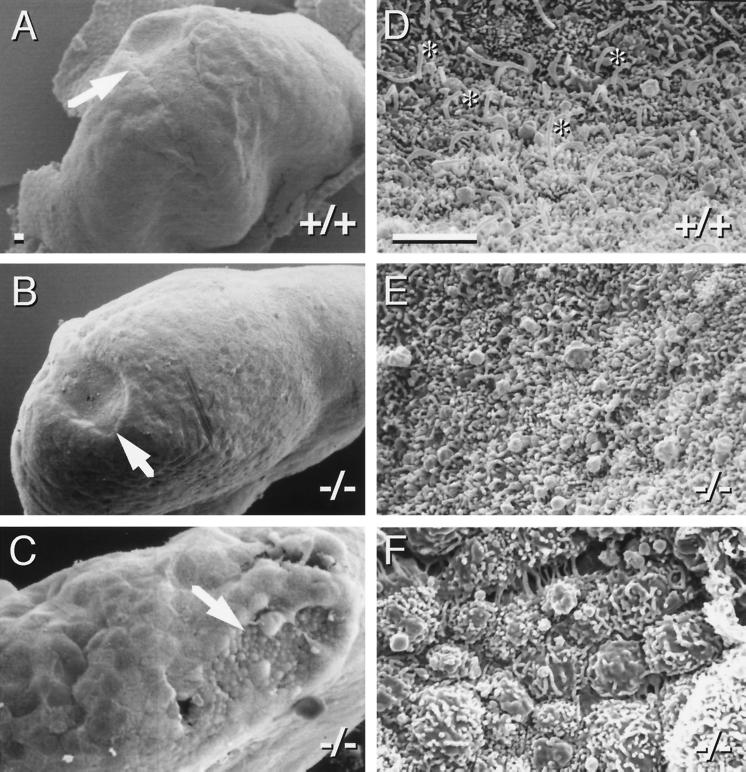
To determine the earliest defects in KIF3A mutant embryos, scanning EM was used to examine the morphology of day 7.0–7.5 p.c. embryos (Fig. 3). The overall morphology of mutant and wild-type embryos at this age appeared similar, with the exception of the embryonic node, which plays a crucial role in L-R patterning (1). Consistent with previous reports (2, 5, 6), each cell of the wild-type embryonic node was found to have a single prominent cilium. Strikingly, KIF3A mutant embryos always lacked cilia on all cells of the node. The position of the mutant node was generally normal, but the nodal plate was often flatter than in wild type, and in many cases overgrowth of surrounding endodermal cells was observed (Fig. 3C). No other obvious morphological defects were observed in homozygous mutant embryos before embryonic day 9.
Figure 3.
KIF3A null mutant embryos lack cilia on cells in the node. (A–C) Scanning electron microscopy of day ≈7.5 p.c. embryos; arrows identify the node. (D–F) High magnification view of nodes. Note the presence of cilia in the wild-type node (D, asterisks) and complete absence from KIF3A mutant nodes (E and F). (Bars = 5 μm.)
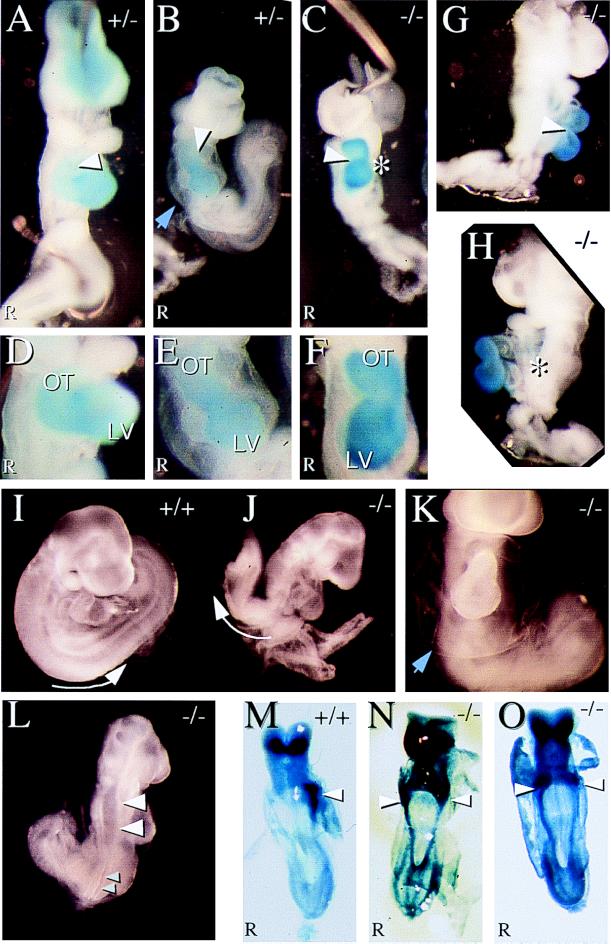
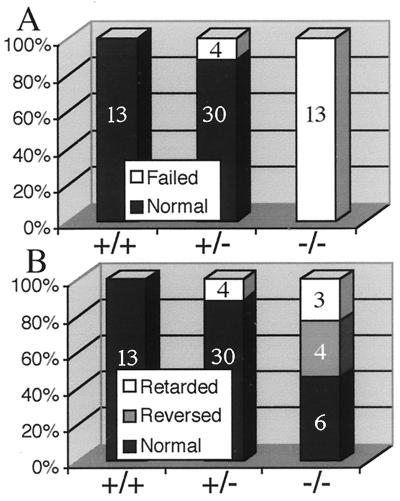
To test whether the L-R genetic control cascade was normal in KIF3A mutants, 8.0 days p.c. KIF3A mutant embryos were examined for expression of Pitx2, which is normally expressed only in the left lateral plate mesoderm (30–34) (Fig. 4M). We found that all null embryos expressed Pitx2 bilaterally (5/5) (Fig. 4 N and O). After day 9 p.c., all null (13/13) and a subset of heterozygous (4/34) embryos appear smaller than all of their wild type (13/13) and most of their heterozygous (30/34) littermates (Fig. 4 A–C). All of these mutant embryos fail to turn from lordotic to fetal position (Figs. 4 I and J and 5A). Mutant embryos are caudally truncated (Fig. 4 E, G, H, and J–L) and exhibit defects in neural tube closure (Fig. 4L), display reduced or no limb primordia and reduced brachial arches; many have a globular pericardium with edema (Fig. 4 B, E, and K). Closer examination of cardiac looping revealed three classes (Fig. 5B): (i) embryos with normal cardiac looping (Fig. 4 A and D); (ii) embryos with reversed cardiac looping (situs inversus, Fig. 4 C and F–H); and (iii) embryos that exhibit retarded but normal cardiac looping that is accompanied by pericardial edema (Fig. 4 B, E, and K). Taken together, these phenotypes and the aberrant Pitx2 expression indicate that the control systems mediating L-R asymmetry are defective in the absence of KIF3A protein. Interestingly, the spontaneous mouse mutant no turning (35) exhibits nearly identical phenotypic abnormalities as both KIF3A and KIF3B mutants, indicating that the same developmental pathways may be disturbed in no turning mutants.
Figure 4.
KIF3A mutant embryos display morphological and asymmetrical defects. Blue staining in A–H is for the ventricle- specific MLC2v message (29). (A) A normal heterozygous 9.5 days p.c. embryo displaying proper cardiac looping. (B) A heterozygous mutant embryo exhibiting retarded cardiac looping. (C) A null mutant embryo displaying reversed cardiac looping. Arrowheads in A–C indicate direction of cardiac looping. (D–F) Closeup of hearts in A–C. (LV = Left Ventricle; OT = Outflow Tract). (G and H) Right and left side views of KIF3A null mutant embryo with reversed cardiac looping. (I and J) A wild-type embryo that underwent normal embryo turning and a KIF3A null embryo that failed to undergo embryonic turning. Arrow indicates the orientation of the embryo posterior. (K) KIF3A null mutant embryo exhibiting edema around the heart. Blue arrowheads in B and K indicate the membrane. (L) KIF3A null mutant embryo that has a neural tube closure defect. Large arrowheads point to open neural tube and small arrowheads point to closed neural tube. (M–O) In situ hybridization of 8.0-days p.c. embryos for Pitx2, ventral views. (M) Normal left side expression of Pitx2 in the lateral plate mesoderm in wild-type embryos. (N and O) Bilateral expression of Pitx2 in KIF3A null mutant embryos. Arrowheads point to strong lateral plate mesoderm staining.
Figure 5.
KIF3A null mutant embryos fail to turn and exhibit randomization of heart looping. (A) Graph showing the percentage and number of embryos and their ability to turn from the lordotic to fetal position for each genotype. (B) Graph showing the percentage and number of embryos for each genotype that exhibit each cardiac looping phenotype.
The phenotypes we observe for KIF3A mutants are similar to those recently reported for KIF3B mutants (2) and establish that both major motor components of kinesin-II are needed to build a cilium in mammalian embryos. These data also support the suggestion that these early cilia are important in some way for establishing the asymmetry of the early L-R signaling pathway. The fact that kinesin-II mouse mutants exhibit developmental and structural abnormalities in addition to L-R asymmetry defects suggests that kinesin-II or the cilia on embryonic nodal cells also perform functions that are not limited to L-R asymmetry determination. It is also formally possible that L-R defects are related in some way to failure of normal anterior–posterior development (36, 37).
It is interesting to note that a subset of KIF3A heterozygous null embryos exhibit identical morphological abnormalities, but reversed cardiac looping was observed only in KIF3A null embryos. This observation suggests that the morphological and structural defects observed in the subset of heterozygous embryos may be unrelated to embryonic nodal ciliary defects, but rather may be caused by other cellular defects that are sensitive to reduced levels of KIF3A protein. Regardless, the presence of defects in heterozygous embryos and observations of mild L-R visceral defects in a low percentage of viable heterozygous animals (data not shown) raise the possibility that mutations in the kinesin-II pathway may be responsible for some visceral defects that are present in human populations including cardiac and cardiovascular defects.
Most recent work on the pathway determining L-R asymmetry has focused on signaling molecules and transcription factors in the genetic control circuit (1). The initial cellular or developmental events initiating this control circuit and establishing its asymmetric character have, however, been less amenable to examination. Several clues to the nature of this initial event are available. First, many years of work on immotile cilia syndrome and Kartagener’s triad in humans has consistently associated bronchial ciliary abnormalities, male infertility, and situs inversus (4). Although many cases are associated with loss of dynein arms, it is also clear that this syndrome is genetically heterogeneous and may involve many different genes and products. Thus, it was suggested that ciliary activity in the early embryo is crucial for L-R patterning (38). Second, recent analysis of the gene identified by the iv mutation in mouse (39) revealed that this locus encoded a dynein molecular motor (called lrd for left-right-dynein). These workers suggested that lrd, in spite of greatest homology to an axonemal or ciliary dynein (as opposed to a cytoplasmic dynein) would function in a cytoplasmic event in cells of the node as opposed to in the cilium. Third, recent work revealed that mouse mutants lacking hepatocyte nuclear factor 4 (a winged helix transcription factor) lacked all cilia in neonatal animals, did not express lrd, and exhibited randomized L-R asymmetry (3). Fourth, previous workers and our work convincingly documented the presence of cilia on cells of the node at the crucial determinative time in development (2, 5, 6) and established that loss of these cilia was correlated with later randomization of the L-R axis. Whether or not these cilia are motile has been controversial (2, 5, 6), although an economical resolution may be that motility is transient and was missed in one of the studies. Further work is needed to corroborate the reports of motility of these cilia since their motility has been suggested to be crucial for establishing an asymmetric L-R gradient of signaling molecules (2).
Our data combined with the previous data point to two likely models for the early cellular events in the cells of the node necessary for proper L-R determination. The first model proposes that dyneins and kinesins needed for ciliary motility or morphogenesis in bronchi and sperm are used in other processes in the cytoplasm of cells of the node to generate cellular polarity or asymmetric transport of determinative factors. Although attractive, this model does not adequately account for the heterogeneity of immotile cilia syndrome associated with situs inversus in humans and the persistent association with ciliary structural defects. A second model proposes that ciliary motility or proper ciliary morphogenesis on cells of the node is an early and crucial event determining or maintaining polarization or signaling of these cells and initiation of an asymmetric pathway of development. The second model is simplest and is consistent with the observations that the failure to form cilia in the KIF3A and KIF3B mutants precedes the observed L-R laterality defects (2). Formally, it remains possible that the ciliary defect in KIF3A and KIF3B mutant cells is either unrelated to or is a secondary manifestation of a different process causing the L-R defect. In this view, the L-R defect might be caused by a cytoplasmic function of KIF3A and KIF3B in cell polarization, which is different than the ciliary morphogenesis function directly observed in mice, sea urchins, and Chlamydomonas. This formal possibility, however, seems unduly complex.
The question of whether the cilia on the cells of the node need to be motile for L-R determination remains open and in need of confirmation (2, 5, 6). The finding that the sequence of lrd is most similar to an axonemal dynein is nonetheless striking (39). How an axonemal-like dynein might function in the 9 + 0 type of cilium found on the nodal cells is unclear. Ordinarily, this type of cilium is immotile, but exceptions have been reported (40), and there are reports that the nodal cilia are in fact motile (2, 6). A simple and ultimately testable possibility is that ciliary motility on cells of the node is transient and required to set the L-R axis.
Our data and the data of others now establish that the kinesin-based transport pathway needed to form flagella in Chlamydomonas (8, 11, 17–20), motile cilia on sea urchin embryos (7), and sensory cilia in Caenorhabditis (10, 21) is conserved to mammals and thus may be used in all types of cilia and flagella (2). Further work with these mutants may help to establish the roles of primary cilia in the many cell types in which they are found and may help in the understanding of some of the transport pathways needed in other types of evolutionarily divergent ciliary structures found on hair cells and in photoreceptors.
Acknowledgments
We thank Ms. Michelle Wilhite for many valuable suggestions about scanning EM preparative methods, Lance Washington for assistance with scanning EM, Charles Graham for help with critical point drying and scanning EM, and Jamey Marth for crucial advice about mutant construction and vectors. This work was supported by National Institutes of Health grants to K.R.C. and an Individual National Research Service award to P.R.L.; a Pharmacology Training grant supported J.R.M. L.S.B.G. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- L-R
left-right
- ES
embryonic stem
- p.c.
postcoitum
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Harvey R P. Cell. 1998;94:273–276. doi: 10.1016/s0092-8674(00)81468-3. [DOI] [PubMed] [Google Scholar]
- 2.Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Knowles H J, Hebert J L, Hackett B P. J Clin Invest. 1998;102:1077–1082. doi: 10.1172/JCI4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afzelius B A. CRC Crit Rev Biochem. 1985;19:63–87. doi: 10.3109/10409238509086788. [DOI] [PubMed] [Google Scholar]
- 5.Bellomo D, Lander A, Harragan I, Brown N A. Dev Dyn. 1996;205:471–485. doi: 10.1002/(SICI)1097-0177(199604)205:4<471::AID-AJA10>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 6.Sulik K, Dehart D B, Iangaki T, Carson J L, Vrablic T, Gesteland K, Schoenwolf G C. Dev Dyn. 1994;201:260–278. doi: 10.1002/aja.1002010309. [DOI] [PubMed] [Google Scholar]
- 7.Morris R L, Scholey J M. J Cell Biol. 1997;138:1009–1022. doi: 10.1083/jcb.138.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozminski K G, Beech P L, Rosenbaum J L. J Cell Biol. 1995;131:1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scholey J M. J Cell Biol. 1996;133:1–4. doi: 10.1083/jcb.133.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabish M, Siddiqui Z K, Nishikawa K, Siddiqui S S. J Mol Biol. 1995;247:377–389. doi: 10.1006/jmbi.1994.0146. [DOI] [PubMed] [Google Scholar]
- 11.Cole D G, Diener D R, Himelblau A L, Beech P L, Fuster J C, Rosenbaum J L. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole D G, Chinn S W, Wedaman K P, Hall K, Vuong T, Scholey J M. Nature (London) 1993;366:268–270. doi: 10.1038/366268a0. [DOI] [PubMed] [Google Scholar]
- 13.Rashid D J, Wedaman K P, Scholey J M. J Mol Biol. 1995;252:157–162. doi: 10.1006/jmbi.1995.0484. [DOI] [PubMed] [Google Scholar]
- 14.Wedaman K P, Meyer D W, Rashid D J, Cole D G, Scholey J M. J Cell Biol. 1996;132:371–380. doi: 10.1083/jcb.132.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamazaki H, Nakata T, Okada Y, Hirokawa N. J Cell Biol. 1995;130:1387–1399. doi: 10.1083/jcb.130.6.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamazaki H, Nakata T, Okada Y, Hirokawa N. Proc Natl Acad Sci USA. 1996;93:8443–8448. doi: 10.1073/pnas.93.16.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walther Z, Vashishtha M, Hall J L. J Cell Biol. 1994;126:175–188. doi: 10.1083/jcb.126.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vashishtha M, Walther Z, Hall J L. J Cell Sci. 1996;109:541–549. doi: 10.1242/jcs.109.3.541. [DOI] [PubMed] [Google Scholar]
- 19.Piperno G, Mead K. Proc Natl Acad Sci USA. 1997;94:4457–4462. doi: 10.1073/pnas.94.9.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piperno G, Mead K, Henderson S. J Cell Biol. 1996;133:371–379. doi: 10.1083/jcb.133.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shakir M A, Fukushige T, Yasuda H, Miwa J, Siddiqui S S. NeuroReport. 1993;4:891–894. doi: 10.1097/00001756-199307000-00013. [DOI] [PubMed] [Google Scholar]
- 22.Kondo S, Sato-Yoshitake R, Noda Y, Aizawa H, Nakata T, Matsuura Y, Hirokawa N. J Cell Biol. 1994;125:1095–1107. doi: 10.1083/jcb.125.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muresan V, Abramson T, Lyass A, Winter D, Porro E, Hong F, Chamberlin N L, Schnapp B J. Mol Biol Cell. 1998;9:637–652. doi: 10.1091/mbc.9.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Z, Goldstein L S. Mol Biol Cell. 1998;9:249–261. doi: 10.1091/mbc.9.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chui D, Oh-Eda M, Liao Y F, Panneerselvam K, Lal A, Marek K W, Freeze H H, Moremen K W, Fukuda M N, Marth J D. Cell. 1997;90:157–167. doi: 10.1016/s0092-8674(00)80322-0. [DOI] [PubMed] [Google Scholar]
- 26.Mereau A, Grey L, Piquet-Pellorce C, Heath J K. J Cell Biol. 1993;122:713–719. doi: 10.1083/jcb.122.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Z, Hanlon D W, Marszalek J R, Goldstein L S. Genomics. 1997;45:123–131. doi: 10.1006/geno.1997.4901. [DOI] [PubMed] [Google Scholar]
- 28.Ruiz-Lozano P, Doevendans P, Brown A, Gruber P J, Chien K R. Dev Dyn. 1997;208:482–490. doi: 10.1002/(SICI)1097-0177(199704)208:4<482::AID-AJA4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 29.Fishman M C, Chien K R. Development (Cambridge, UK) 1997;124:2099–2117. doi: 10.1242/dev.124.11.2099. [DOI] [PubMed] [Google Scholar]
- 30.Ryan A K, Blumberg B, Rodriguez-Esteban C, Yonei-Tamura S, Tamura K, Tsukui T, de la Pena J, Sabbagh W, Greenwald J, Choe S, et al. Nature (London) 1998;394:545–551. doi: 10.1038/29004. [DOI] [PubMed] [Google Scholar]
- 31.Logan M, Pagan-Westphal S M, Smith D M, Paganessi L, Tabin C J. Cell. 1998;94:307–317. doi: 10.1016/s0092-8674(00)81474-9. [DOI] [PubMed] [Google Scholar]
- 32.Piedra M E, Icardo J M, Albajar M, Rodriguez-Rey J C, Ros M A. Cell. 1998;94:319–324. doi: 10.1016/s0092-8674(00)81475-0. [DOI] [PubMed] [Google Scholar]
- 33.St. Amand T R, Ra J, Zhang Y, Hu Y, Baber S I, Qiu M, Chen Y. Biochem Biophys Res Commun. 1998;247:100–105. doi: 10.1006/bbrc.1998.8740. [DOI] [PubMed] [Google Scholar]
- 34.Yoshioka H, Meno C, Koshiba K, Sugihara M, Itoh H, Ishimaru Y, Inoue T, Ohuchi H, Semina E V, Murray J C, et al. Cell. 1998;94:299–305. doi: 10.1016/s0092-8674(00)81473-7. [DOI] [PubMed] [Google Scholar]
- 35.Melloy P G, Ewart J L, Cohen M F, Desmond M E, Kuehn M R, Lo C W. Dev Biol. 1998;193:77–89. doi: 10.1006/dbio.1997.8787. [DOI] [PubMed] [Google Scholar]
- 36.Lohr J L, Danos M C, Yost H J. Development (Cambridge, UK) 1997;124:1465–1472. doi: 10.1242/dev.124.8.1465. [DOI] [PubMed] [Google Scholar]
- 37.Danos M C, Yost H J. Development (Cambridge, UK) 1995;121:1467–1474. doi: 10.1242/dev.121.5.1467. [DOI] [PubMed] [Google Scholar]
- 38.Afzelius B A. Science. 1976;193:317–319. doi: 10.1126/science.1084576. [DOI] [PubMed] [Google Scholar]
- 39.Supp D M, Witte D P, Potter S S, Brueckner M. Nature (London) 1997;389:963–966. doi: 10.1038/40140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Odor D L, Blandau R J. Am J Anat. 1985;174:437–453. doi: 10.1002/aja.1001740407. [DOI] [PubMed] [Google Scholar]