Abstract
longitudinals lacking (lola) is a complex Drosophila gene encoding at least 20 protein isoforms,each bearing the same N-terminal constant region linked to a different C-terminal variable region. Different isoforms specify different aspects of axon growth and guidance. We show here that lola mRNAs are generated by alternative trans-splicing of exons sequentially encoded by the same DNA strand. Chromosomal pairing facilitates interallelic trans-splicing,allowing complementation between mutations in the constant and those in the variable exons. We demonstrate that at least one variable exon is transcribed from its own promoter,and trans-spliced to the constant exons transcribed separately.
Keywords: Pre-mRNA processing, BTB-Zn finger transcription factor, single nucleotide polymorphism
Alternative splicing is a crucial source of diversity in the proteomes of higher eukaryotes (for reviews, see Black 1998; Graveley 2001; Maniatis and Tasic 2002). Some genes encode an extraordinarily large number of alternatively spliced variants. For example, the mammalian neurexin genes produce over 1000 variants of a presynaptic cell-adhesion molecule linking the pre- and postsynaptic compartments of synapses and essential for Ca2+-triggered neurotransmitter release (Missler et al. 2003). More remarkably, Dscam, a Drosophila axon guidance receptor gene, encodes over 38,000 variants (Schmucker et al. 2000). These genes are implicated in nervous system functions and/or development, suggesting a crucial role for alternative splicing in establishing the highly complex organization of animal tissues. Given that each isoform has its own function, alternative splicing must be strictly controlled to express proper isoforms in proper cells at the proper timing.
Splicing can occur either in cis or in trans: cis-splicing joins exons within a pre-mRNA, whereas trans-splicing joins exons of independently transcribed pre-mRNAs. Several cases of trans-splicing have been reported in mammalian cells (Shimizu and Honjo 1993; Fujieda et al. 1996; Caudevilla et al. 1998; Frantz et al. 1999; Chatterjee and Fisher 2000; Takahara et al. 2000; Finta and Zaphiropoulos 2002; Flouriot et al. 2002; Tasic et al. 2002; Wang et al. 2002). However, these occur with very low frequency, and their functional significance has not been demonstrated, leading to the proposal that they may represent “splicing noise” (Tasic et al. 2002; Wang et al. 2002). In contrast, trans-splicing, as an essential mechanism for generation of mRNAs, most likely occurs in the Drosophila modifier of mdg4 [mod(mdg4)] locus, because of its unusual genomic organization: Some of the alternatively spliced exons are encoded by the opposite DNA strand (Dorn et al. 2001; Labrador et al. 2001; Mongelard et al. 2002). mod(mdg4) encodes 26 isoforms, each consisting of identical N termini containing a BTB domain but variable C termini, 20 of which contain the conserved C2H2-type zinc finger motifs (Dorn et al. 2001). Surprisingly, seven isoforms are encoded by the antiparallel DNA strand, and are indeed expressed in vivo (Dorn et al. 2001). Therefore, generation of mRNA by trans-splicing is obvious in the mod-(mdg4) locus. No case of trans-splicing has been reported to date in genes with typical genomic organization, in which all exons are sequentially encoded by the same DNA strand.
The Drosophila longitudinals lacking (lola) gene encodes a family of BTB-Zn finger transcription factors required for a number of axon guidance decisions in the developing Drosophila nervous system (Giniger et al. 1994; Madden et al. 1999; Crowner et al. 2002; Goeke et al. 2003). lola is a large and complex locus, spanning over 60 kb and consisting of 32 exons aligned on the same DNA strand, and it produces at least 80 splicing variants through multiple promoter activities and alternative splicing (Fig. 1A; Ohsako et al. 2003). There are four promoters initiating transcription at distinct exons corresponding to different positions within the 5′UTR (exons 1–4). Each splicing variant has a constant sequence (exons 5–8) encoding the N-terminal region containing a BTB dimerization domain (Bardwell and Treisman 1994) and one or two exons alternatively selected from 20 groups of exons (exons 9–32) encoding a C-terminal variable region, 17 of which have unique zinc finger motifs. Almost all of the 17 putative DNA binding domains have unique combinations of predicted DNA contact residues, suggesting that they are likely to have unique DNA target specificities. Functional diversity of lola may be further increased through the BTB domain-mediated dimerization of isoforms. The lola isoforms are expressed in a complex pattern of tissues in the embryo, with some forms present in broad and overlapping domains and others in single tissues or dynamic patterns of isolated cells (Goeke et al. 2003). Furthermore, mutations inactivating single lola isoforms show defects in specific subsets of lola-dependent axon guidance choice points (Goeke et al. 2003). These findings suggest that alternative splicing of lola pre-mRNA plays critical roles in specifying a diverse variety of axon guidance decisions.
Figure 1.
Interallelic complementation among five lethal mutations in lola. (A) Schematic representation of the genomic organization of the lola locus. The entire locus consists of 32 exons; gray, white, and black boxes indicate 5′ variable, constant, and 3′ variable exons, respectively. Transcription starts at either of the 5′ variable exons, and 3′ variable exons are alternatively spliced to the constant exons, generating variants encoding 20 Lola isoforms with distinct C-terminal domains containing Zn finger motifs (Ohsako et al. 2003). (B) Locations of point mutations in EMS-induced mutant alleles lolaORE120, lolaORC46, lolaORE50, lolaORC4, and lolaORE119. Only the exons relevant to this study are indicated (C5–8, constant exons; V10–11, V21–22, V23, and V32, variable exons for isoforms B, K, L, and T, respectively). The arrow at the left end indicates the transcription start site and direction. (C) Relative viability (%) of F1 flies of the indicated genotype. The values were calculated based on the number of flies against expected ones, which was half of the number of flies with CyO chromosome, a balancer for all lola mutant alleles. Bars indicate the limits of 95% confidence.
Although the number of isoforms produced in this locus is not of the extent of those in Neurexin or Dscam, it suffers several mechanistic complexities. First, the alternatively spliced exons are the terminal ones in the mRNA, and therefore are associated with as many as 20 potential polyadenylation sites. Thus, proper expression of isoforms requires strict regulation of all these potential polyadenylation sites in coordination with the alternative splicing. Second, it is difficult to see how multiple splice variants are coexpressed within a single cell. Somehow, only a single 3′ end can be linked to the common exons in a single processed transcript, even though other 3′ ends are competent to be joined in the same cell. Trans-splicing may be one possibility to overcome these potential problems. However, despite similarities in molecular features of lola and mod(mdg4), the possibility of trans-splicing is less obvious in this case, because all lola exons are sequentially encoded in the same DNA strand. Therefore, careful experimental analysis is necessary to conclude whether or not trans-splicing is involved in the generation of lola mRNA.
In this report, we show that at least some lola isoforms are generated through alternative trans-splicing of the constant and variable exons. Genetic tests demonstrate that mutations in lola common exons can complement mutations in variable exons, and analysis of single nucleotide polymorphisms (SNPs) shows that this reflects the presence of chimeric, wholly wild-type mature transcripts in these mutant animals. This trans-splicing is highly efficient, with about half of the mature transcripts for some isoforms arising from trans-splicing events. Efficient trans-splicing is prevented by chromosome rearrangements, suggesting that the process requires chromosome pairing. Finally, we find that the final lola exon is expressed autonomously from a promoter in the preceding intron. The combination of internal promoters and trans-splicing may simplify the problem of achieving the complex pattern of lola isoform expression during development.
Results and Discussion
To dissect the genetic properties of the lola locus, we performed interallelic complementation tests among five ethylmethane sulfonate (EMS)-induced mutant alleles, lolaORE120, lolaORC46, lolaORC4, lolaORE50, and lolaORE119, whose molecular lesions are already identified (Fig. 1B; see Supplemental Material). Exons C5–8 constitute the constant region for all isoforms, and V10–11, V21–22, V23, and V32 correspond to the variable region of isoforms B, K, L, and T, respectively (Ohsako et al. 2003). All are homozygous lethal with severe defects in embryonic axon guidance (Crowner et al. 2002; Goeke et al. 2003). Viabilities of F1 flies from various crosses are summarized in Figure 1C. Flies heterozygous for alleles mutated in the same exon unit (lolaORE120/lolaORC46 for C5–8 and lolaORE50/lolaORC4 for V21–22) were lethal. In contrast, animals heterozygous for mutations in exons V21–22 and V23 (lolaORE50/lolaORE119 and lolaORC4/lolaORE119) showed nearly normal viability. The interallelic complementation between alleles mutated in different variable exons is reasonable, as a functional version of each isoform is supplied by one of the homologous chromosomes. To our surprise, however, combinations of mutations in the constant exons with those in the variable exons (lolaORC46/lolaORE50, lolaORE120/lolaORE119, and so on) also resulted in viable flies.
To explain these unexpected results, we postulated three models: (1) Functional mRNAs are produced through interallelic trans-splicing or any other mechanisms recombining two pre-mRNAs; (2) a wild-type locus was generated through recombination or gene conversion; or (3) functional complementation between the two mutant proteins. Model 3 is rather unlikely, because lolaORC46 contains a nonsense mutation within a constant exon, so that its mRNA would be degraded quickly via nonsense-mediated mRNA decay; even if it could survive, the translated protein lacks the entire variable region. In models 1 and 2, wild-type mRNA variants should be present in the hybrid animals. To examine this possibility, we determined the sequences of C5–8/V21–22 and C5–8/V23 cDNAs amplified by reverse transcriptase PCR (RT–PCR) from total RNA fractions of flies of the genotype lolaORE120/lolaORC4 and lolaORC46/lolaORE119, respectively. Indeed, we found C5–8/V21–22 and C5–8/V23 cDNA clones with no molecular lesions. In model 2, DNA recombination or gene conversion at the lola locus should occur at an extraordinarily high frequency at a very early stage of development. We examined whether it happened in the germ-line cells, which start dividing at a very early stage. lolaORE120/lolaORC4 and lolaORC46/lolaORE119 flies were crossed to Df(2R)stan2, a deficiency uncovering the entire lola region. None of the hemizygous offspring were viable, suggesting that DNA recombination or gene conversion at the lola locus is unlikely. Therefore, model 1 seemed the most likely mechanism to explain the interallelic complementation between mutations in the constant and those in the variable regions and the presence of chimeric mRNAs. In all sequenced clones including those described below, joining of exons occurs invariably at consensus splicing sites, suggesting that recombinant mRNAs are generated through trans-splicing, rather than any other mechanism.
We next quantified the frequency of interallelic trans-splicing events in the lola locus using SNPs as markers for each homologous chromosome in the hybrids. Several SNPs and restriction fragment length polymorphisms (RFLPs) were identified within the regions of C5–8, V10–11, V23, and V32 among three D. melanogaster strains, Oregon R (OR), yellow white (YW) and Shanghai 83K-6 (83K; see Supplemental Material). These molecular markers allowed us to determine the chromosomal origin of mRNA for the constant and variable regions separately. Figure 2 shows the result of RFLP analysis of lola C5–8/V32 variant cDNA. Total RNA fraction was prepared from adult males of F1 hybrids between OR and YW (OR/YW F1). A 3045-bp fragment of C5–8/V32 cDNA was amplified by RT–PCR under conditions that minimize possible template switching (Tasic et al. 2002; see Materials and Methods). The PCR products were subjected to agarose gel electrophoresis after allele-specific restriction digestion with BstZ17I and FspI (Fig. 2, lane 5). Two DNA fragments (3045 and 1195 bp) were detected, indicating that interallelic hybrid mRNAs were indeed produced. The intensities of these two DNA bands were approximately equal to those of DNA fragments derived from the same chromosome. No significant template switching seemed to occur under these conditions, because no such hybrid cDNA fragments were detected in the reaction containing a mixture of equal amounts of total RNA prepared from strains OR and YW (Fig. 2, lane 4). The results demonstrate that trans-splicing occurs at the constant/variable junction of lola mRNA.
Figure 2.
RFLP analysis of interallelic trans-splicing events. The C5–8/V32 variant cDNAs were RT–PCR-amplified from total RNA fraction of D. melanogaster strains Oregon R (OR) and yellow white (YW), and their hybrids (F1), and analyzed by 1.5% agarose gel electrophoresis after restriction digestion with FspI and BstZ17I. (Lane 1) M indicates 1 kb ladder markers. (Lane 2) O indicates OR (Lane 3) Y denotes YW. (Lane 4) O+Y indicates equal amounts of OR and YW RNAs mixed before the RT–PCR. (Lane 5) O/Y F1 denotes OR/YW F1. (Lane 6) U indicates undigested RT–PCR products from OR. Structures of C5–8/V32 variant cDNAs are illustrated to the right. The segments derived from OR and YW are indicated in the rectangle and ellipsoid with designations of O and Y, respectively. The thick vertical bars represent the allele-specific restriction sites. The fragment lengths obtained after restriction digestion are indicated in base pairs (bp).
To confirm the frequency of interallelic trans-splicing events, we cloned the PCR products into pGEM-T vector, sequenced, and determined the C/V haplotypes (Table 1). With respect to the C5–8/V10–11 variant (isoform B), only one out of 94 clones contained the sequences derived from both alleles (cDNA variant haplotypes OR/YW and YW/OR), indicating that the variant is predominantly generated through alternative cis-splicing. In contrast, for the C5–8/V32 variant (isoform T), 45 of 95 clones (47%) contained the cDNA variants with interallelic hybrid haplotypes. Similar results were obtained with F1 hybrids between 83K and YW. The frequency of cDNA variants with interallelic hybrid haplotypes was 47% (38/81) and 44% (36/82) for C5–8/V23 (isoform L) and C5–8/V32 (isoform T), respectively. The results demonstrate that these isoforms are generated though alternative trans-splicing of the constant and variable exons.
Table 1.
Quantitative analysis of interallelic trans-splicing events
| Genotype | Variants | cDNA variant haplotypea No. of clones | Frequencyb | |||
|---|---|---|---|---|---|---|
| OR/YW | OR/OR
|
YW/YW
|
OR/YW
|
YW/OR
|
||
| C5-8/V10 | 42 | 51 | 0 | 1 | 0.01 | |
| C5-8/V32 | 26 | 24 | 29 | 16 | 0.47 | |
| 83K/YW | 83K/83K
|
YW/YW
|
83K/YW
|
YW/83K
|
||
| C5-8/V23 | 22 | 21 | 20 | 18 | 0.47 | |
| C5-8/V32 | 26 | 20 | 26 | 10 | 0.44 | |
| YW/CKG30 | YW/YW
|
C/C
|
YW/C
|
C/YW
|
||
| C5-8/V23 | 40 | 45 | 13 | 11 | 0.22 | |
| 83K/CKG30 | 83K/83K
|
C/C
|
83K/C
|
C/83K
|
||
| C5-8/V32 | 43 | 46 | 9 | 10 | 0.18 | |
| OR/T(2;3)X3 | OR/OR
|
T/T
|
OR/T
|
T/OR
|
||
| C5-8/V32 | 59 | 48 | 1 | 0 | 0.01 | |
| 83K/T(2;3)X3 | 83K/83K
|
T/T
|
83K/T
|
T/83K
|
||
| C5-8/V32 | 48 | 42 | 0 | 0 | 0.00 | |
Variant haplotype is expressed as a combination of allele names for constant/variable exons. CKG30 and T(2;3)X3 are abbreviated as C and T, respectively
Number of cDNA clones with interallelic hybrid haplotypes per total number of sequenced clones
It is important to note that the variable exons (V23 and V32) assort independently of the constant exons; they are trans-spliced to the constant exons derived from either homologous chromosome at nearly equal frequency. This suggests that the splicing must occur when the pre-mRNAs from the two homologous chromosomes are in close proximity. In Drosophila and other dipteran insects, genes on homologous chromosomes are thought to be physically proximal to each other through pairing (Metz 1916). Transvection and trans-sensing effects are severely affected when homologous chromosome pairing is disrupted by chromosomal rearrangements (Lewis 1954; Gelbart 1982). To examine the influence of chromosomal rearrangements on the frequency of interallelic trans-splicing in the lola locus, we performed SNP analyses of F1 hybrids with a CKG30 balancer chromosome with multiple inversions, and those with T(2;3)X3, a translocation involving the lola locus (Table 1). The frequency of cDNA clones with interallelic hybrid haplotypes was significantly reduced by CKG30[24 of 109 (22%) for C5–8/V23 (YW/C and C/YW), and 19 of 108 (18%) for C5–8/V32 (83K/C and C/83K)] and by T(2;3)X3 [only one of 108 (1%) for C5–8/V32 (OR/T and T/OR), and none of 90 for C5–8/V32 (83K/T and T/83K)]. These results clearly indicate that interallelic trans-splicing occurs frequently when the two chromosomal loci are positioned such that they can pair effectively.
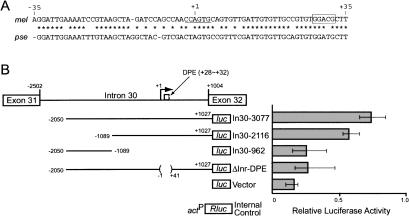
Next we examined whether pre-mRNAs encoding the variable exons are transcribed independently of the constant exons. We attempted a poly-d(C)-mediated 5′ rapid amplification of cDNA ends (RACE) analysis using total RNA fraction of embryos with a primer corresponding to exon V32. The 5′ RACE products were cloned and sequenced. In most of the clones (36/40), the 5′ ends matched one of the 5′ ends of the mature mRNAs previously determined by Ohsako et al. (2003). In the remaining four clones, the 5′ ends were the adenine located 1003 bp upstream of the splicing junction of exon V32 (defined as +1 in Fig. 3A, nucleotide position 58,035). The sequence between -2 and +4 (CCAGTG) partially matched Drosophila consensus of initiator (Inr) element (T-C-A+1-G/T-T-T/C), where A+1 is the transcription start site (Arkhipova 1995). No obvious TATA box was found, but the downstream promoter element (DPE), GGACG, was present between +28 and +32 (Drosophila consensus is A/G-G-A/T-C/T-G; Kutach and Kadonaga 2000). We compared the sequence of this region with that of the corresponding region of the Drosophila pseudoobscura genome (Drosophila Genome Project at the Human Genome Sequencing Center, http://www.hg-sc.bcm.tmc.edu/projects/drosophila). The exon/intron structure of the lola locus in D. pseudoobscura was estimated based on the D. melanogaster sequence (Ohsako et al. 2003). The sequence of the region containing Inr and DPE (-2 to +32) was highly conserved (82.4% identity) between the two species (the overall identity for the sequence of intron 30 was 52.7%). Transcription start sites located within intron 30 were supported by region-specific semiquantitative RT–PCR analysis using total RNA fraction prepared from wild-type and transgenic flies carrying an intron 30-lacZ fusion construct: The region downstream of the putative Inr was expressed at higher levels than the region upstream of it (Supplementary Fig. 1).
Figure 3.
Detection of V32 exon-specific core promoter element. (A) Nucleotide sequence comparison around the V32-specific core promoter element between D. melanogaster (mel) and D. pseudoobscura (pse). The transcription start site was estimated based on the 5′ RACE, and defined as +1. The putative Inr and DPE elements are underlined and boxed, respectively. Asterisks indicate identical nucleotides. (B) Reporter assay for V32-specific promoter activity. Structures of constructs containing intron 30–luciferase fusions are schematically shown to the left. The putative transcription initiation site and direction are indicated by the bent arrow. The open box indicates the putative DPE element. The reporter constructs were transfected into Drosophila S2 cells, and luciferase activities were determined. The act–Rluc construct was cotransfected as an internal control. Results represent the mean ± S.E. for three independent experiments.
Promoter activity of the DPE (+28GGACG+32) located within intron 30 was confirmed by luciferase reporter assay in the Drosophila S2 cell line (Fig. 3B). Four DNA segments of intron 30 (In30-3077, In30-2116, In30-962, and ΔInr–DPE) were inserted upstream of the promoterless luc gene encoding firefly luciferase. The constructs were introduced into S2 cells, and the luciferase activities were determined 48 h after transfection. The act-Rluc construct was cotransfected to normalize the transfection efficiency. Two DNA segments, In30-3077 and In30-2116, containing the DPE had significant promoter activity, whereas no significant activities were detected in In30-962 and ΔInr–DPE segments lacking the DPE. Therefore, the DPE (+28GGACG+32) within intron 30 is likely to be a core promoter element for transcription of exon V32. Although there might be additional regulatory sequences elsewhere, these data strongly support that exon V32 (the variable region of isoform T) is an independent transcription unit. Because exons V21–22 and V23 are also trans-spliced to the constant exons, the genomic regions containing these exons may also be independent transcription units.
In summary, therefore, alternative trans-splicing at the constant/variable junction generates a significant portion of the mRNA from the lola locus. For some isoforms, nearly half of the mature RNAs were chimeras between pre-mRNAs derived from the homologous chromosomes, indicating that the mRNAs were generated through trans-splicing exclusively. Finally, in one case, we found a variable exon being expressed from a dedicated promoter in the preceding intron. lola provides the first demonstration of abundant mRNAs generated by alternative trans-splicing of exons sequentially encoded by the same DNA strand. The frequent interallelic trans-splicing events support the pairing of homologous chromosomes in Drosophila. In contrast, mammalian hybrid alpha/gamma protocadherin mRNAs, which are most likely generated by trans-splicing, can be spliced only intrachromosomally (Tasic et al. 2002). This difference may be due to a lack of chromosomal pairing during transcription in mammalian cells, and/or a high competence of Drosophila pre-mRNAs to be trans-spliced.
There are important implications regarding the mechanism of trans-splicing. First, because the efficiency of interchromosomal trans-splicing was dramatically reduced by chromosomal rearrangements, we infer that trans-splicing occurs locally either cotranscriptionally or posttranscriptionally. Cotranscriptional trans-splicing is consistent with a recent view of gene expression in which all steps of mRNA processing take place cotranscriptionally (Maniatis and Reed 2002; Orphanides and Reinberg 2002; Proudfoot et al. 2002). It is also supported by reported examples of trans-splicing events detected in mammalian cells in which trans-splicing occurred between pre-mRNAs derived from the same genes or closely linked ones (Shimizu and Honjo 1993; Fujieda et al. 1996; Caudevilla et al. 1998; Frantz et al. 1999; Chatterjee and Fisher 2000; Takahara et al. 2000; Finta and Zaphiropoulos 2002; Flouriot et al. 2002; Tasic et al. 2002; Wang et al. 2002). Dorn et al. (2001) showed that trans-splicing of mod(mdg4) pre-mRNAs could occur between those derived from the original locus and a transgene inserted in a different chromosome. However, this may not represent the natural situation, because a high level of transgene expression was induced by the GAL4–UAS system, and RT–PCR not controlled for template switching was used to detect trans-spliced mRNAs. Therefore, it may be reasonable to assume that natural trans-splicing occurs locally, most likely cotranscriptionally, as is the case for cis-splicing. An additional requirement for trans-splicing would be a mechanism allowing integration of pre-mRNAs encoding variable exons into the spliceosome operating in the constant region. The outron sequences (transcribed region preceding the variable exons) may contain cis-elements required for this process.
Second, variable regions may consist of multiple independent transcription units whose pre-mRNAs are each trans-spliced to the constant exons. With internal promoter(s) for variable exons and cotranscriptional alternative trans-splicing, the regulation of lola isoform expression could be much simplified. Unlike alternative cis-splicing, simultaneous expression of multiple isoforms can be achieved without complex regulation of splicing and polyadenylation sites of a single mRNA. Multiple pre-mRNAs containing different variable exons could be expressed in the same cells depending on their own promoter activities, which would then be trans-spliced to the constant exons, possibly in a concentration-dependent manner. Furthermore, the internal promoter(s) for variable exons may be tissue-specific, and thus it could also simplify the mechanism of tissue-specific expression of particular variants as well.
lola shares many molecular features with mod(mdg4), and it is therefore possible that mod(mdg4) isoforms sequentially encoded in the same DNA strand could also be generated through trans-splicing. Furthermore, there are other fly genes encoding BTB-Zn finger proteins, such as fruitless (Usui-Aoki et al. 2000) and Br-C, some of which show complex patterns of intragenic complementation reminiscent of what we see with lola (Bayer et al. 1997). These genes produce multiple splice variants, often with a complex pattern of expression. Trans-splicing may be a general means to generate splice variants in this family of genes.
Materials and methods
SNP analysis and RFLP analysis
Total RNA fraction was prepared from male adult flies using an RNeasy mini-kit (QIAGEN). cDNAs were synthesized with oligo-d(T) primer using You-prime first-strand beads (Amersham Bioscience), and amplified under conditions whereby the concentration of PCR product would not exceed 0.5 ng/μL (94°C for 5 min, 20–30 cycles of 94°C for 30 sec, 57°C for 45 sec, and 72°C for 2 min), which minimizes template switching (Tasic et al. 2002). The PCR products were purified from an agarose gel with a Gel extraction kit (QIAGEN).
For SNP analysis, cDNAs were amplified by PCR with the following primers: TH174 (AGCCGGAACCAGCAGCCAAGCGTCG) and TH204 (ATTGAACCCAAAGCCGAATACGATG) corresponding to the C5–8 sequence, TH205 (GCCGATGATGCGCTTGTGGATGTGG) corresponding to the V10 sequence, TH206 (TTGTGGGCTTTCCACTGCCATTTCC) corresponding to the V23 sequence, and TH064 (TTCGTCGTCCTCGGTCGAATAGCCC) and TH207 (ATGCTCCGTGGACGATGCCGCTGTC) corresponding to the V32 sequence. PCR products were cloned into pGEM-T easy vector (Promega). Individual clones were sequenced with BigDye terminator (Applied Biosystems).
For the RFLP analysis, cDNA of the C5–8/V32 variant was amplified by PCR with a C5–8-specific primer TH130(CACTGGCGGCGGAGTAGCTCCCAAG) and a V32-specific primer TH062 (GTTAATAATTTTTAAGATACAATAAACTCAGTC). PCR product was digested with FspI and BstZ17I (NEB), electrophoresed on 1.5% agarose gel in TAE buffer (40 mM Trisacetate and 1 mM EDTA), and visualized by ethidium bromide staining.
Other Materials and Methods are described in Supplemental Material.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas “Genome Science” from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No.12202002) to T.A., and by grant IBN 9904519 from the National Science Foundation (USA) to E.G. T.H. is a research fellow of the Japan Society for the Promotion of Science.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1137303.
Supplemental Material is available at http://www.genesdev.org.
References
- Arkhipova I.R. 1995. Promoter elements in Drosophila melanogaster revealed by sequence analysis. Genetics 139: 1359-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell V.J. and Treisman, R. 1994. The POZ domain: A conserved protein–protein interaction motif. Genes & Dev. 8: 1664-1677. [DOI] [PubMed] [Google Scholar]
- Bayer C.A., von Kalm, L., and Fristrom, J.W. 1997. Relationships between protein isoforms and genetic functions demonstrate functional redundancy at the Broad-Complex during Drosophila metamorphosis. Dev. Biol. 187: 267-282. [DOI] [PubMed] [Google Scholar]
- Black D.L. 1998. Splicing in the inner ear: A familiar tune, but what are the instruments? Neuron 20: 165-168. [DOI] [PubMed] [Google Scholar]
- Caudevilla C., Serra, D., Miliar, A., Codony, C., Asins, G., Bach, M., and Hegardt, F.G. 1998. Natural trans-splicing in carnitine octanoyltransferase pre-mRNAs in rat liver. Proc. Natl. Acad. Sci. 95: 12185-12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee T.K. and Fisher, R.A. 2000. Novel alternative splicing and nuclear localization of human RGS12 gene products. J. Biol. Chem. 275: 29660-29671. [DOI] [PubMed] [Google Scholar]
- Crowner D., Madden, K., Goeke, S., and Giniger, E. 2002. Lola regulates midline crossing of CNS axons in Drosophila. Development 129: 1317-1325. [DOI] [PubMed] [Google Scholar]
- Dorn R., Reuter, G., and Loewendorf, A. 2001. Transgene analysis proves mRNA trans-splicing at the complex mod(mdg4) locus in Drosophila. Proc. Natl. Acad. Sci. 98: 9724-9729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finta C. and Zaphiropoulos, P.G. 2002. Intergenic mRNA molecules resulting from trans-splicing. J. Biol. Chem. 277: 5882-5890. [DOI] [PubMed] [Google Scholar]
- Flouriot G., Brand, H., Seraphin, B., and Gannon, F. 2002. Natural trans-spliced mRNAs are generated from the human estrogen receptor-alpha (hER alpha) gene. J. Biol. Chem. 277: 26244-26251. [DOI] [PubMed] [Google Scholar]
- Frantz S.A., Thiara, A.S., Lodwick, D., Ng, L.L., Eperon, I.C., and Samani, N.J. 1999. Exon repetition in mRNA. Proc. Natl. Acad. Sci. 96: 5400-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujieda S., Lin, Y.Q., Saxon, A., and Zhang, K. 1996. Multiple types of chimeric germ-line Ig heavy chain transcripts in human B cells: Evidence for trans-splicing of human Ig RNA. J. Immunol. 157: 3450-3459. [PubMed] [Google Scholar]
- Gelbart W.M. 1982. Synapsis-dependent allelic complementation at the decapentaplegic gene complex in Drosophila melanogaster. Proc. Natl. Acad. Sci. 79: 2636-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E., Tietje, K., Jan, L.Y., and Jan, Y.N. 1994. lola encodes a putative transcription factor required for axon growth and guidance in Drosophila. Development 120: 1385-1398. [DOI] [PubMed] [Google Scholar]
- Goeke S., Greene, E.A., Grant, P.K., Gates, M.A., Crowner, D., Aigaki, T., and Giniger, E. 2003. Alternative splicing of lola generates 19 transcription factors controlling axon guidance in Drosophila. Nature Neurosci. 6: 917-924. [DOI] [PubMed] [Google Scholar]
- Graveley B.R. 2001. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 17: 100-107. [DOI] [PubMed] [Google Scholar]
- Kutach A.K. and Kadonaga, J.T. 2000. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell. Biol. 20: 4754-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrador M., Mongelard, F., Plata-Rengifo, P., Baxter, E.M., Corces, V.G., and Gerasimova, T.I. 2001. Protein encoding by both DNA strands. Nature 409: 1000. [DOI] [PubMed] [Google Scholar]
- Lewis E.B. 1954. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am. Nat. 88: 225-239. [Google Scholar]
- Madden K., Crowner, D., and Giniger, E. 1999. LOLA has the properties of a master regulator of axon–target interaction for SNb motor axons of Drosophila. Dev. Biol. 213: 301-313. [DOI] [PubMed] [Google Scholar]
- Maniatis T. and Reed, R. 2002. An extensive network of coupling among gene expression machines. Nature 416: 499-506. [DOI] [PubMed] [Google Scholar]
- Maniatis T. and Tasic, B. 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418: 236-243. [DOI] [PubMed] [Google Scholar]
- Metz C.W. 1916. Chromosome studies on the Diptera II. The paired association of chromosomes in the Diptera and its significance. J. Exp. Zool. 21: 213-279. [Google Scholar]
- Missler M., Zhang, W., Rohlmann, A., Kattenstroth, G., Hammer, R.E., Gottmann, K., and Sudhof, T.C. 2003. α-Neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature 423: 939-948. [DOI] [PubMed] [Google Scholar]
- Mongelard F., Labrador, M., Baxter, E.M., Gerasimova, T.I., and Corces, V.G. 2002. Trans-splicing as a novel mechanism to explain interallelic complementation in Drosophila. Genetics 160: 1481-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsako T., Horiuchi, T., Matsuo, T., Komaya, S., and Aigaki, T. 2003. Drosophila lola encodes a family of BTB-transcription regulators with highly variable C-terminal domains containing zinc finger motifs. Gene 311: 59-69. [DOI] [PubMed] [Google Scholar]
- Orphanides G. and Reinberg, D. 2002. A unified theory of gene expression. Cell 108: 439-451. [DOI] [PubMed] [Google Scholar]
- Proudfoot N.J., Furger, A., and Dye, M.J. 2002. Integrating mRNA processing with transcription. Cell 108: 501-512. [DOI] [PubMed] [Google Scholar]
- Schmucker D., Clemens, J.C., Shu, H., Worby, C.A., Xiao, J., Muda, M., Dixon, J.E., and Zipursky, S.L. 2000. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101: 671-684. [DOI] [PubMed] [Google Scholar]
- Shimizu A. and Honjo, T. 1993. Synthesis and regulation of trans-mRNA encoding the immunoglobulin epsilon heavy chain. FASEB J. 7: 149-154. [DOI] [PubMed] [Google Scholar]
- Takahara T., Kanazu, S.I., Yanagisawa, S., and Akanuma, H. 2000. Heterogeneous Sp1 mRNAs in human HepG2 cells include a product of homotypic trans-splicing. J. Biol. Chem. 275: 38067-38072. [DOI] [PubMed] [Google Scholar]
- Tasic B., Nabholz, C.E., Baldwin, K.K., Kim, Y., Rueckert, E.H., Ribich, S.A., Cramer, P., Wu, Q., Axel, R., and Maniatis, T. 2002. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol. Cell 10: 21-33. [DOI] [PubMed] [Google Scholar]
- Usui-Aoki K., Ito, H., Ui-Tei, K., Takahashi, K., Lukacsovich, T., Awano, W., Nakata, H., Piao, Z.F., Nilsson, E.E., Tomida, J., et al. 2000. Formation of the male-specific muscle in female Drosophila by ectopic fruitless expression. Nat. Cell Biol. 2: 500-506. [DOI] [PubMed] [Google Scholar]
- Wang X., Su, H., and Bradley, A. 2002. Molecular mechanisms governing Pcdh-γ gene expression: Evidence for a multiple promoter and cis-alternative splicing model. Genes & Dev. 16: 1890-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]