Abstract
The Brain Tumor (Brat) protein is recruited to the 3′ untranslated region (UTR) of hunchback mRNA to regulate its translation. Recruitment is mediated by interactions between the Pumilio RNA-binding Puf repeats and the NHL domain of Brat, a conserved structural motif present in a large family of growth regulators. In this report, we describe the crystal structure of the Brat NHL domain and present a model of the Pumilio–Brat complex derived from in silico docking experiments and supported by mutational analysis of the protein–protein interface. A key feature of the model is recognition of the outer, convex surface of the Pumilio Puf domain by the top, electropositive face of the six-bladed Brat β-propeller. In particular, an extended loop in Puf repeat 8 fits in the entrance to the central channel of the Brat β-propeller. Together, these interactions are likely to be prototypic of the recruitment strategies of other NHL-containing proteins in development.
Keywords: Brain Tumor, Pumilio, mRNA translation, NHL domain, β-propellor, crystal structure
Translation regulation of maternally encoded mRNAs is essential in generating the early protein gradients that govern embryonic patterning in Drosophila. In particular, normal development of the anterior–posterior axis proceeds only when translation of maternal hunchback (hb) mRNA is repressed at the posterior (Hülskamp et al. 1989; Irish et al. 1989; Struhl 1989). Repression is mediated by binding of Pumilio (Pum) to specific sequences (Nanos response elements, NREs) in the 3′ untranslated region (UTR) of hb mRNA (Wharton et al. 1998; Wang et al. 2002), and the subsequent recruitment of two cofactors into a quaternary complex (Sonoda and Wharton 1999, 2001). The first of these is Nanos (Nos), a zinc-finger protein that provides the spatial cue for abdominal segmentation (Wang and Lehmann 1991). The second cofactor is Brain Tumor (Brat; Arama et al. 2000), which is recuited jointly via interactions with Pum and Nos.
In addition to its role as a translational repressor in the early embryo, Brat also acts as a tumor suppressor during later development. Loss of function brat alleles are associated with a dramatic overgrowth of the larval brain (Hankins 1991). Although they do not metastasize in situ, transplantation of brat mutant brain or imaginal disc cells do so upon injection into larvae (Woodhouse et al. 1998). When analyzed in somatic clones in the wing imaginal disc, brat cells are larger than wild type, with larger nucleoli and more rRNA, suggesting a role in growth or size regulation (Frank et al. 2002). Although the mechanism by which Brat regulates cell size is not yet clear, its biological activity is conserved; expression of Drosophila Brat in Caenorhabditis elegans rescues the enlarged nucleolar phenotype associated with loss of ncl-1 function. Ncl-1 and Brat are the closest relatives encoded by their respective genomes, exhibiting particularly high homology in the conserved NHL domain that defines an emerging family of growth regulators, described below.
The NHL domain was defined by amino acid sequence homologies among Ncl-1, HT2A (a human protein that binds to HIV Tat), and Lin-41 (a translational regulator in C. elegans; Slack and Ruvkun 1998). The sequence repeats that comprise NHL domains have been proposed to form β-propeller structures, similar to those of WD40 repeat-bearing molecules. Many NHL proteins have additional motifs—a ring finger (R), B-box zinc fingers (B), and coiled-coil (CC)—and therefore fall into a larger family of RBCC-NHL proteins. Brat is an imperfect member of the RBCC family, lacking the RING domain (Fig. 1A). The NHL domain is central to the activity of Brat in regulation of both growth in larval tissues and translation in the embryo. Missense mutations in the NHL domain or stop codons that eliminate its synthesis entirely cause overgrowth of larval tissues (Arama et al. 2000). Some of the same missense mutations block recruitment into a repression complex in vitro and compromise hb regulation in the early embryo, suggesting that the same surface of the NHL domain is used in both processes (Sonoda and Wharton 2001). Most importantly, expression of the isolated NHL domain is sufficient to provide Brat function to otherwise brat embryos, demonstrating that the translational effector function is embedded within the domain (Sonoda and Wharton 2001).
Figure 1.
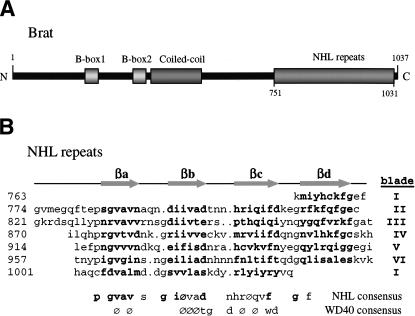
Brat primary and secondary structure. (A) Cartoon showing the relative locations of the conserved domains (B boxes, coiled-coil, and NHL domain) in full-length Brat. (B) Structure based alignment of the Brat NHL repeats. The secondary structure elements (β-strands βa–βd) are shown above the alignment. The consensus NHL sequence is shown below with residues conserved in Brat in bold. The WD40 consensus is shown for comparison. (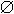 ) Hydrophobic residue.
) Hydrophobic residue.
To better understand the mechanism by which Brat regulates both embryonic patterning and cell growth, we first determined a high-resolution crystal structure of the isolated NHL domain. We then generated a model of the NHL–Pum complex in molecular docking experiments, supported by mutational analysis of the NHL and Pumilio surfaces. The model reveals a novel role for the NHL domain, and provides a framework to understand the activities of β-propeller-containing proteins in a wide range of organisms.
Results and Discussion
Structure determination
The NHL domain from Drosophila Brat, encompassing residues 756 to 1037, was expressed in Escherichia coli and purified to homogeneity. Orthorhombic crystals were obtained from solutions of ammonium phosphate and MPD, containing two molecules per asymmetric unit (AU). For phasing, multiwavelength anomalous dispersion (MAD) data were measured from a selenomethionine (SeMet) derivative (Supplementary Table 1) that yielded an interpretable electron density map (2.4 Å resolution) and allowed partial building of both monomers in the AU. A more complete NHL domain monomer was built by averaging the density of the two molecules related by noncrystallographic symmetry (NCS). Later, a native data set to 1.95 Å was collected; however, this was slightly nonisomorphous with respect to the SeMet data and required molecular replacement to locate both molecules in the native AU. Solvent flattening along with NCS averaging were used to build a complete model of both molecules with the native data. The refined structure contains residues 759–1037 for each protomer, and 663 water molecules.
Rcryst and Rfree are 20.02% and 25.23%, respectively, for data between 15 and 1.95 Å resolution. The rms deviations for bonds and angles are 0.006 Å and 1.36°, respectively.
Structure of the NHL domain
The Brat NHL domain is a six-bladed β-propeller, with the blades arrayed in a radial fashion around the central pseudo-sixfold axis, and each blade composed of a highly twisted four-stranded antiparallel β-sheet. The β-strands are labeled a–d from the inside to the outside of the molecule, whereby each βa lines a solvent-filled channel in the interior and βd delineates the outer perimeter of the propeller. Each NHL repeat specifies a single blade of the propeller derived from contiguous sequence in the primary structure, with the exception of the first blade, where βa–βc are provided by C-terminal sequence and βd derives from the most N-terminal residues (Figs. 1, 2). This “Velcro” maintains the circular arrangement of the blades, connecting the ends of the NHL primary sequence into the doughnut-shaped molecule (∼47 Å in diameter and ∼34 Å in height).
Figure 2.
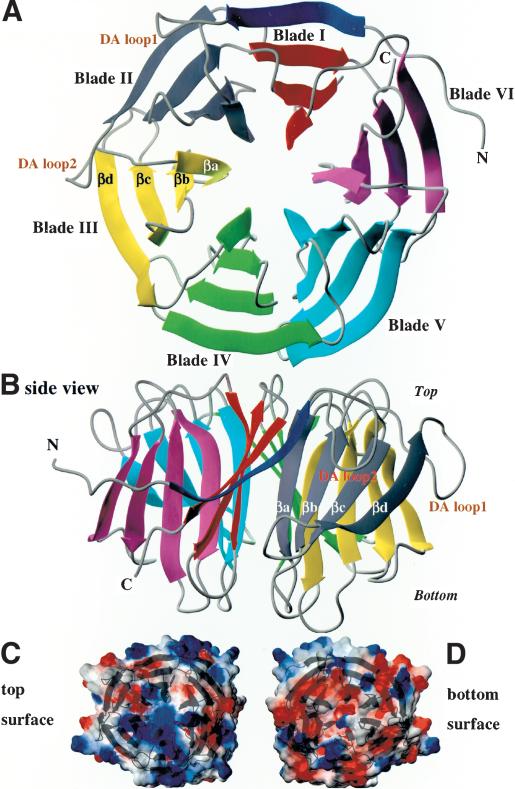
Brat NHL domain structure and surface. (A) The NHL domain is a six-bladed β-propeller as seen looking at the “top” surface, with flexible loops connecting the β-strands of each blade (B) on the top and bottom surfaces. Calculation of electrostatics at the surface of the NHL domain reveals the generally electropositive nature of the top surface (C) compared with the electronegative bottom surface (D).
The Brat blades are regular in structure, their β-sheets superimposing with root-mean-square deviations (rmsd) of 0.97–1.25 Å. The blades diverge in structure primarily in the loops radiating from the “top” and “bottom” faces of the propeller, connecting the β-strands. By convention, the top face of a β-propeller is defined as containing loops connecting βb to βc within a blade, and βd from one blade to βa of the next blade (the DAloop; Fig. 2). Conversely, the bottom side contains loops connecting βa to βb and βc to βd (Lambright et al. 1996). The only parts of the Brat NHL domain that do not conform to the “ideal” NHL repeat are the two extralong DA loops connecting blades 1, 2, and 3 (Figs. 1, 2). A computation of the electrostatic potential of the molecule reveals that the top and bottom surfaces bear a considerably different charge. The top is mostly electropositive whereas the bottom is much more electronegative (Fig. 2C).
The β-propeller is a multifunctional domain, which, in the right context, can mediate protein:protein interactions, bind metals, catalyze enzymatic reactions, and possibly transport small molecules through the central channel. β-Propellers with six, seven, and eight blades have been observed in proteins both with and without obvious repeats in their primary sequence. The most notable repeated motif is the WD40 repeat that specifies a seven- or eight-bladed β-propeller in proteins such as the β-subunits of heterotrimeric G proteins; that is, β-transducin (Sondek et al. 1996) and the transcription corepressor Tup1 (Sprague et al. 2000), both seven blades, and Cdc-4 (Orlicky et al. 2003) with eight blades, among others. Although, NHL repeats do not match the WD consensus, and fold into to a six- instead of a seven- or eight-bladed β-propeller, there are similarities in the pattern of buried hydrophobic residues. In particular, an “F” in the βc strand of Brat blades 2, 4, 5, and 6 is at a structurally analogous position to the “W” in WD40 repeats (Fig. 3). However, unlike the WD40 repeats in β-transducin, which have a structural triad (Asp–His–Ser) that forms a hydrogen bonding network extending to the signature W of the WD40 motif (Sondek et al. 1996), the Brat NHL repeats contain no such hydrogen-bonding network, in part because there is no equivalent trio of polar residues and the conserved F lacks the hydrogen bonding potential of a W. Instead, the conserved F is part of a broad hydrophobic cluster that folds and holds the NHL blades together, making extensive van der Waals contacts with conserved I/V residues in strand βc. Thus, much of the conservation of sequence between the NHL blades and across NHL-containing proteins is due to buried hydrophobic residues, located roughly in the middle of each β-strand. There are conserved prolines at the start of strand βa and glycines at the start of strands βb and βd that may allow for tight turns between the strands. Overall, the Brat β-propeller is slightly bigger than the β-transducin propeller, even though it has one blade less. This larger size, 47 Å in diameter versus 44 Å in β-transducin, may reflect the slightly longer β-strands in Brat (43–48 residues per NHL repeat as compared with 40 residues per WD40 in β-transducin).
Figure 3.
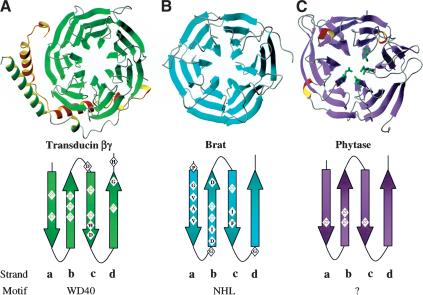
β-Propeller structures with consensus repeats. Three β-propeller structures are shown with the consensus sequence of the repeat motif mapped onto secondary structure elements below. (A) Transducin β (shown bound to its partner Transducin γ) is a seven-bladed β-propeller constructed from WD40 repeats. (B) Brat is a six-bladed propeller with each blade an NHL repeat. (C) Phytase is also a six-bladed propeller, but with very little sequence conservation across blades (residues highlighted in green are the active site residues). (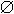 ) Hydrophobic residue.
) Hydrophobic residue.
Six-bladed β-propellers have been seen in diverse proteins, ranging from neuramindase in influenza virus, quinoprotein glucose dehydrogenase (gluDH), phytase, and the periplasmic protein TolB in bacteria, to the recycling portion of the LDL receptor in humans. Interestingly, only TolB (Abergel et al. 1999) and the LDL receptor (Jeon et al. 2001) contain an easily identifiable repetitive sequence: the P[A/S][W/F][A/S]PDG and YWTD repeats, respectively. The Brat six-bladed β-propeller is more regular than these previously determined structures with an average rmsd of 1.15 Å among the Brat blades versus 1.31 Å among the blades of TolB, for example. This uniformity in structure may reflect the extended homology of NHL repeats, where up to 19residues are at conserved positions. Curiously, in spite of unrelated sequence and function, the closest structural homolog to Brat (in the protein database) is phytase (Fig. 3; Ha et al. 2000). The β-propellers of the two proteins superimpose with an rmsd of 2.5 Å (for 300 Cα atom pairs), which is significantly lower than the rmsd values obtained between Brat and the other six-bladed β-propellers (with rmsd ranging between 3.0 and 3.2 Å for gluDH, transducinβ, neuraminidase, and TolB, for instance). Phytase catalyzes the hydrolysis of phytic acid to phosphate in a calcium loaded state (five or six calcium ions) with the DA loop1 and the loops lining the central channel mediating calcium binding (Ha et al. 2000). There is no indication in our structure that the equivalent loops in Brat partake in analogous metal binding. The central channel in Brat is completely filled with ordered waters and is closed at both ends by the side chains of arginine residues emanating from the βa strands. In all, highly disparate sequences appear capable of forming β-propellers that differ in size, shape, and charge, commensurate with different functions. The NHL β-propeller is the essential scaffold for both the translational repression and cell growth inhibitory activities of Brat in Drosophila.
Pum–NHLinteraction
To understand how the NHL domain is specifically targeted to hb mRNA, we attempted to cocrystallize the entire quaternary complex (Pum, Nos, NHL domain, and NRE), as well as various subassemblies. None of these strategies has so far succeeded, including an attempt to cocrystallize Pum and the NHL domain, consistent with the idea that interactions with both Pum and Nos are required for Brat recruitment (Sonoda and Wharton 2001). The Nos structure is unknown, and preliminary evidence suggests it is largely disordered in solution, folding as it joins the complex (T. Edwards and A. Aggarwal, unpubl.).
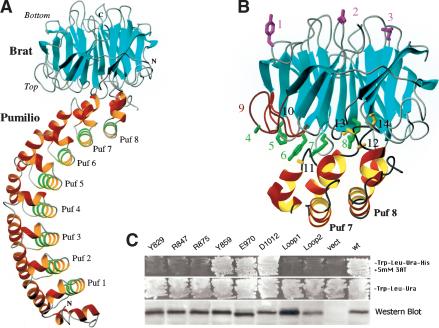
Therefore, we used the docking program BiGGER (Palma et al. 2000) to generate models of the Pum–NHL domain binary complex starting with the structures of the isolated Pum RNA-binding domain (Edwards et al. 2001) and the NHL domain (Fig. 2). The program generates a family of related models with the convex edge of the Brat propeller inserted into the concave surface of the Puf domain, which is known to bind RNA (Edwards et al. 2001; Wang et al. 2002). Athough these models scored high for buried surface area (4500 Å2), because the search was carried out in the absence of RNA, they scored less well for other criteria, such as desolvation of surface hydrophobic residues (-4.1 kcal/mole) and side chain contacts (-27 kcal/mole). However, a model that maximizes side chain complementarity (-37 kcal/mole) and scores high for relative desolvation energy (-9.1 kcal/mole) places the top of the NHL β-propeller against the convex surface of Puf repeats 7 and 8 (Fig. 4). In particular, the extralong loop between helices H1 and H2 of Puf repeat 8 fits in the entrance to the central channel of the NHL domain. This model scores high in two of the four categories used to evaluate results, and was therefore used as a starting point from which to investigate interactions between Brat and Pumilio.
Figure 4.
Predicted structure of the Brat NHL domain bound to the Pumilio Puf domain. (A) The docked structure predicted by the program BiGGER. The RNA-binding surface of Pumilio is lined by the helices highlighted in green. (B) Mutagenesis supports this model. Substitutions at residues highlighted in green (on Brat) or gold (on Pumilio) disrupt recruitment in the yeast four-hybrid assay, whereas substitutions at magenta residues do not. Substitutions are as follows (1–10 Brat mutations, 11–14 Pumilio): (1) Y859A, (2) D1012A, (3) E970A, (4) G774D, (5) H802L, (6) Y829A, (7) R847A, (8) R875A, (9) DA Loop1 (G774-F780) deletion, (10) DA Loop2 (G821-L827) deletion, (11) G1330D, (12) C1365R, (13) T1366D, (14) N1368S. (C) Brat recruitment assayed in a yeast four-hybrid experiment. Interaction between all four partners is required for growth in the absence of histidine (top two panels). (Bottom panel) A portion of a Western blot of yeast extracts (expressing the Brat mutants labeled as above) probed with rabbit antiserum raised against the NHL domain to detect activation domain–Brat fusion proteins. All of the mutants accumulate to approximately the same level, suggesting that the mutants are soluble and folded. Each lane contains the same amount of protein, measured by Bradford assays.
Although the modeling program received no input other than the isolated structures of Pum and the NHL domain, this solution agrees remarkably well with extant biochemical and genetic data. Substitutions in Brat at G774 (to D) and H802 (to L) abrogate recruitment into the hb repression complex (Sonoda and Wharton 2001); the mutant side chains can presumably no longer make required contacts. In addition, the G1330D substitution in the Pum680 mutant that recruits Nos but cannot recruit Brat (Sonoda and Wharton 2001) would clash with acidic residues in the NHL domain (Glu 782 and Asp 1021). Similarly, substitutions in the extended H1-H2 loop of Puf repeat 8 (C1365, T1366, N1368) that disrupt interactions with Brat but not Nos (Edwards et al. 2001) also map to the predicted interface. Overall, the only minor disagreement between the model and earlier work concerns the role of PumF1367, which in our current model interacts with residues around the central channel of the NHL domain, but was suggested earlier to mediate interaction with Nos (Edwards et al. 2001). Given the role of flanking Pum residues, it seems most likely that the apparent effect on Nos recruitment is indirect; for example, mutation of F1367 may disrupt the structure of the nearby Nos binding site.
To further test the model, we prepared mutant NHL derivatives with single amino acid substitutions along both the top (interacting) surface of the domain as well as derivatives with substitutions along the opposite (bottom) surface that points away from Pum as a control. Mutants were designed either to change the electrostatics of each surface or to remove large aromatic side chains. In addition, we prepared derivatives in which the exceptionally long DA loops connecting blades 1 and 2 as well as 2 and 3 of the β-propeller were deleted, because they make extensive contacts with Pum in the model. Each mutant was expressed in yeast, and its ability to join a ternary Pum/NRE/Nos complex was assayed by the yeast four-hybrid assay as previously described (Sonoda and Wharton 2001). As predicted by the model, substitutions on the top surface (Y829A, R847A, R875A) and DA loop deletions block recruitment, whereas substitutions on the opposite surface (Y859A, E970A, D1012A) do not (Fig. 4B,C). These surface substitutions have no apparent effect on the stability of the NHL domain, as each mutant protein accumulates to approximately the same level in yeast (Fig. 4C).
In summary, extensive interactions along one surface of the β-propeller, organized around its central channel, appear to mediate recruitment to the 3′ UTR of hb mRNA. The model of Figure 4 is supported by three independent lines of evidence: the in silico molecular interactions from which it derives, the distribution of Pum surface mutants that interfere specifically with Brat recruitment without affecting RNA binding or Nos recruitment, and the mutational analysis of the NHL domain. In theory, substitutions along the top surface of Brat might block interaction not with Pum but with Nos; however, this seems rather unlikely given the agreement of the in silico model with the analysis of Pum mutants.
Conclusions
One feature of the Brat–Pum model is common to protein complexes formed by other β-propellers—interaction along the top surface, particularly around the central channel. The WD40 domain of the transcriptional corepressor Tup1 interacts with the DNA-binding factor Matα2 to regulate mating-type genes in budding yeast (Sprague et al. 2000). Although the structure of this complex is unknown, all the mutations in Tup1 that interfere with Tup1–Matα2 interaction are located on the top surface of its seven-bladed β-propeller around the central channel, analogous to the mutations described here for Brat. The ubiquitin-conjugating enzyme Cdc4 similarly uses the top surface of its eight-bladed β-propeller to bind a peptide ligand derived from the Cdk inhibitor, Sic1 (Orlicky et al. 2003). In the case of Cdc4, the peptide-binding site is relatively small (buried surface area of ∼750 Å2) when compared to the large interface in the docked Brat–Pum complex (2,900 Å2). However, in each case, a flexible peptide (or a loop in the case of Pumilio) docks in and around the central pore, suggesting an emerging recognition theme for β-propeller molecules.
Brat is normally recruited not to Pum alone, but to a ternary complex of Pum and Nos bound to the NRE. Our model of the Pum–Brat subassembly suggests that the “edge” of the Brat β-propeller is available to interact with Nos, much as Gβ and the scaffolding protein clathrin use the sides of their seven-bladed β-propellers to bind cofactors (Gaudet et al. 1996; ter Haar et al. 1998; Gβ may be the best analogy as it uses both the sides and the top of its β-propeller to recruit partner proteins). Although its location in the repressor complex is not yet well defined, Nos is probably recruited to the Pum–RNA complex via contacts made by the C terminus of the Pum RNA-binding domain (Edwards et al. 2001). The proximity of the NHL domain to the presumptive Nos-binding site on Pum extends the likelihood of cooperative Brat–Nos interactions, and may explain why Brat is only recruited subsequent to Pum and Nos binding to the hb 3′ UTR (Sonoda and Wharton 2001).
The Drosophila proteome contains two additional NHL domain proteins [MeiP-26 and Dappled (Dpld)] that, based on genetic evidence, appear to be tumor suppressors and growth regulators like Brat. It is tempting to speculate they interact with cofactors or regulatory targets much as Brat interacts with Pum in Figure 4. However, neither seems likely to use Pum as a cofactor. The NHL domains of Brat and MeiP-26 are very similar: There are no major insertions or deletions in the DA loops of MeiP-26, and the top surface of its β-propeller, like that of Brat, is electropositive. Thus, based on structural considerations, MeiP-26 might interact with Pum; however, genetic experiments suggest it does not do so in vivo (L. Kadyrova and R.P. Wharton, unpubl.). In contrast, structural considerations suggest the NHL domain of Dpld, which governs the growth of larval organs, is unlikely to bind Pum due to substantial differences in its predicted surface charge distribution and the presence of large insertions in the DA loops on the top surface. Therefore, although Brat, MeiP-26, and Dpld may use their NHL domains in a similar manner, each probably binds to distinct partners.
Based on the analysis of loss- and gain-of-function experiments, Brat appears to regulate abdominal segmentation (via hb translation), brain size, cell size in the imaginal discs, and the accumulation of rRNA. Strikingly, substitutions that abrogate many of these processes map to the “same” top surface of the NHL domain, near the central channel. This suggests that Brat may recognize protruding, flexible loops in a number of protein cofactors or regulatory targets, much as it recognizes the loop in Puf repeat 8 that constitutes the core of the Brat–Pum interaction surface.
Materials and methods
Protein expression and purification
Residues 756–1037 containing the NHL domain of Brat from Drosophila melanogaster (accession no. AAF53771) were expressed as a His-tagged fusion from the vector pET19b (Novagen) in E. coli BL21 (DE3) pLysS cells. Cells from a 4-L culture induced with 1 mM IPTG at O.D.600 = 0.6 for 5 h at 30°C were harvested by centrifugation, then sonicated in buffer A (20 mM Tris, 500 mM NaCl at pH 7.9) plus 0.1% NP40. The soluble fraction was loaded onto a nickel-charged Hi-Trap chelating column (Pharmacia) and washed with buffer A. The column was washed with 10% and 18% buffer B (buffer A + 1 M imidazole), and the His-tagged protein eluted at 40% B. The N-terminal His-tag was removed by proteolysis and the Brat NHL domain was dialyzed against buffer C (20 mM Bis Tris, 50 mM NaCl, 1 mM DTT at pH 6). The NHL domain was loaded onto a MonoS column (Pharmacia) and eluted with an increasing concentration of salt (generally at around 300 mM NaCl). The pure Brat NHL domain was concentrated by ultrafiltration to 25 mg/mL and stored in aliquots at -70°C. Selenomethionine-substituted Brat was prepared as above using the methionine auxotrophic cells B834 (Novagen) in defined media with selenomethionine instead of methionine. Additional DTT was included during the purification of SeMet protein.
Crystallization and structure determination
Crystals were obtained by hanging drop vapor diffusion at 20°C, with a well solution containing 53% di-basic ammonium phosphate and 4% MPD. The crystals appear after 24–48 h and belong to space group p212121 with cell dimensions of a = 47.68 Å, b = 94.31 Å, and c = 130.24 Å. There are two molecules in the crystallographic asymmetric unit. All data were collected at 110°K after the crystals had been soaked in cryo-protectant solution composed of well solution plus 15% MPD. Initial native data were collected at CHESS (A1 beamline).
MAD data at three wavelengths were collected from the SeMet crystals at the Advanced Photon Source (APS, SBC-CAT beamline) to a maximum Bragg spacing of 2.2 Å at wavelengths corresponding to the edge and peak of the selenium K edge absorption profile, plus a high energy remote point (Supplementary Table 1). Selenium positions were identified using Solve (Terwilliger and Berendzen 1999) and refined in mlphare (Otwinowski 1991). CNS (Crystallography and NMR system; Brunger et al. 1998) was used for initial phasing to 2.8 Å, which was then extended with solvent flattening to 2.4 Å. This yielded an electron density map into which the majority of both molecules in the asymmetric unit could be built, using the program O (Jones et al. 1991). The noncrystallographic symmetry axis relating the two molecules was then determined, and the map was averaged to allow the building of a complete monomer. Subsequently, a higher-resolution native data set (1.95 Å) was collected at the National Synchrotron Light Source (Brookhaven beamline X25). However, this crystal was slightly nonisomporphous with respect to the SeMet crystal, with cell dimensions of a = 45.77 Å, b = 94.58 Å, and c = 130.5 Å. A molecular replacement solution was obtained using the monomer built above with the SeMet data. The model was refined against the high-resolution native data using CNS, with iterative rounds of rebuilding in O. The final refined model has a crystallographic R-factor of 0.2002 and R-free of 0.2523 (Supplementary Table 1). This model includes residues 759–1037 for molecule A and 756–1037 for molecule B and 663 water molecules, and has good stereochemistry with 98.0% of residues in the most favored and allowed regions of the Ramachandran plot (Laskowski et al. 1993).
Docking
Docking was performed with the program BiGGER algorithm (Palma et al. 2000) using a monomer of the Brat NHL domain and the 313 residue Puf domain of Pumilio (Edwards et al. 2001). Default settings were used, and the top 100 solutions were analyzed.
Mutagenesis
Site-directed mutagenesis of Brat was performed by standard methods. The yeast interaction assay was described by Sonoda and Wharton (2001).
Accession number
The coordinates and structure factors have been deposited with the PDB with the ID code 1Q7F.
Acknowledgments
We are grateful to Keith Brister (APS), Marian Szebenyi, Irina Kriksunov (CHESS), and Lonnie Berman and Michael Becker (NSLS) for facilitating X-ray data collection. We thank Katie Wang, Jose Trincao, and Carlos Escalante for useful discussions. This work was supported by grants from the NIH to A.K.A. (GM62947) and R.P.W. (G64726). R.P.W. is also an Associate Investigator of the HHMI. B.D.W. was supported by a training grant from the NIH (T32-HD-40372).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1119403.
References
- Abergel C., Bouveret, E., Claverie, J.M., Brown, K., Rigal, A., Lazdunski, C., and Benedetti, H. 1999. Structure of the Escherichia coli TolB protein determined by MAD methods at 1.95 A resolution. Structure Fold. Des. 7: 1291-1300. [DOI] [PubMed] [Google Scholar]
- Arama E., Dickman, D., Kimchie, Z., Shearn, A., and Lev, Z. 2000. Mutations in the β-propeller domain of the Drosophila brain tumor (brat) protein induce neoplasm in the larval brain. Oncogene 19: 3706-3716. [DOI] [PubMed] [Google Scholar]
- Brunger A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 (Pt 5): 905-921. [DOI] [PubMed] [Google Scholar]
- Edwards T.A., Pyle, S.E., Wharton, R.P., and Aggarwal, A.K. 2001. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell 105: 281-289. [DOI] [PubMed] [Google Scholar]
- Frank D.J., Edgar, B.A., and Roth, M.B. 2002. The Drosophila melanogaster gene brain tumor negatively regulates cell growth and ribosomal RNA synthesis. Development 129: 399-407. [DOI] [PubMed] [Google Scholar]
- Gaudet R., Bohm, A., and Sigler, P.B. 1996. Crystal structure at 2.4 angstroms resolution of the complex of transducin βγ and its regulator, phosducin. Cell 87: 577-588. [DOI] [PubMed] [Google Scholar]
- Ha N.C., Oh, B.C., Shin, S., Kim, H.J., Oh, T.K., Kim, Y.O., Choi, K.Y., and Oh, B.H. 2000. Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states. Nat. Struct. Biol. 7: 147-153. [DOI] [PubMed] [Google Scholar]
- Hankins G.R. 1991. “Analysis of a Drosophila neuroblastoma gene.” Ph.D. thesis, pp. 107. Department of Biology, University of Virginia, Charlottesville.
- Hülskamp M., Schröder, C., Pfeifle, C., Jäckle, H., and Tautz, D. 1989. Posterior segmentation of the Drosophila embryo in the absence of a maternal posterior organizer gene. Nature 338: 629-632. [DOI] [PubMed] [Google Scholar]
- Irish V., Lehmann, R., and Akam, M. 1989. The Drosophila posterior-group gene nanos functions by repressing hunchback activity. Nature 338: 646-648. [DOI] [PubMed] [Google Scholar]
- Jeon H., Meng, W., Takagi, J., Eck, M.J., Springer, T.A., and Blacklow, S.C. 2001. Implications for familial hypercholesterolemia from the structure of the LDL receptor YWTD–EGF domain pair. Nat. Struct. Biol. 8: 499-504. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 47 (Pt 2): 110-119. [DOI] [PubMed] [Google Scholar]
- Lambright D.G., Sondek, J., Bohm, A., Skiba, N.P., Hamm, H.E., and Sigler, P.B. 1996. The 2.0 A crystal structure of a heterotrimeric G protein. Nature 379: 311-319. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26: 283-291. [Google Scholar]
- Orlicky S., Tang, X., Willems, A., Tyers, M., and Sicheri, F. 2003. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112: 243-256. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. 1991. Maximum likelihood refinement of heavy atom parameters. In Isomorphous replacement and anomalous scattering (eds. W. Wolf, et al.), pp. 80-86. Science and Engineering Research Council, Daresbury Laboratory, Warrington, UK.
- Palma P.N., Krippahl, L., Wampler, J.E., and Moura, J.J. 2000. BiGGER: A new (soft) docking algorithm for predicting protein interactions. Proteins 39: 372-384. [PubMed] [Google Scholar]
- Slack F.J. and Ruvkun, G. 1998. A novel repeat domain that is often associated with RING finger and B-box motifs. Trends Biochem Sci 23: 474-475. [DOI] [PubMed] [Google Scholar]
- Sonoda J. and Wharton, R.P. 1999. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes & Dev 13: 2704-2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2001. Drosophila Brain Tumor is a translational repressor. Genes & Dev 15: 762-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondek J., Bohm, A., Lambright, D.G., Hamm, H.E., and Sigler, P.B. 1996. Crystal structure of a G-protein β γ dimer at 2.1 A resolution. Nature 379: 369-374. [DOI] [PubMed] [Google Scholar]
- Sprague E.R., Redd, M.J., Johnson, A.D., and Wolberger, C. 2000. Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast. EMBO J. 19: 3016-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G. 1989. Differing strategies for organizing anterior and posterior body pattern in Drosophila embryos. Nature 338: 741-744. [DOI] [PubMed] [Google Scholar]
- ter Haar E., Musacchio, A., Harrison, S.C., and Kirchhausen, T. 1998. Atomic structure of clathrin: A β propeller terminal domain joins an α zigzag linker. Cell 95: 563-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen, J. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55 (Pt 4): 849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. and Lehmann, R. 1991. Nanos is the localized posterior determinant in Drosophila. Cell 66: 637-647. [DOI] [PubMed] [Google Scholar]
- Wang X., McLachlan, J., Zamore, P.D., and Hall, T.M.T. 2002. Modular recognition of RNA by a human pumilio-homology domain. Cell 110: 501-512. [DOI] [PubMed] [Google Scholar]
- Wharton R.P., Sonoda, J., Lee, T., Patterson, M., and Murata, Y. 1998. The Pumilio RNA-binding domain is also a translational regulator. Mol. Cell 1: 863-872. [DOI] [PubMed] [Google Scholar]
- Woodhouse E., Hersperger, E., and Shearn, A. 1998. Growth, metastasis, and invasiveness of Drosophila tumors caused by mutations in specific tumor suppressor genes. Dev. Genes Evol. 207: 542-550. [DOI] [PubMed] [Google Scholar]