Abstract
The embryonic lethal abnormal visual system (ELAV) is a gene-specific regulator of alternative pre-mRNA processing in neurons of Drosophila. Here we define a functional in vivo binding site for ELAV in neurons through the development of a reporter gene system in transgenic animals in combination with in vitro binding assays. ELAV binds to erect wing (ewg) RNA 3′ of a polyadenylation site in the terminal intron 6. At this polyadenylation site, ELAV inhibits 3′-end processing in vitro in a dose-dependent and sequence-specific manner, and ELAV binding is necessary in vivo to promote splicing of ewg intron 6. Further, the AAUAAA poly(A) complex recognition sequence, together with ELAV, is required to regulate neural 3′ splice site choice in vivo. In addition, the use of segmentally labeled RNA substrates in UV cross-linking assays suggest that ELAV does not inhibit or redirect binding of cleavage factor dCstF64 at the regulated polyadenylation site on ewg RNA. These data indicate that binding of 3′-end processing factors, together with ELAV, can regulate alternative splicing.
Keywords: Alternative splicing, alternative polyadenylation, neuron-specific RNA-binding protein, posttranscriptional regulation, elav-regulated genes
Alternative pre-mRNA processing is a key mechanism to increase proteomic diversity and to expand regulatory mechanisms of gene expression. An astounding 40%-60% of human genes are alternatively spliced in at least one exon (Modrek and Lee 2002). Alternative pre-mRNA processing is especially widespread in the nervous system and substantially increases neuronal diversity and functional complexity (Dredge et al. 2001; Grabowski and Black 2001).
Although considerable progress has been made in elucidating the mechanism of general splicing (e.g., see Hastings and Krainer 2001) understanding alternative splicing remains a major challenge. In several systems, specific regulatory proteins have been identified and were shown to either positively or negatively affect spliceosome assembly at regulated introns by binding to nearby splice sites (for reviews, see Lopez 1998; Smith and Valcarcel 2000; Maniatis and Tasic 2002). Either differential concentrations of general splicing factors and/or the presence of tissue-specific factors have been attributed regulatory roles. In a few cases, the interaction of tissue-specific factors with pre-mRNA processing machinery could be determined (e.g., Labourier et al. 2001; Lallena et al. 2002). In other cases, however, elucidation of clear mechanisms is made difficult due to multiple binding sites for tissue-specific factors distributed in the vicinity of processing signals, as observed for Sxl-binding sites on its own transcript (Sakamoto et al. 1992; Horabin and Schedl 1993; Wang and Bell 1994), Nova-1-binding sites in a GlyRα2 intron (Jensen et al. 2000), and embryonic lethal abnormal visual system (ELAV)-binding sites in the regulated intron of nrg (Lisbin et al. 2001).
ELAV family proteins, characterized by containing three RNA recognition motifs (RRMs), are among candidates for neuron-specific regulators of RNA processing. ELAV, originally identified in Drosophila as a panneurally expressed protein (Robinow et al. 1988), has been shown to affect neuronal pre-mRNA processing as a gene-specific regulator (Koushika et al. 2000) and has also been implicated in autoregulation (Samson 1998). In contrast, the neuron-specific members of the human ELAV family, HuB (Hel-N1), HuC, HuD, similar to the ubiquitously expressed HuA (HuR), have been ascribed only cytoplasmic RNA-processing functions (Myer et al. 1997; Levy et al. 1998; Peng et al. 1998, Antic et al. 1999; Ford et al. 1999; Gallouzi and Steitz 2001; Kullmann et al. 2002).
ELAV is essential for producing neuroglian (nrg) and erect wing (ewg) neuronal mRNAs that require splicing of the terminal exon resulting in distinct 3′ ends (Koushika et al. 1996). Thus, ELAV could stimulate splicing of the last intron and/or inhibit 3′-end formation within the last intron. Most general factors operating in 3′-end formation have been identified (for review, see Zhao et al. 1999). Although models of terminal exon definition exist (Niwa and Berget 1991), the regulation of alternative 3′-end formation and its coordination with splicing is poorly understood. Situations comparable to ewg and nrg have been described for alternative splicing of CT/CGRP (Crenshaw et al. 1987) and for IgM alternative 3′-end processing (Peterson 1994). In CT/CGRP premRNA processing, inclusion of exon 4 and premature 3′-end formation occurs in thyroid cells, whereas exon 4 is skipped in neurons. Regulation at the level of 3′-end formation in exon 4 has been suggested, because an enhancer sequence located 3′ of the polyadenylation (pA) site stimulates 3′-end formation (Lou et al. 1996). Although neuron-specific factors have not been identified, overexpression of PTB stimulates exon 4 inclusion and 3′-end formation (Lou et al. 1999). In IgM pre-mRNA processing, splicing of the last intron resulting in membrane bound IgM is prevented by use of an intronic pA site to produce secreted IgM in later stages of B-cell development (for review, see Peterson 1994). Increased activity and concentrations of CstF64(cleavage stimulation factor) together with low hnRNP F concentrations have been attributed to premature 3′-end formation versus splicing (Edwalds-Gilbert and Milcarek 1995; Takagaki et al. 1996; Takagaki and Manley 1998; Veraldi et al. 2001).
In this study, we have investigated how alternative pre-mRNA processing of ewg intron 6 is regulated by ELAV and defined the in vivo binding site for ELAV on ewg RNA in neurons using in vivo and in vitro approaches. We find that ELAV binds 3′ of a pA site and inhibits 3′-end formation in the terminal ewg intron 6 to promote alternative splicing. Further, we tested if ELAV binding interferes with pA site recognition by cleavage and polyadenylation specificity factor (CPSF) by analyzing the effects of a deletion of the AAUAAA pA siterecognition sequence on neural splice site choice, and if ELAV competes with binding of CstF. We demonstrate that the absence of the AAUAAA pA sequence leads to increased nonneuronal 3′ splice site choice in neurons. We also provide evidence that ELAV does not inhibit binding of CstF at the regulated pA site. Our studies show that neural 3′ splice site choice is influenced by both ELAV and 3′-end processing factors.
Results
In the absence of ELAV, transcripts terminate in ewg-terminal intron in neurons
Previous work has shown that ewg is broadly expressed, but splicing of the terminal intron 6 occurs only in neurons and is dependent on ELAV. Further, ectopic expression of ELAV in nonneural wing discs is sufficient to induce intron 6 splicing and expression of EWG protein (Koushika et al. 2000). These results raise the possibility that ewg transcripts terminate in intron 6 in the absence of ELAV using the intronic consensus pA sites (Fig. 1B).
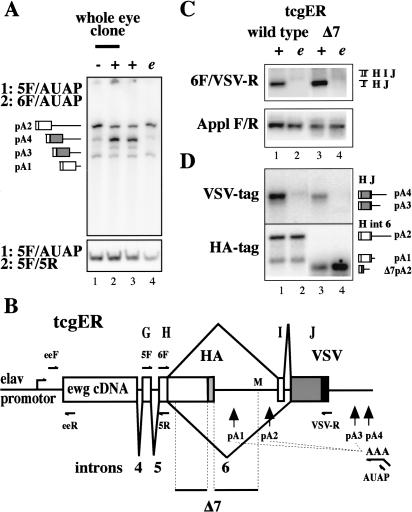
Figure 1.
ELAV switches polyadenylation of ewg pre-mRNA to splicing. (A) 3′ ends of ewg transcripts amplified by nested RT-PCR with RNA extracted from eye discs with no (-), reduced (e), or wild-type (+) ELAV levels. (e) elavedr. The top panel shows the amplified ends of ewg transcripts, and the bottom panel shows a common part of ewg transcripts amplified as a standard. Primers used in the two PCR reactions are indicated on the left and depicted in B. Graphic illustrations of PCR products are shown on the left and indicate the different 3′ ends and pA sites as depicted in B. PCR products were separated on 5% denaturing polyacrylamide gels. (B) Schematic of the ewg rescue reporter gene (tcgER). The transcriptional start is indicated by an arrow in the elav promotor. ORFs are shown as boxes with either a HA or VSV tag at the end. Deletion Δ7 is indicated at the bottom [starts 89 nt after the 5′ splice site, extends to the MfeI site (M) in intron 6, and also contains an HA tag]. pA used are indicated by vertical arrows. RT and return 3′ RACE primers are shown for the pA4 site only. (C) Splicing of ewg intron 6 analyzed by semi-quantitative RT-PCR with eye disc RNA from wild-type and Δ7 tcgER in wild-type (+) or elavedr background (e). A fragment from Appl transcripts was amplified as standard. Graphic illustrations of ewg splice products are shown on the right. Uniformly 32P-dCTP-labeled PCR products were separated on 8% polyacrylamide gels. (D) Southern blot probed with either 32P-end-labeled HA or VSV oligonucleotides to visualize ends of transcripts from wild-type and Δ7 tcgER transgenes amplified by 3′ RACE from eye disc RNA in wild-type (+) and elavedr mutant background (e) as indicated in C. Graphic illustrations of PCR products are shown on the right and indicate the different 3′ ends and pA sites. The shorter fragments detected in lanes 3 and 4 on the HA-Southern are due to the deletion Δ7 and 3′-end formation at pA2.
To define 3′ ends of ewg transcripts, we used 3′ rapid amplification of cDNA ends (3′ RACE). Briefly, after reverse transcription (RT) with an oligo dT primer containing an anchor sequence, two PCR amplifications were done using nested primers and the amplified fragments were sequenced. To assesses the relative usage of pA sites semi-quantitative 3′ RACE was done with 32P-labeled primers in the second PCR reaction (Fig. 1A, 6F or 5R to amplify the ends of all transcripts or a common part of all transcripts as standard, respectively; Fig. 1B).
In wild-type eye discs, most poly(A)+ transcripts are spliced and terminate at pA4(Fig. 1A, lanes 2,3). In the absence of ELAV, poly(A)+ transcripts predominantly terminate in intron 6 at pA2 (Fig. 1A, lane 1). Loss of ELAV from eye discs was achieved by generating mosaic animals using a mitotic recombination technique (Stowers and Schwarz 1999; see Materials and Methods). Reducing ELAV levels, as in the synthetic elavedr mutant (Koushika et al. 1996), is sufficient for predominant 3′-end formation in intron 6 at pA2 (Fig. 1A, cf. lanes 4 and 3,1). In wing imaginal discs, the same switch as in eye discs is observed with respect to 3′-end processing in the absence or presence of ELAV (data not shown). Thus, in the absence of ELAV, ewg poly(A)+ transcripts terminate in intron 6, whereas the presence of ELAV promotes splicing of intron 6.
A chimeric cDNA/genomic rescue reporter gene recapitulates in vivo 3′-end processing
Next, to determine the cis elements necessary for ELAV-dependent splicing of ewg intron 6 in neurons, we used a transgenic approach and developed a reporter construct (tcgER) shown in Figure 1B. We chose the elav promotor, because it expresses exclusively in neurons at a moderate to low level and because it resembles expression levels of the ewg gene. Furthermore, an elav-promotor-driven ewg cDNA transgene rescues the lethality of ewg null alleles (DeSimone et al. 1996). To focus on 3′-end processing, the 5′ end of ewg cDNA was fused to the genomic 3′ end (Fig. 1B). To distinguish reporter and endogenous ewg transcripts, a hemagglutinin tag (HA) and a vesicular stomatitis virus (VSV) tag were added at the end of the ORFs resulting from spliced and unspliced intron 6. This tagged cDNA/genomic reporter transgene (tcgER) fully rescues the lethality of an ewg null allele (90%-100%) and, identical to the endogenous gene, the major protein resulting from splicing of intron 6 is expressed in neurons (data not shown; Koushika et al. 1999).
RNA processing of tcgER-generated transcripts was analyzed using semi-quantitative reverse transcriptase PCR (RT-PCR) with RNA from wild-type and elavedr eye discs. In wild-type neurons, splicing of intron 6 is observed with almost no inclusion of microexon I (Fig. 1C, lane 1; data not shown). Thus, the minor splice product that includes exon I observed in nonneuronal tissue represents the nonneuronal mode of splicing (Koushika et al. 2000). In elavedr, intron 6 splicing is severely reduced and thus usually not detected (Fig. 1C, lane 2). Further, 3′ RACE was used to determine the 3′ ends of mRNAs originating from tcgER by detection on Southern blots with either 32P-labeled HA or VSV oligonucleotides (Fig. 1D). In wild type, both intron 6 spliced (VSV tag) and unspliced (HA tag) mRNAs are detected, whereas in elavedr, intron 6 is mostly not spliced (Fig. 1D, lanes 1,2). In brief, tcgER behaves as the endogenous ewg gene.
To further delimit the region of intron 6 necessary for ELAV-dependent regulation, a set of deletions, Δ1-Δ7 in intron 6, were made in the parent tcgER construct and ewg RNA processing was analyzed in eye discs of transgenic flies. The largest deletion, Δ7, includes all smaller deletions, Δ1-6, and deletes ∼75% of intron 6 (Fig. 1B). Δ7 mimics ewg 3′-end processing of parental tcgER transgenes (Fig. 1C,D, cf. lanes 1,3 and 2,4; the shorter fragments detected in Fig. 1D, lanes 3,4, on the HA Southern are due to the deletion Δ7). Thus, cis elements necessary for ELAV-dependent posttranscriptional regulation of ewg are within 25% of intron 6 and/or to the 3′ untranslated region (UTR).
ELAV binds 3′ of a poly(A) signal in ewg intron 6
Next, we used UV cross-linking assays to determine if ELAV binds directly to ewg RNA Δ8 (Δ7 without the HA tag) and/or the 3′ UTR (Fig. 2A). “Neuronal” and nonneuronal nuclear extracts were prepared from Drosophila heads and Drosophila Kc cells. [32P]ATP- or UTP-labeled substrate RNA was UV cross-linked, digested with RNase, and cross-linked proteins were resolved on SDS gels.
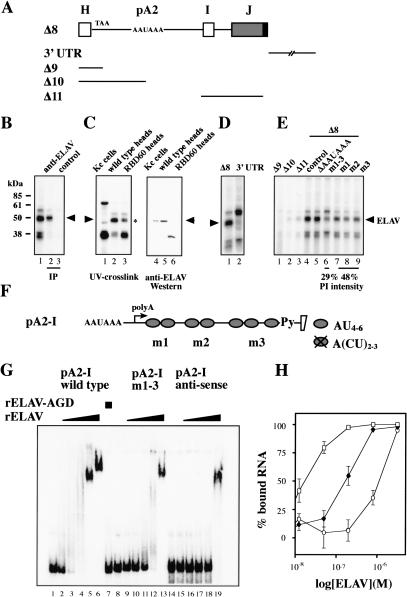
Figure 2.
ELAV binds 3′ of the pA2 site on ewg premRNA. (A) Schematic of ewg RNAs used for UV crosslinking assays. Δ8 derives from Δ7, but lacks the HA tag. (B) UV cross-linking of uniformly [32P]ATP-labeled Δ8 RNA in neuronal nuclear extract. Proteins were separated on 10% SDS-polyacrylamide gels after crosslinking and RNase digestion (lane 1) or after IP with anti-ELAV antibodies (lane 2). The control IP (lane 3) was done with protein A/G beads alone. Arrowhead indicates ELAV. (C) UV cross-linking of uniformly [32P]ATP-labeled Δ8 RNA in nuclear extract from Kc cells and heads of wild-type flies or flies expressing the N-terminal truncation mutant RBD60 (lanes 1-3), and anti-ELAV Western of those nuclear extracts (lanes 4-6). A 50-kD protein present in addition to ELAV is marked by a star next to lane 3. Arrowheads indicate ELAV. (D) UV cross-linking of uniformly [32P]ATP-labeled ewg Δ8 and 3′ UTR RNA. (E) UV cross-linking of uniformly [32P]ATP-labeled Δ9-Δ11 RNAs and mutated Δ8 RNAs. AU4-6 motif mutations are depicted in F. At the bottom, labeling intensity of the 50-kD signal from mutant RNAs is compared with parental Δ8 RNA (control, lane 4) and shown as PhosphorImager intensity (PI) from three experiments. Arrowheads indicate ELAV. (F) Schematic of ewg RNA pA2-I (pA2 signal to intron I) showing AU4-6 motif locations (ovals). Crossed oval represents mutated AU4-6 motifs to A(CU)2-3. For sequence see Figure 7A. (Py) Pyrimidine tract. (G) EMSA of RNA pA2-I. The indicated uniformly [32P]ATP-labeled RNAs (100 pM) were incubated with recombinant HA-ELAV (12.5 nM, 50 nM, 0.2 μM, 0.8 μM, and 3.2 μM) or HA-ELAV-AGD (3.2 μM, lane 7). The first lane of each RNA (lanes 1,8,14) shows the input RNA without protein. (H) Graphic representation of EMSA data. The percent of bound RNA (input RNA -unbound RNA/input RNA × 100) is plotted against the concentration of recombinant ELAV (in molar) presented as log from three experiments. (Open squares) Wild-type RNA pA2-I; (filled rectangles) pA2-I RNA with mutation m1-3; (open circles) anti-sense RNA pA2-I.
A set of proteins cross-linked to Δ8 with a predominant band at 50 kD, the size of ELAV (Fig. 2B, lane 1). Subsequently, ELAV's identity was verified by immunopreciptation (IP) after cross-linking and RNase digestion (Fig. 2B, lane 2). Both [32P]ATP- and UTP-labeled RNAs cross-link equally well to ELAV (data not shown).
To determine if other proteins in the 50 kD range cross-link to Δ8 RNA, nuclear extracts were prepared from flies expressing a functional N-terminal deletion mutant of ELAV (RBD60; Yao et al. 1993), but no endogenous ELAV. The RBD60 protein is detected as a 40 kD band not seen with wild-type extracts (Fig. 2C, lane 3). The signal in the 50-kD range is reduced but not absent (Fig. 2C, lane 3), indicating an additional RNA-binding protein. In Western analysis, no 50-kD band is detected in the RBD60 extract (Fig. 2C, lane 6). No ELAV crosslinking to Δ8 RNA was detected with nonneuronal extract (Fig. 2C, lane 1) that contains only a small amount of ELAV (Fig. 2C, lane 4).
We also tested if ELAV can be UV cross-linked to the 3′ UTR. The ELAV band observed with Δ8 is absent with the 3′ UTR RNA, although other cross-linking proteins are observed with a prominent band at 55 kD (Fig. 2D). To further restrict ELAV binding on RNA Δ8, the shorter RNAs Δ9-Δ11 (Fig. 2A) were used for UV cross-linking. ELAV did not cross-link to RNAs Δ9-Δ11 (Fig. 2E, lanes 1-3).
ELAV family proteins have previously been shown to bind preferentially to AU-rich sequences (Levine et al. 1993; Gao et al. 1994; Abe et al. 1996). Examination of the sequence between the pA signal and intron I revealed several AU-rich elements consisting of three and a partial fourth repeat of a tandem AU4-6 motif separated by a short spacer (Fig. 2F). These AU motifs can be aligned to a putative consensus sequence (see Fig. 7A, below). The signifigance, however, needs to be determined. The most 3′ AU-rich motif partially overlaps with the polypyrimidine tract. To test if tandem AU4-6 motifs are involved in ELAV binding, U-to-C substitutions were introduced in Δ8 [A(CU)2-3, Fig. 2F]. m1, m2, and m3 represent mutations in the first, second, and third with a partial fourth tandem AU4-6 motif, whereas the polypyrimidine tract was not mutated. A fourth mutation, m1-3, that includes all the mutant nucleotides in m1, m2, and m3 was also created.
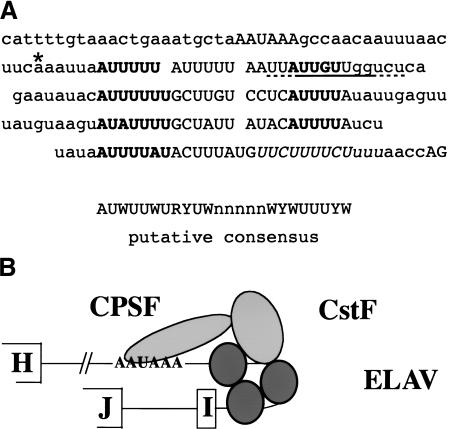
Figure 7.
(A) Sequence context around ewg pA2 up to the 3′ splice site of exon I. (B) Schematic of CPSF, CstF, and ELAV binding to ewg pre-mRNA. In A, the AAUAAA sequence, tandem AU4-6 motifs, and the AG dinucleotide from the 3′ splice site at exon I are in capital letters. A G/U-rich element is underlined and the polypyrimidine tract is in italics. The cleavage site is indicated with an asterisk. AU4-6 motifs are in bold and tandem AU4-6 motifs are aligned to a putative consensus sequence evident within the ELAV binding sites.
Mutations m1, m2, and m3 each reduced the UV crosslinking of [32P]ATP-labeled substrate RNA to the 50 kD band by ∼50% (Fig. 2E, cf. lanes 7-9 and 4), whereas the m1-3 mutation reduced it by ∼70% compared with Δ8 RNA (Fig. 2E, cf. lanes 6 and 4). These values on the effect of mutations on ELAV cross-linking are likely to be underestimates as proteins other than ELAV also contribute to the signal at 50 kD (Fig. 2C, lane 3). Deletion of the AAUAAA pA recognition sequence did not alter the cross-linking pattern (Fig. 2E, lane 5).
The interaction of ELAV with ewg target RNA was further demonstrated in electrophoretic mobility shift assays (EMSA) using recombinant ELAV and in vitro transcribed substrate RNA. Recombinant ELAV was produced in Escherichia coli as a GST-fusion protein and the GST moiety was cleaved off. Recombinant ELAV binds to substrate RNA pA2-I (the sequence from the pA signal to exon I; Fig. 2F) in a concentration-dependent manner with a Kd of ∼17 nM (Fig. 2G, lanes 2-6, 2H). To show that binding to ewg RNA resides in the RRM domains, point mutations in RNP1 of each RRM were introduced, changing the conserved Y/VGF to AGD. Recombinant ELAV-AGD protein does not bind ewg RNA (Fig. 2G, lane 7) and does not rescue elav mutant flies (Lisbin et al. 2000). To further demonstrate that tandem AU4-6 motifs in ewg RNA mediate ELAV binding, EMSAs were performed with substrate RNA pA2-I m1-3. The affinity to bind ELAV is ∼10-fold reduced (Fig. 2G, lanes 9-13, 2H). Using substrate RNAs with single mutations m1, m2, or m3 or double combinations showed intermediate effects in EMSAs (data not shown). As a control for binding specificity, we tested binding of recombinant ELAV to the antisense RNA of RNA pA2-I. Although the Kd for this binding reaction is low (∼1.1 μM; Fig. 2G, lanes 15-19, 2H), it indicates that ELAV has a general affinity for RNA. As evident from lanes 5, 6, 13, and 19 in Figure 2G, recombinant ELAV most likely forms a stable complex on substrate RNA.
ELAV binding inhibits 3′-end formation both in vitro and in vivo
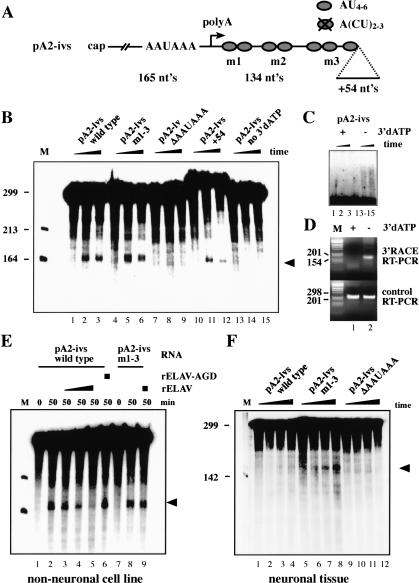
Because ELAV binds in the proximity of the intronic pA2 site, ELAV-dependent splicing of ewg intron 6 could be explained by an inhibtory function of ELAV on 3′-end formation at pA2. To test this hypothesis, an in vitro cleavage/pA assay was established using nuclear extracts from a nonneuronal Drosophila cell line (Kc cells) that have only traces of ELAV (Fig. 2C, lane 4). As a substrate, capped RNA pA2-ivs (299 nt) that includes the necessary signals for 3′-end processing (Fig. 3A) was used. To make the cleavage product detectable, a chain-terminating ATP analog (3′dATP) was included to inhibit pA. Under these conditions, the 5′ cleavage fragment is detected as a shorter RNA with an expected size of 165 nt (Fig. 3B, lanes 2,3), whereas the 3′ fragment is rapidly degraded in nuclear extracts and thus not detectable (Wahle and Keller 1994). To verify that the 5′ cleavage fragment is authentic and is distinct from degradation products, several controls were performed. First, deletion of the AAUAAA pA signal-recognition sequence abolished cleavage and the 5′ cleavage fragment is not detected (Fig. 3B, lanes 8,9). Second, adding unrelated sequence at the 3′ end of the substrate RNA (Fig. 3A, pA2-ivs +54) did not affect the size of the cleavage product and thus verified the position of the cleavage site (Fig. 3B, lanes 11,12). Third, omitting 3′dATP from the reaction resulted in the absence of the 165-nt cleavage product (Fig. 3B, lanes 14,15) because poly(A) tails of heterogeneous length are added. The 5′ cleavage product is therefore shifted to a larger size partly overlapping with the substrate RNA (Fig. 3C, lanes 14,15; this figure shows a longer exposure of parts from the gel in Fig. 3B). Fourth, to demonstrate the addition of a poly(A) tail to the 5′ cleavage product, oligo dT-mediated reverse transcription and 3′ RACE amplification were used. A poly(A) tail is added to the 5′ cleavage product in the absence of 3′dATP (Fig. 3C, lanes 14,15), resulting in the amplification of a correctly sized 3′ RACE product (Fig. 3D, top panel, lane 2). No poly(A) tails are added in the presence of 3′dATP (Fig. 3C, lanes 2,3) and consequently no 3′ RACE product is observed (Fig. 3D, top panel, lane 1). As a standard, RT-PCR was done with a substrate-specific primer in the 3′ region of pA2-ivs (Fig. 3D, bottom panel). Fifth, sequencing of the cleavage product obtained in the absence of 3′dATP (band in lane 2, Fig. 3D, top panel) revealed no difference to cleavage products obtained in vivo (data not shown). With some substrates, poly(A) polymerase and degradation of the 3′ cleavage fragment can be inhibited by inclusion of EDTA whereas the cleavage reaction is still permitted in the absence of Mg2+ (Wahle and Keller 1994). Cleavage of the ewg substrate pA2-ivs, however, is inhibited by inclusion of EDTA in the assay (data not shown). With ewg substrate RNAs, 5′ cleavage products did not always accumulate over the time of the assay, likely because cleavage rates did not exceed RNA degradation rates.
Figure 3.
ELAV inhibits 3′-end formation at pA2 in vitro. (A) Schematic of ewg 299-nt substrate RNA pA2-ivs used for in vitro cleavage/pA. The m1-3 mutation is as in Figure 2F; ΔAAUAAA deletes those nucleotides and +54 adds 54 nt at the 3′ end. (B) Cleavage/pA assay of ewg wild-type and mutant pA2-ivs RNAs as indicated above the panel in nonneuronal nuclear extract. Time points were 0, 25, and 50 min. The 5′ cleavage product is marked with an arrowhead. (M) RNA size marker. (C) Longer exposure of upper parts of the gel from B shows the addition of poly(A) tails in the absence of 3′dATP and results in RNAs longer than the substrate (lanes 14,15). (D, top panel) Amplification of 5′ cleavage fragments from in vitro cleavage/pA assays in the presence (+) or absence (-) of 3′dATP by oligo dT-mediated RT-PCR (3′ RACE). (Bottom panel) Control RT-PCR to show equal input was done with a RT/return primer in the 3′ part of pA2-ivs substrate RNA to amplify uncleaved substrate RNA. Fragments were separated on 3% agarose gels. (M) DNA size marker. (E) Cleavage/pA assay of ewg wild-type and mutant pA2-ivs RNAs as indicated above the panel in nonneuronal nuclear extract. Concentration of recombinant ELAV is 0.2, 0.4, and 0.8 μM and of ELAV-AGD is 0.8 μM. (F) Cleavage/pA assay of wild-type and mutant substrate pA2-ivs RNAs in neuronal nuclear extract. Time points were 0, 20, 40, and 60 min.
To test if ELAV is sufficient to inhibit cleavage of the wild-type ewg substrate RNA pA2-ivs, we added recombinant ELAV to cleavage reactions containing nuclear extracts prepared from “nonneuronal” Kc cells. Addition of recombinant ELAV inhibits cleavage of the pA2-ivs RNA in a concentration-dependent manner (Fig. 3E, lanes 3-5). Next we tested if the ability of ELAV to inhibit the cleavage reaction also depends on ELAV's capacity to bind RNA. Addition of recombinant ELAV-AGD mutant, unable to bind RNA (Fig. 2G, lane 7), does not inhibit the cleavage reaction (Fig. 3E, cf. lanes 6 and 5). To test if ELAV's ability to inhibit the cleavage reaction is sequence specific, mutant substrate RNA pA2-ivs m1-3 was used, previously shown to bind ELAV with reduced affinity (Fig. 2G,H). Cleavage of the pA2-ivs m1-3 RNA is not affected at ELAV concentrations that inhibit cleavage of the wild-type pA2-ivs RNA (Fig. 3E, cf. lanes 9 and 8,5).
Next, we wanted to verify that ewg substrate RNA is processed in a neural mode in neuronal nuclear extracts. Consistent with our expectation, wild-type substrate pA2-ivs is not cleaved in neuronal nuclear extract prepared from Drosophila heads (Fig. 3F, lanes 2-4), whereas cleavage of mutant pA2-ivs m1-3 substrate RNA is restored due to reduced ELAV binding (Fig. 3F, lanes 6-8). Introducing mutation m1-3 into pA2-ivs does not increase cleavage efficiency (Fig. 3B, cf. lanes 2,3 and 5,6). Deleting the AAUAAA pA complex recognition sequence in pA2-ivs abolishes cleavage (Fig. 3F, lanes 10-12). ELAV immunodepletion of neuronal nuclear extract resulted in loss of cleavage activity (data not shown). This could be explained by a further reduction of the low processing activity for the pA2-ivs substrate, or, alternatively, ELAV immunodepletion might also deplete pA factors (see Fig. 6E, below).
Figure 6.
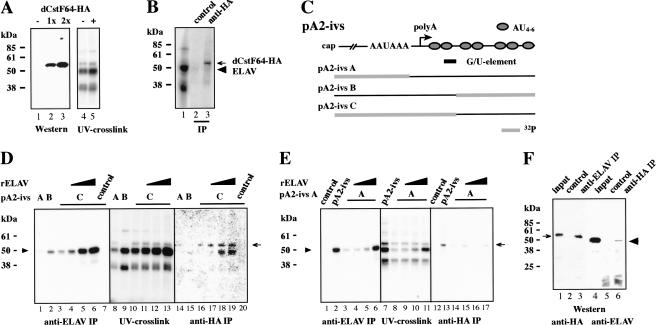
dCstF64 binds to ewg pA2 in the presence of ELAV. (A) Western showing expression of dCstF64-HA in eye discs using GMR-GAL4 driver from 0, 1, or 2 UAS transgenes (lanes 1-3, respectively), and UV cross-linking of pA2-ivs RNA in neuronal nuclear extract prepared from wild-type fly heads (-, lane 4) and fly heads expressing dCstF64-HA with elav-GAL4 driver (+, lane 5). (B) UV cross-linking of Δ8 RNA in neuronal nuclear extract containing dCstF64-HA and IP with anti-HA antibodies (lane 3). Control IP was done with protein A/G beads alone. The arrow marks dCstF64-HA and the arrowhead points toward ELAV. (C) Schematic of segmentally 32P-labeled pA2-ivs RNAs used in UV cross-linking assays in D and E. A G/U-rich sequence element as a putative dCstF64-binding site is indicated below pA2-ivs. (D,E) UV cross-linking assays with partially 32P-labeled pA2-ivs RNAs in neuronal nuclear extract containing dCstF64-HA, and IP with anti-ELAV and anti-HA antibodies. Added recombinant ELAV (0.16 μM, 0.32 μM, and 0.64 μM) to UV cross-linking extracts and labeled RNA are indicated above the panel. As a control, UV cross-linking reactions with pA2-ivs C or pA2-ivs were incubated with protein A/G beads alone (D, lanes 7,20; E, lanes 1,12). The arrow marks dCstF64-HA and the arrowhead points toward ELAV. (F) Coimmunopreciptation of ELAV and dCstF64 from neuronal nuclear extract expressing UAS dCstF64-HA with elav-GAL4 driver. (Lanes 1-3) IP with anti-ELAV antibodies and detection of dCstF64-HA (arrow) with anti-HA antibodies. (Lanes 4-6) IP with anti-HA antibodies and detection of ELAV (arrowhead). (Lanes 2,5) Control IP was with protein A/G beads alone. Twenty-five percent input of nuclear extract proteins is shown in lanes 1 and 4.
Taken together, our results from the in vitro cleavage assays clearly demonstrate a direct role for ELAV in inhibiting 3′-end processing. To test if ELAV binding to the ewg transcript in vivo is critical for regulating splicing of intron 6, mutation m1-3 was introduced in the tcgER construct and transgenic fly lines were established.
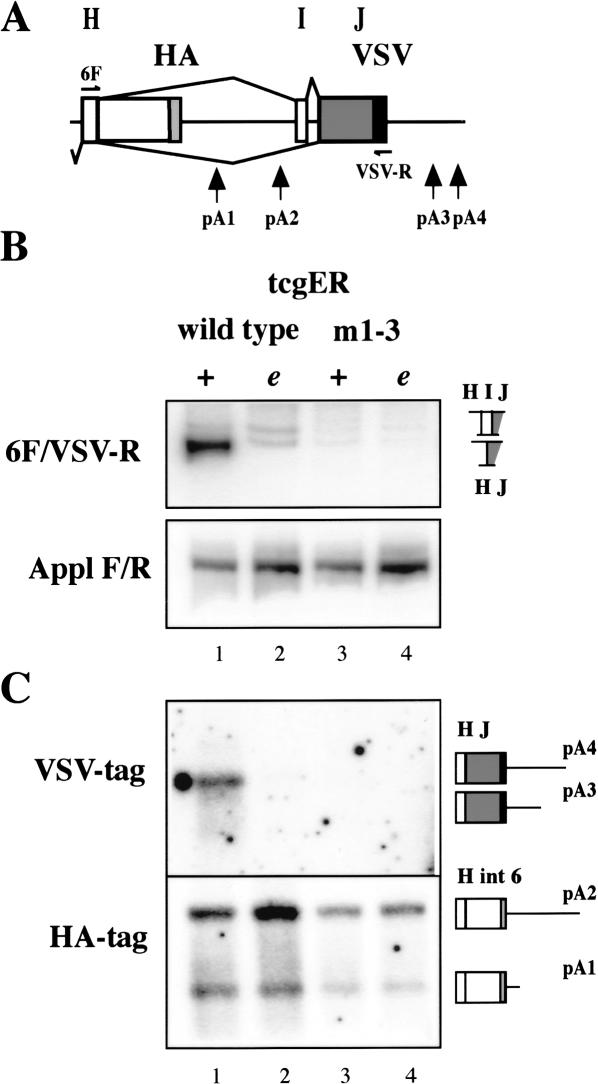
Splicing of intron 6 in the mutant m1-3 is abolished in photoreceptors (Fig. 4B, cf. lanes 1 and 3) and in adult brains (data not shown) as assayed with semi-quantitative RT-PCR. Complete loss of splicing and concomitant increased use of pA2 in mutant m1-3 is further verified with 3′ RACE and detection of VSV or HA tags by Southern analysis (Fig. 4C, cf. lanes 1 and 3).
Figure 4.
Inhibition of 3′-end formation at pA2 in neurons depends on ELAV binding in vivo. (A) Schematic of ewg intron 6 as present in tcgER reporter gene. (B) Splicing of ewg intron 6 analyzed by RT-PCR with eye disc RNA from wild-type and m1-3 (mutations in AU4-6 motifs) tcgER in wild-type (+) or elavedr mutant background (e). A fragment from Appl transcripts was amplified as standard. Graphic illustrations of ewg splice products are shown on the right. (C) Southern blot probed with either a 32P-end-labeled HA or VSV oligonucleotide to visualize 3′ ends of transcripts from wild-type and m1-3 tcgER amplified by 3′ RACE from eye disc RNA in wild-type (+) or elavedr mutant background (e) as indicated in Figure 4B. Graphic illustrations of PCR products are shown on the right and indicate the different 3′ ends and pA sites.
The ability of m1-3 mutant transgenes to rescue viability is only ∼42 ± 5.6% (n = 7 independent insertions ± S.E.) and is comparable to a deletion of exon J (43 ± 7.65 rescue, n = 8). These data demonstrate genetically that ELAV function in ewg intron 6 processing is abolished in the mutant m1-3.
Deletion of AAUAAA at pA2 permits nonneuronal 3′ splice site choice in neurons
Binding of ELAV near the pA2 site could interfere with CPSF/AAUAAA-mediated recognition of the pA site (Willusz et al. 1990; Gilmartin and Nevins 1991; Keller et al. 1991). We therefore wanted to test if abolishing 3′-end formation at the pA2 site was sufficient to yield the neuronal splicing pattern of intron 6, which is splicing of exon H to J without inclusion of exon I. Thus, we established transgenic fly lines containing a deletion of the conserved AAUAAA sequence in the tcgER reporter, which would eliminate poly(A)-complex formation and 3′-end formation.
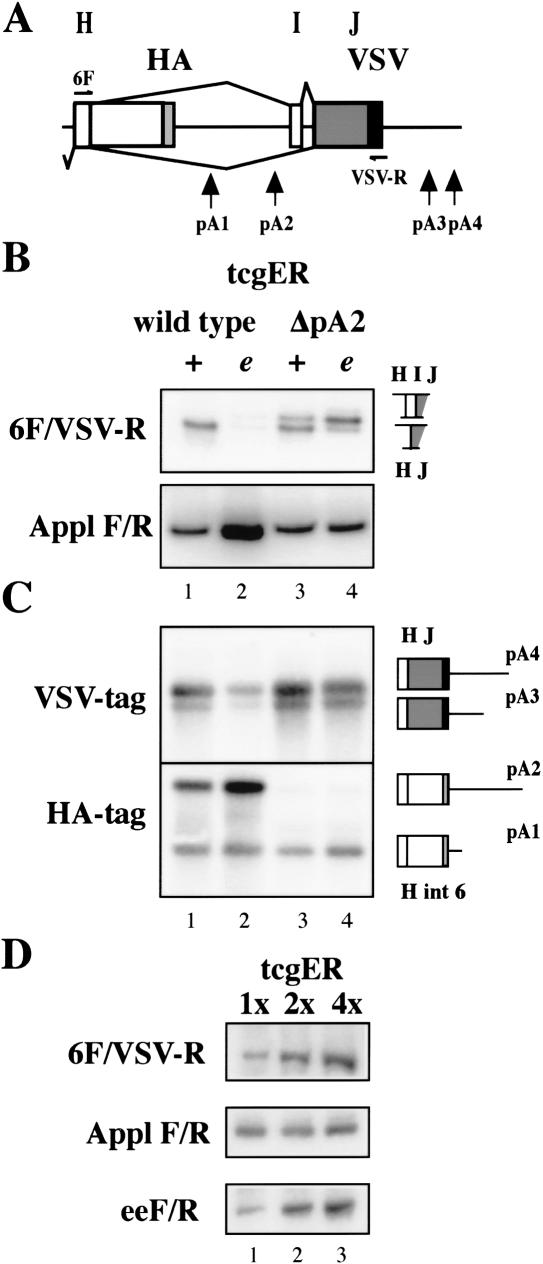
Because usage of a pA site is also determined by sequence context, we verified that deletion of the AAUAAA sequence in tcgER ΔpA2 reporter renders it nonfunctional by Southern analysis after 3′ RACE. No transcripts terminating at pA2 were detected in the ΔpA2 mutant (Fig. 5C; no upper band in lanes 3,4was detected with the HA probe).
Figure 5.
The AAUAAA pA recognition sequence is required for neural 3′ splice site choice. (A) Schematic of ewg intron 6 as present in tcgER reporter gene. (B) Splicing of ewg intron 6 analyzed by RT-PCR with eye disc RNA from wild-type and ΔpA2 (AAUAAA deleted at pA2) tcgER in wild-type (+) or elavedr mutant background (e). A fragment from Appl transcripts was amplified as standard. Graphic illustrations of ewg splice products are shown on the right. (C) Southern blot probed with either a 32P-end-labeled HA or VSV oligonucleotide to visualize 3′ ends of transcripts from wild-type and ΔpA2 tcgER amplified by 3′ RACE from eye disc RNA in wild-type (+) or elavedr mutant background (e) as indicated in B. Graphic illustrations of PCR products are shown on the right and indicate the different 3′ ends and pA sites. (D) RT-PCR from eye disc RNA from wild-type flies containing one, two, or four copies of wild-type tcgER transgenes. A fragment from Appl transcripts was amplified as a standard, and a fragment amplified from the 5′ end of the reporter transcript (eeF/R) shows the amount of transcripts.
Next, we compared splicing of intron 6 in parental and ΔpA2 tcgER-expressing transgenes in photoreceptors. In neurons, intron 6 is spliced, and transcripts that include exon I are extremely rare and mostly below detection limits (Fig. 5B, lane 1; but see also lane 1 in Figs. 1C, 4B). With reduced ELAV levels, splicing of intron 6 is reduced to very low or undetectable levels (Fig. 5B, lane 2; but see also lane 2 in Figs. 1C, 4B). Deletion of the AAUAAA sequence from pA2 results in a significant population of transcripts that include exon I in wild-type photoreceptor neurons (Fig. 5B, lane 3). When ELAV levels were reduced, inclusion of exon I became the major splice product in the pA2 deletion mutant (Fig. 5B, lane 4). Replacing the AAUAAA sequence with an unrelated sequence had exactly the same effect as the deletion (data not shown).
These experiments show that the inhibition of pA-complex formation in itself is not sufficient to completely mimic ELAV-mediated effects on intron 6 splicing. One reason for the increased inclusion of exon I could be the increase in the amount of transcripts available for splicing caused by the removal of the pA2 site. In this scenario, ELAV levels could become limiting and allow the inclusion of exon I. To examine this possibility, splicing of intron 6 was analyzed in developing photoreceptor neurons by RT-PCR in flies with one, two, or four copies of the tcgER transgene. No inclusion of exon I could be detected with increased transcript levels (Fig. 5D). Also, removing of pA1, the proximal intronic polyadenylation site as in tcgER Δ7, did not result in increased inclusion of exon I (Fig. 1C, lane 3). Thus transcript saturation is unlikely to be the cause for exon I inclusion in wild-type photoreceptors. Another possibility is that ELAV does not inhibit recognition of the pA2 site, but rather components of pA complex together with ELAV are necessary to achieve authentic neuronal intron 6 splicing. The following experiments were undertaken to test if ELAV binding competes with one of the poly(A) complex components involved in pA-site recognition.
ELAV binding to ewg RNA does not compete with dCstF64 binding
Processing of the 3′ end is initiated by binding of CPSF to the AAUAAA sequence and stabilization by CstF (Willusz et al. 1990; Gilmartin and Nevins 1991; Keller et al. 1991). Because ELAV-binding sites potentially overlap with the expected position of dCstF64 binding to ewg RNA at pA2 (G/U-element; Fig. 6C), we wanted to test if ELAV physically inhibits dCstF64-binding. We therefore cloned CstF64 from Drosophila into a UAS vector, added a HA tag at the C terminus, and established transgenic fly lines. Expression of correct-sized dCstF64-HA (54 kD) was verified by overexpressing transgenes in eye imaginal discs with a GMR-GAL4 driver (Fig. 6A).
Next, we tested whether an additional protein can be detected after UV cross-linking in nuclear extracts from flies expressing dCstF64-HA (from heads of elav-GAL4/UAS dCstF64 flies) compared with wild-type flies. No clearly distinguished additional protein at the size of dCstF64-HA can be detected when resolving the 32P protein adducts (Fig. 6A, cf. lanes 5 and 4), although a protein with the same molecular weight size is detected. Therefore, we used IP with an anti-HA antibody to identify 32P adducts on dCstF64 as a result of binding to ewg RNA (Fig. 6B, lane 3).
To verify that dCstF64 binds ewg RNA pA2-ivs at the expected position (G/U-element, Fig. 6C), segmentally labeled substrate RNAs were generated and used for UV cross-linking with neuronal nuclear extract containing dCstF64-HA. Segmentally labeled substrate RNAs were generated by dividing pA2-ivs into two parts and transcribed separately by T7 RNA polymerase in vitro from suitable sites, which were found immediately after the AAUAAA sequence and after the first tandem AU4-6 motif in pA2-ivs. One of the transcripts was 32P-labeled and ligated to an unlabeled part resulting in full-length pA2-ivs substrate RNA (Fig. 6C).
No major differences are observed when comparing the cross-linking patterns of pA2-ivs RNAs A-C, except that the 55-kD band is absent with substrate B (Fig. 6D, lanes 8-10). As noted previously, however, cross-linking of both ELAV and dCstF64-HA is obscured by similarsized proteins (Figs. 2C, 6A) and thus IP was used to identify binding to RNA.
IP with anti-HA antibodies identifies 32P adducts on dCstF64 from RNA pA2-ivs C (Fig. 6D, lane 16), which includes a G/U-rich sequence element expected to bind dCstF64(MacDonald et al. 1994). ELAV cross-links prominently to pA2-ivs B (Fig. 6D, lane 2), to a lesser extent to pA2-ivs C (Fig. 6D, lane 3), and weakly to pA2-ivs A (Fig. 6D, lane 1).
Next, RNA pA2-ivs C was used in UV cross-linking assays to test whether addition of recombinant ELAV to nuclear extracts competes with dCstF64 binding to RNA pA2-ivs. A fivefold increase of ELAV in the nuclear extract did not reduce 32P adducts on dCstF64(Fig. 6D, lane 19; final concentration of endogenous and recombinant ELAV together is ∼0.8 μM). Despite RNase treatment after UV cross-linking, we observed coimmunopreciptation of ELAV with dCstF64 when increasing ELAV levels (Fig. 6D, lanes 18,19). Addition of recombinant ELAV did not change cross-linking of major proteins (Fig. 6D, cf. lanes 10 and 11-13).
When increasing ELAV levels with RNA pA2-ivs C, it is not possible to distinguish between dCstF64-HA binding to its expected position and redirection to sites 5′ of the AAUAAA sequence. We thus tested whether elevating ELAV levels leads to an increase of dCstF64 crosslinking to RNA pA2-ivs A, which would indicate redirection. Although variably weak cross-linking of dCstF64-HA to RNA pA2-ivs A can be detected, no correlation with ELAV levels is observed (Fig. 6E, cf. lanes 3-6 and 14-17).
Overexpression of dCstF64-HA or ELAV with GMRGAL4 in eye discs did not affect levels of intron-6-spliced transcripts, suggesting again that dCstF64 is not competing with ELAV binding and that ELAV levels are saturated (data not shown).
To further confirm the interaction of ELAV and dCstF64 observed in Figure 6D (lanes 18,19), coimmunopreciptation with either anti-ELAV or anti-HA antibodies from nuclear extracts was performed in the presence of RNase to eliminate RNA-mediated interactions. HA-tagged dCstF64 can be pulled down from nuclear extract with ELAV and vice versa (Fig. 6F, lanes 3,6). The lower pull-down efficiencies for ELAV are likely because of competition with endogenous dCstF64.
Discussion
Although the ewg gene is ubiquitously transcribed, a salient feature is the unusual posttranscriptional regulation of this transcription factor. The last intron 6 is only spliced in the presence of ELAV, as in neurons, or when ELAV is provided ectopically. This, in turn, leads to the expression of the major EWG protein isoform sufficient for full rescue of viability and neuronal function (Koushika et al. 1999, 2000). In this study, we developed a rescue reporter transgene, tcgER, that recapitulates ELAV-mediated regulation of ewg transcripts in neurons of developing and adult Drosophila flies. We show that ELAV binds directly to ewg RNA close to an intronic pA site and inhibits 3′-end formation at this site to promote neuronal splicing of ewg intron 6.
Direct ELAV binding to AU4-6 motifs in ewg RNA
Several lines of in vitro and in vivo evidence converge to identify the AU4-6 motifs 3′ of pA2 in ewg intron 6 as a functional ELAV-binding site. Deletions introduced in tcgER reporter transgenes show that only ∼25% of intron 6 is sufficient for ELAV-dependent regulation. Within the remaining RNA, ELAV UV cross-links in neuronal nuclear extracts to AU4-6 motif containing region pA-I, but not to the flanking sequence or the 3′ UTR. In addition, EMSAs show that recombinant ELAV binds with nanomolar affinity to ewg RNA pA-I. Mutational analysis further substantiated ELAV's binding to AU4-6 motifs in vitro, as U-to-C substitutions considerably reduced ELAV binding in UV cross-linking assays as well as in EMSAs. Moreover, AU4-6 motifs are necessary to inhibit cleavage of substrate RNA in in vitro cleavage/pA assays with neuronal nuclear extracts or when recombinant ELAV is added to nonneuronal extract. Finally, tcgER reporter transgenes with mutated AU4-6 motifs fail to show the neuronal processing mode of ewg intron 6, demonstrating the importance of these motifs to ELAV regulation in vivo.
The ELAV-binding site on ewg RNA consists of several AU4-6 motifs, consistent with previously reported binding preferences of ELAV/Hu proteins to AU-rich sequences (Levine et al. 1993; Gao et al. 1994; Abe et al. 1996). Whithin this site individual tandem AU4-6 motifs contribute to ELAV binding, indicating that several ELAV molecules bind to ewg RNA. Recently, Hu proteins were found to interact with each other in yeast two-hybrid assays and coimmunopreciptations, and could thus potentially form a complex on binding to target RNA (Kasashima et al. 2002). This is consistent with our own observations and might indicate that ELAV/Hu proteins associate cooperatively on target RNA to form a complex (M. Soller and K. White, unpubl.).
ELAV inhibits cleavage associated with an intronic pA site
In this article, we demonstrate that ELAV inhibits cleavage of ewg substrate RNA in in vitro cleavage assays in a sequence-specific and concentration-dependent manner. We further show that the inhibitory activity of ELAV resides in its ability to bind RNA. Thus, ELAV is not inhibitory via titrating any essential component. This is of particular importance, as ELAV was also found to interact with dCstF64 in nuclear extracts. Although we do not yet know if the interaction of ELAV and dCstF64 is direct, inhibition of pA by ELAV cannot be explained by sequestering pA factors (e.g., dCstF64) from binding to ewg RNA in vitro. Rather, specificity in the substrate RNA and assembly of ELAV and CstF64 on ewg RNA might play a critical role in inhibiting 3′-end processing.
How does ELAV binding mediate inhibition of 3′-end processing at ewg pA2? In IgM processing, binding of hnRNP F was found to compete with CstF64 binding to the regulated pA site, together with a change in RNA-binding activity of CstF64 during B-cell development (Edwalds-Gilbert and Milcarek 1995; Veraldi et al. 2001). Thus, hnRNP F interferes with pA site recognition. Although ELAV- and dCstF64-binding sites potentially overlap 3′ of the ewg pA2 site, competition for binding to ewg RNA was not observed in this study. Furthermore, deletion of the AAUAAA pA recognition sequence did not result in ELAV-independent neural splice site choice; instead, the neural specificity in the 3′ splice choice was lost. That the AAUAAA sequence of the pA2 site is important for neuron-specific splicing suggests that pA factors bind to the ewg pA2 site in neurons and that this binding also influences 3′ splice site choice. Additionally, overexpression of neither ELAV nor dCstF64 in neurons changed ewg 3′-end processing, although only about half the amount of all transcripts splice intron 6 in neurons. This suggests that levels of both ELAV and dCstF64 are saturating, and that 3′-end formation and the presence of ELAV are not mutually exclusive. In contrast, levels of CstF64 have been shown to be critical for IgM processing in vivo using cell culture systems (Takagaki et al. 1996; Takagaki and Manley 1998). In summary, our results argue against a role of ELAV in competing with pA site recognition by CPSF and CstF.
ELAV and poly(A) factors promote alternative splicing of ewg RNA
In neurons, splicing of ewg intron 6 is achieved through inhibition of intronic 3′-end formation at pA2 and distal 3′ splice site selection. Figure 7A depicts the sequence around ewg pA2 with the regulatory elements. A model of ELAV binding 3′ of pA2 together with a partial pA complex consisting of CPSF and CstF is shown in Figure 7B.
By what mechanism does ELAV inhibit 3′-end processing to allow splicing? ELAV's binding in the proximity of the cleavage site could slow the recruitment of cleavage factors (CF I and CF II) and/or poly(A) polymerase (PAP) resulting in a delay of the cleavage reaction. Alternatively, execution of the cleavage reaction could involve a structural rearrangement that is affected by ELAV binding. In either case, this intermediate pA complex consisting of at least CPSF and CstF together with ELAV alters the timing of 3′-end processing to allow for the assembly of the splicosome to the neuronal 3′ splice site of intron 6 and for splicing to proceed.
Transcription and RNA processing are coupled through the C-terminal domain of the largest subunit of RNA polymerase II (pol II; for review, see Proudfoot et al. 2002). Low processivity of RNA pol II could occlude the availability of a 3′ splice site and thus favor intronic 3′-end processing. The short distance of only 164 nt from the AAUAAA sequence to the 3′ splice site of exon I, however, makes this an unlikely scenario. Furthermore, ELAV's ability to inhibit cleavage in vitro in a concentration- and sequence-dependent manner argues against a role in stimulating RNA pol II processivity to make the neural 3′ splice site available for splicing before 3′-end processing occurred.
What drives the choice of the neural splice site in ewg intron 6? An interesting alliance between ELAV and components of the pA complex in choosing the neural 3′ splice site was revealed when analyzing mutations of the AAUAAA pA complex recognition sequence (ΔpA2). In ΔpA2, inclusion of exon I can occur even in the presence of ELAV, whereas in the absence of ELAV, inclusion of exon I is the major splice product. Thus, the ability of the pA site to initiate the assembly of pA factors in the presence of ELAV is key to the tight regulation of usage of the distal 3′ splice site in neurons. As a consequence, exon I is not included in wild-type neurons. In nonneuronal tissue, inclusion of exon I is observed at low frequency, as the few transcripts that escape 3′-end formation at pA2 are spliced to exon I (Koushika et al. 2000). Thus, ELAV and factors bound to the pA2 site together block the 3′ splice site of exon I.
In summary, we show that the RNA-binding protein ELAV can inhibit 3′-end formation without affecting recognition of the pA site by CPSF and CstF64. ELAV and components of the pA complex then direct exclusive use of the distal 3′ splice site to promote the neural processing mode. Because bona fide pA sites are frequently found in introns, binding of pA complex components could contribute to localize splice sites, and, as shown in this study, can regulate alternative splicing.
Materials and methods
Fly genetics and recombinant DNA technology
Fly breeding, genetics, transformation, and DNA mutagenesis were according to standard procedures as detailed in Koushika et al. (2000), Lisbin et al. (2001), and Sambrook and Russel (2001). Eye discs devoid of ELAV were generated in mosaic animals by mitotic recombination using an elav null mutation (elave5) according to Stowers and Schwarz (1999). At the end of larval development, the elave5 homozygous clones comprise ∼90%-95% of photoreceptors (data not shown).
DNA clones were fully sequenced before subcloning into the reporter construct. Details of generation of the reporter constructs and mutants are available on request. The following oligonucleotides (antisense) were used to generate the mutants used in this study: Δ7-l (CGCCGGCTTAGGACTGACGATAAGCTTCCCGCTCTCGAGATCAGCGAATAGCACTG), Δ7-HinDIII-MfeI linker (AGCTTGTAGGATCCATAGTC), m1 (CAACAATAATTAGAGATGAGAGTTAATTTGAAGTTAAATTGTTGGC), m2 (TCAATATAGAGTGAGGACAAGCAGAGAGTGAATATTCTGAGAC), m3 (GAACATAAAGTTTAAAGTTATAAGATAGAGTGTATAATAGCAGAGTATACTTACATAAAC), ΔpA2-del (GAAGTTAAATTGTTGGCTAGCATTTCAGTTTACAAAATGTACAAGC), and ΔpA2-ins (GAAGTTAAATTGTTGGCAACGTTAGTAGCATTTCAGTTTACAAAATGTACAAGC). Two to three independent insertion lines fulfilling the expected rescue ability were analyzed for each construct.
dCstF64 was amplified from genomic DNA (400 ng/reaction) as two separate overlapping pieces using primers 64F1 (CGAAGATCTGTTTGTCAACGTTTGCTAAGATTTTGTCG)/64R2 (GGGTAGAGAAGCGCGCAAATCCTGGTCC) and 64F2 (GCGCTCGGGATGCTCTTCAAGGCCAACC)/64R1 (GGACTAGTTGCTCGAGCGCTGGGTACTCTTCGCAATCTGC) with Pwo polymerase (Roche) for 30 cycles (94°C for 30 sec, 58°C for 30 sec, and 68°C for 1 min with an initial denaturation step at 94°C for 30 sec and a final extension at 68°C for 1 min). The fragments were then added as pieces from a 1% agarose gel to a second PCR with flanking primers 64F1/64R1 and amplified with an initial denaturation step at 94°C for 5 min in water followed by an annealing and extension step at 68°C for 10 min to synthesize full-length dCstF64 ORF. dCstF64 DNA was then cloned into a modified pUAST adding a C-terminal HA tag and the ewg 3′ UTR from intron 6 using BglII, XhoI, and SpeI sites and sequenced before transformation of flies.
RNA extraction and RT-PCR
RNA extraction and RT-PCR were done as in Koushika et al. (2000). RT for 3′ RACE on RNA from 30 eye discs was done with 10 pmole of primer AP [GGCCACGCGTCGACTAGTAC(T)17; Invitrogen], and nested PCR was on 10% of the RT reaction with anchor primer AUAP (GGCCACGCGTCGACTAGTAC) and 5F for 22 cycles (94°C for 30 sec, 60°C for 40 sec, and 72°C for 120 sec with an initial denaturation step at 94°C for 30 sec and a final extension at 72°C for 5 min) on 10% of the RT reaction and with a second nested primer (6F or 5R) for 10 cycles on 10% of the first PCR reaction. RT for 3′ RACE on RNA from in vitro cleavage reactions was done the same except that denaturation was at 60° C for 5 min, annealing of the AP-G primer (10 fmole, containing a 3′ guanosine) was at 50°C for 10 min, and extension was at 46°C for 30 min. For nested PCR, primers KS T3F (GGGAACAAAAGCTGGGTACAATTGC)/AUAP were used with 10% of the RT reaction for 30 cycles (94°C for 30 sec, 54°C for 30 sec, and 72°C for 20 sec with an initial denaturation step at 94°C for 30 sec) and then F6i2 (CAAGTCAATTGCAAAAGAGGGAGAATGAAAAAGCAAC)/AUAP for 15 cycles on 4% of the first PCR reaction. Sequencing was done with primer F6i3 (GCGATTGTTGTGATTTTAATATAATTTTACCTT). Control RT was done with 10 fmole of primer R6int8 (AACATAAAGTATAACTCGAGGACAAGCAAAAAATGTATATTCTGAG) and PCR for 30 cycles with primers KS T7F/R6int8.
Primers were eeF (GGGAGAAAATCGAAGCAGAGAGC), eeR (GGAAGTGTTGCCGGCACTTTGC), VSV-R (ACTTGCCCAGGCGGTTCATCTC), ApplF2 (GCGGCCAATCCCAACCTAGAGACC), ApplR2 (AAACTGCAGCACAATGGGATGGTGTGTTGTGAC), or as in Koushika et al. (2000).
Electrophoretic mobility shift assays
GST-fusion proteins were produced in E. coli according to the manufacturer's instructions and cleaved off the GST moiety with PrecissionProtease (Amersham) and stored in protease cleavage buffer after removal of the protease. Gelpurified RNA in 50 μg/mL tRNA (Roche) and 1 U/μL RNasin (Roche) was heated for 5 min at 65°, renatured at RT, then mixed with an equal volume of recombinant protein to a final concentration of 50 mM Tris-HCl (pH 7.5), 40 mM KCl, 20 mM KGlutamate, 15 mM NaCl, 25 μg/mL tRNA, 0.5 mM DTT, 50 μg/mL BSA (acetylated) in a total of 10 μL, and incubated at room temperature for 20 min. Five microliters reaction with 2 μL 50% glycerol were loaded on 4% (80:1 acrylamide/bisacrylamide) polyacrylamide gels and run at 4°C at 200 V in 0.5× TBE.
Nuclear extract preparation
Fly heads were collected by freezing flies in liquid nitrogen, shaking vigorously, and then sieving (Fisher). Heads were stored at -80°C.
The protocol for nuclear extract preparation from fly heads is adapted from Spikes and Bingham (1992) and So and Rosbash (1997). All procedures are done on ice or at 4°C. One gram of heads was homogenized in 7 mL of buffer A [15 mM HEPES at pH 7.6, 10 mM KCl, 5 mM MgCl2, 350 mM sucrose, 0.1 mM EDTA, 0.5 mM EGTA, 1 mM DTT, 1 mM PMSF (0.2 M in isopropanol), 1 μg/mL leupeptin] with 10 strokes with the loose pestle and 1 stroke with the tight pestle in a Dounce homogenizer. The homogenate was then passed through a 10-mL Polyprep column (Bio-Rad), adjusted to 10 mL total volume, overlaid on 3 mL of buffer B (same as A, but with 800 mM sucrose) in a 14-mL polypropylene centrifuge tube and spun for 5 min at 16,000g in a Sorvall HB-4(swing out) rotor. The supernatant was completely removed, avoiding the top of the two layers.
Nuclei were then dissolved in 300 μL buffer C [20 mM HEPES at pH 7.6, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 1 mM PMSF, 1 μg/mL leupeptin, 25% (v/v) glycerol (Ultrapure, GIBCO)] by vortexing, a small magnetic stirrer was added, and nuclei extracted by vigorous stirring for 30 min on ice. The supernatant was adjusted to 0.5 mL, nuclei were pelleted 10 min at 16,000g and the supernatant removed, avoiding the top layer and the pellet. Dialysis was done for 3 h with three changes in buffer D [20 mM HEPES at pH 7.6, 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 1 mM PMSF, 1 μg/mL leupeptin, 20% (v/v) glycerol], the extract spun for 10 min at 16,000g, and aliquots were flash frozen in liquid nitrogen for storage at -80°C. Protein concentration was ∼5 mg/mL. DTT, PMSF, and leupeptin were added immediately before use.
Kc cells were grown in D22 media and nuclear extract was prepared according to Dignam et al. (1983).
UV cross-linking assays and immunoprecipitations
In a total volume of 10 μL, 1-10 fmoles of capped RNA was incubated in 40% (v/v) nuclear extract, 1 mM ATP, 5 mM creatine phosphate, 2 mM MgAcetate, 20 mM KGlutamate, 1 mM DTT, 20 U RNasin (Roche), and 5 μg/mL tRNA at room temperature for 25 min. Heparin was added to 25 μg/mL, incubated for 5 min, and UV cross-linked on ice at 254 nm for 20 min in a Stratalinker (Stratagene), followed by digestion with RNase A/T1 mix (Ambion) at room temperature for 15 min.
IPs were done in a final volume of 120 μL using 10 μL monoclonal anti-ELAV antibodies 7D or anti-HA antibodies 5D3 (Roche), 20 μL protein A/G beads (Santa Cruz Biotechnology), and 10 μL nuclear extract or UV cross-linking mix in NET 150 or 500 (150 or 500 mM NaCl, 0.5 M Tris-HCl at pH 7.5, 0.01% NP-40) at room temperature for 2 h.
Splinted ligation of RNAs
Gel-purified PCR products generated with Pwo polymerase (Roche) were used for in vitro transcription of RNAs by T7 polymerase. After gel purification RNAs were ligated according to Moore and Query (2000) using DNA bridging oligos Glig2 (GAGGACAAGCAAAAAATGTATATTCTGAGACCAACAATAATTAAAAATAAAAATTAATTTGAAGTTAAATTGTTGGC) and Glig3 (TAAAAATTAATTTGAAGTTAAATTGTTGGCTTTATTTAGCATTTCAGTTTACAAAATGTACAAGCAAATATTTTAAGG; start of the 3′ fragment is underlined).
In vitro cleavage/polyadenylation assays
In a total volume of 20 μL 1-5 fmoles of capped and gel-purified RNA were incubated in 40% (v/v) nuclear extract, 0.1 mM ATP, 0.5-2 mM 3′dATP (Sigma; neutralized with KOH), 5 mM creatine phosphate, 0.6 mM MgAcetate, 1.5 mM DTT, 40 U RNasin (Roche), 5 μg/mL tRNA, and 1.5% (v/v) PEG8000 (from frozen aliquots) at room temperature (24°C-27°C). Recombinant ELAV [diluted into 40% (v/v) PrecissionProtease cleavage buffer] was added to the cleavage assay to a final volume of 5% (v/v). Reactions were quenched in stop buffer (100 mM Tris at pH 7.5, 10 mM EDTA, 1% SDS, 150 mM NaCl, 300 mM NaAc), phenol-chloroform extracted, and ethanol precipitated.
Acknowledgments
We thank S. Stowers and T. Schwarz for fly stocks prior to publication; I. Haussmann and D. Motola for help with cloning; J.-R. Néptune for help with fly transformations; P. Parmenter for cell culture; and I. Haussmann, M. Moore, M. Rosbash, and many colleagues at Brandeis for advice, discussions, and comments on the manuscript. We also thank referees for their comments and suggestions. This work was supported by National Institute of Health Grant NS 37169 and NS 44232. M.S. was supported by a Fellowship from the Swiss National Science Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1106703.
Corresponding authors.
References
- Abe R., Sakashita, E., Yamamoto, K., and Sakamoto, H. 1996. Two different RNA binding activities for the AU-rich element and the poly(A) sequence of the mouse neuronal protein mHuC. Nucleic Acids Res. 24: 4895-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antic D., Lu, N., and Keene, J.D. 1999. ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes & Dev. 13: 449-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crenshaw III E.B., Russo, A.F., Swanson, L.W., and Rosenfeld, M.G. 1987. Neuron-specific alternative RNA processing in transgenic mice expressing a metallothionein-calcitonin fusion gene. Cell 49: 389-398. [DOI] [PubMed] [Google Scholar]
- DeSimone S.M., Coelho, C. Roy, S., VijayRaghavan, K., and White, K. 1996. ERECT WING, the Drosophila member of a family of DNA binding proteins is required in imaginal myoblasts for flight muscle development. Development 122: 31-39. [DOI] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz, R.M., and Roeder, R.G. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11: 1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dredge B.K., Polydorides, A.D., and Darnell, R.B. 2001. The splice of life: Alternative splicing and neurological disease. Nat. Rev. Neurosci. 2: 43-50. [DOI] [PubMed] [Google Scholar]
- Edwalds-Gilbert G. and Milcarek, C. 1995. Regulation of poly(A) site use during mouse B-cell development involves a change in the binding of a general polyadenylation factor in a B-cell stage-specific manner. Mol. Cell. Biol. 15: 6420-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford L.P., Watson, J., Keene, J.D., and Wilusz, J. 1999. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes & Dev. 13: 188-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi I.E. and Steitz, J.A. 2001. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 294: 1895-1901. [DOI] [PubMed] [Google Scholar]
- Gao F.B., Carson, C.C., Levine, T., and Keene, J.D. 1994. Selection of a subset of mRNAs from combinatorial 3′ untranslated region libraries using neuronal RNA-binding protein Hel-N1. Proc. Natl. Acad. Sci. USA 91: 11207-11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin G.M. and Nevins, J.R. 1991. Molecular analysis of two poly(A) site processing factors that determine the recognition and efficiency of cleavage of the pre-mRNA. Mol. Cell. Biol. 11: 2432-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski P.J. and Black, D.L. 2001. Alternative RNA splicing in the nervous system. Prog. Neurobiol. 65: 289-308. [DOI] [PubMed] [Google Scholar]
- Hastings M.L. and Krainer, A.R. 2001. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell. Biol. 13: 302-309. [DOI] [PubMed] [Google Scholar]
- Horabin J.I. and Schedl, P. 1993. Regulated splicing of the Drosophila sex-lethal male exon involves a blockage mechanism. Mol. Cell. Biol. 13: 1408-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K.B., Dredge, B.K., Stefani, G., Zhong, R., Buckanovich, R.J., Okano, H.J., Yang, Y.Y., and Darnell, R.B. 2000. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron 25: 359-371. [DOI] [PubMed] [Google Scholar]
- Kasashima K., Sakashita, E., Saito, K., and Sakamoto, H. 2002. Complex formation of the neuron-specific ELAV-like Hu RNA-binding proteins. Nucleic Acids Res. 30: 4519-4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W., Bienroth, S., Lang, K.M., and Christoferi, G. 1991. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3′ processing signal AAUAAA. EMBO J. 10: 4241-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika S.P., Lisbin, M.J., and White, K. 1996. ELAV, a Drosophila neuron-specific protein, mediates the generation of an alternatively spliced neural protein isoform. Curr. Biol. 6: 1634-1641. [DOI] [PubMed] [Google Scholar]
- Koushika S.P., Soller, M., DeSimone, S.M., Daub, D.M., and White, K. 1999. Differential and inefficient splicing of a broadly expressed Drosophila erect wing transcript results in tissue-specific enrichment of the vital EWG protein isoform. Mol. Cell. Biol. 19: 3998-4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika S.P., Soller, M., and White, K. 2000. The neuron-enriched splicing pattern of Drosophila erect wing is dependent on the presence of ELAV protein. Mol. Cell. Biol. 20: 1836-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullmann M., Göpfert, U., Siewe, B., and Hengst, L. 2002. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′ UTR. Genes & Dev. 16: 3087-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labourier E., Adams, M.D., and Rio, D. C. 2001. Modulation of P-element pre-mRNA splicing by a direct interaction between PSI and U1 snRNP 70K protein. Mol. Cell 8: 363-373. [DOI] [PubMed] [Google Scholar]
- Lallena M.J., Chalmers, K.J., Llamazares, S., Lamond, A.I., and Valcarcel, J. 2002. Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell 109: 285-296. [DOI] [PubMed] [Google Scholar]
- Levine T.D., Gao, F., King, P.H., Andrews, L.G., and Keene, J.D. 1993. Hel-N1: An autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol. Cell. Biol. 13: 3494-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy N.S., Chung, S., Furneaux, H., and Levy, A.P. 1998. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J. Biol. Chem. 273: 6417-6423. [DOI] [PubMed] [Google Scholar]
- Lisbin M.J., Gordon, M., Yannoni, Y.M., and White, K. 2000. Function of RRM domains of Drosophila melanogaster ELAV: RNP1 mutations and RRM domain replacements with ELAV family proteins and SXL. Genetics 155: 1789-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisbin M.J., Qiu, J., and White, K. 2001. The neuron-specific RNA binding protein ELAV regulates neuroglian alternative splicing in neurons and binds directly to its pre-mRNA. Genes & Dev. 15: 2546-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez A.J. 1998. Alternative splicing of pre-mRNA: Developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 32: 279-305. [DOI] [PubMed] [Google Scholar]
- Lou H., Gagel, R.F., and Berget, S.M. 1996. An intron enhancer recognized by splicing factors activates polyadenylation. Genes & Dev. 10: 208-219. [DOI] [PubMed] [Google Scholar]
- Lou H., Helfman, D.M., Gagel, R.F., and Berget, S.M. 1999. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol. Cell. Biol. 19: 78-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald C.C., Wilusz, J., and Shenk, T. 1994. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNA downstream of the cleavage site and influences cleavage site location. Nucleic Acids Res. 14: 6647-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T. and Tasic, B. 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418: 236-243. [DOI] [PubMed] [Google Scholar]
- Modrek B. and Lee, C. 2002. A genomic view of alternative splicing. Nat. Genet. 30: 13-19. [DOI] [PubMed] [Google Scholar]
- Moore M.J. and Query, C.C. 2000. Joining of RNAs by splinted ligation. Methods Enzymol. 317: 109-123. [DOI] [PubMed] [Google Scholar]
- Myer V.E., Fan, X.C., and Steitz, J.A. 1997. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 16: 2130-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M. and Berget, S.M. 1991. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes & Dev. 5: 2086-2095. [DOI] [PubMed] [Google Scholar]
- Peng S.S., Chen, C.Y., Xu, N., and Shyu, A.B. 1998. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 17: 3461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M.L. 1994. RNA processing and expression of immunoglobulin genes. In Handbook of B and T lymphocytes (ed. E.C. Snow), pp. 321-342. Academic Press, San Diego, CA.
- Proudfoot N.J., Furger, A., and Dye, M.J. 2002. Integrating mRNA processing with transcription. Cell 108: 501-512. [DOI] [PubMed] [Google Scholar]
- Robinow S., Campos, A.R., Yao, K.M., and White, K. 1988. The elav gene product of Drosophila, required in neurons, has three RNP consensus motifs. Science 242: 1570-1572. [DOI] [PubMed] [Google Scholar]
- Sakamoto H., Inoue, K., Higuchi, I., Ono, Y., and Shimura, Y. 1992. Control of Drosophila Sex-lethal pre-mRNA splicing by its own female-specific product. Nucleic Acids Res. 20: 5533-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook R.P. and Russel, D.V. 2001. Molecular cloning—A laboratory handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Samson M.L. 1998. Evidence for 3′ untranslated region-dependent autoregulation of the Drosophila gene encoding the neuronal nuclear RNA-binding protein ELAV. Genetics 150: 723-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.W. and Valcarcel, J. 2000. Alternative pre-mRNA splicing: The logic of combinatorial control. Trends Biochem. Sci. 25: 381-388. [DOI] [PubMed] [Google Scholar]
- So W.V. and Rosbash, M. 1997. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 16: 7146-7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spikes D. and Bingham, P.M. 1992. Analysis of spliceosome assembly and the structure of a regulated intron in Drosophila in vitro splicing extracts. Nucleic Acids Res. 20: 5719-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers R.S. and Schwarz, T.L. 1999. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics 152: 1631-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y. and Manley, J.L. 1998. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol. Cell 2: 761-771. [DOI] [PubMed] [Google Scholar]
- Takagaki Y., Seipelt, R.L., Peterson, M.L., and Manley, J.L. 1996. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87: 941-952. [DOI] [PubMed] [Google Scholar]
- Veraldi K.L., Arhin, G.K., Martincic, K., Chung-Ganster, L.H., Wilusz, J., and Milcarek, C. 2001. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell. Biol. 21: 1228-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E. and Keller, W. 1994. 3′-end processing of mRNA. In RNA processing: A practical approach (eds. S.J. Higgins and B.D. Hames), pp. 1-34. Oxford University Press, Oxford, UK.
- Wang J. and Bell, L.R. 1994. The Sex-lethal amino terminus mediates cooperative interactions in RNA binding and is essential for splicing regulation. Genes & Dev. 8: 2072-2085. [DOI] [PubMed] [Google Scholar]
- Willusz J., Shenk, T., Takagaki, Y., and Manley, J.L. 1990. A multicomponent complex is required for the AAUAAA-dependent cross-linking of a 64 kilodalton protein to polyadenylation substrates. Mol. Cell. Biol. 10: 1244-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao K.-M., Samson, M.-L., Reeves, R., and White, K. 1993. Gene elav of Drosophila melanogaster: A prototype for neuronal-specific RNA binding protein gene family that is conserved in flies and humans. J. Neurobiol. 24: 723-739. [DOI] [PubMed] [Google Scholar]
- Zhao J., Hyman, L., and Moore, C. 1999. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63: 405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]