Abstract
atonal (ato) encodes a basic helix–loop–helix protein and is required for the specification of R8 photoreceptor cells in Drosophila. In the eye imaginal discs, expression of Ato protein is initially in a dorsoventral stripe of cells anterior to the morphogenetic furrow (MF). In the MF, this stripe expression is resolved into regularly spaced clusters of Ato-positive cells, the proneural clusters, which are intervened with Ato-negative cells. Another basic helix–loop–helix protein, Daughterless (Da), dimerizes with Ato and is expressed at an enhanced level in Ato-expressing cells. Here we show that during the late stages of proneural clusters, the mitogen-activated protein kinase (MAPK) is activated in proneural clusters. Normal ato or da activity is required for maintenance of MAPK activation. Furthermore, in ato or da mutants, Ato expression is expanded to all cells in the MF, suggesting that ato and da are required for Ato repression in cells between proneural clusters. By changing the MAPK activity in proneural clusters, we show that MAPK activation mediates Ato repression nonautonomously. Consistently, hyperactivation of the MAPK in a stripe of cells posterior to or overlapping the Ato stripe eliminates the formation of proneural clusters. Taken together, these results suggest that a negative regulatory loop involving MAPK activation and Ato repression is required for the generation of evenly spaced proneural clusters.
The Drosophila compound eye is composed of 750 ommatidia, each consisting of eight photoreceptor cells, R1–R8, four cone cells, and several other accessory cells (1). In the third-instar larvae, neuronal differentiation of the retina is initiated at the most posterior tip of the eye imaginal disc and progresses anteriorly as a wave, which is marked by the morphogenetic furrow (MF) (2). Anterior to the MF, cells are undifferentiated. After the sweep of the MF, the first cells to differentiate are R8 photoreceptor cells, the founder cells of individual ommatidia. Other photoreceptor cells (R1–R7), cone cells, and pigment cells are recruited in a precise sequence into growing clusters around the specified R8 cells, thus forming the ommatidia (1, 3).
The proneural gene atonal (ato) specifies R8 photoreceptor cells (4, 5). The Ato expression pattern in the eye imaginal disc delineates the R8 specification process (4–6). This pattern includes a low level of expression in a dorsoventral stripe of undifferentiated cells abutting the anterior margin of the MF. Gradually in the MF, this stripe is resolved into equally spaced clusters of Ato-positive cells intervened with gaps of Ato-negative cells. Each Ato-positive cluster is further restricted into a single R8 precursor cell that expresses Ato for 3–4 rows after the MF. The restriction process by which R8 precursors are selected from clusters of Ato-positive cells depends on the mechanism of lateral inhibition, which is mediated by the Notch (N) signaling pathway (7) consisting of components such as Su(H) and E(spl)-C (8). However, the mechanism that resolves the initial Ato stripe into evenly spaced Ato-positive clusters in the MF is not clear.
Following the specification of R8 precursors, neighboring cells are recruited sequentially to form growing clusters, which eventually consist of all photoreceptor cells, cone cells, and pigment cells. The recruitment process requires the epidermal growth factor receptor (Egfr) signaling pathway (3, 9, 10) mediated through the Ras/Raf/mitogen-activated protein kinase (MAPK) cascade. Inactivation of the Egfr activity impedes the recruitment process (9). In addition to the blockade of cell differentiation, a profound effect on eye development is observed, including cell proliferation defects, induction of cell death posterior to the MF, and minor defects in the spacing of R8 precursors (11).
Gabay et al. (12) showed by anti-dp-ERK antibody (13) staining that the MAPK signaling is highly activated in evenly spaced clusters of cells in the MF. These dp-ERK clusters appear slightly anterior to the rosette-like clusters (10) and colocalize with Ato proneural clusters in the MF (14). Inactivation of Egfr by using the temperature-sensitive allele Egfrts results in reduction of MAPK activation in the MF only for the initial 2 hr (10). The level of MAPK activation is restored thereafter, suggesting that Egfr is not the only component responsible for MAPK activation in the MF. There is also little defect on furrow progression and Ato expression in Egfrts mutants. In another report (11), there is some, but not significant, disturbance on the spacing of R8 precursors in Egfr or Ras1 mutant clones across the MF. However, MAPK activation within these clones has not been examined. Taken together, these data suggest that more than one signaling pathway is required for MAPK activation in the evenly spaced clusters within the MF.
The pattern of dp-ERK distribution in the eye discs has been extensively examined (10, 14). In this study, we show that in the MF the dynamic expression of dp-ERK corresponds to Ato proneural clusters. We further show that ato and daughterless (da) are required for two events in the MF: MAPK activation in proneural clusters and Ato repression in gaps between proneural clusters. Ato repression in the gaps is likely mediated by the MAPK signaling in proneural clusters because changes in the level of MAPK activation negatively regulate Ato expression nonautonomously. Two mutants that hyperactivate the MAPK in the MF and fail to form proneural clusters provide further evidence for the negative regulation of Ato by the MAPK signaling.
MATERIALS AND METHODS
Immunostaining.
The eye discs were fixed in 4% formaldehyde solution before antibody binding. Primary antibodies were rabbit anti-Ato (gift from Y. Sun and Y. N. Jan, University of California, San Francisco; 1:1,000), mouse anti-dp-ERK (Sigma; 1:250), rat anti-Elav (Developmental Studies Hybridoma Bank, University of Iowa; 1:500), mouse anti-Boss (gift from S. L. Zipursky, University of California, Los Angeles; 1:1,000), mouse anti-myc (Santa Cruz Biotechnology; 1:250), mouse anti-β-galactosidase (Sigma; 1:250), and rabbit anti-β-galactosidase (Cappel, 1:1,000). Secondary antibodies (Jackson ImmunoResearch) were Cy3 anti-rabbit (1:1,000) and Cy5 anti-mouse (1:500).
ato1 and dakx136 Mutant Clone.
Second- or early third-instar larvae of the hsFLP/+; FRT82 ato1/FRT82 p[w+ hs-π-myc] (15), hsFLP/+; FRT82 ato1/FRT82 p[w+ construct D], or hsFLP/+; FRT40 dakx136/FRT40 p[w+ construct D] (16) genotype were incubated at 37°C for 1 hr. The eye discs of late third-instar larvae were fixed for immunostaining.
Nts Temperature Shift Assay.
w-Nts/w-Nts larvae were incubated at 32°C for 2 hr before immunostaining.
hs-rho and hs-aos Experiment.
For dp-ERK and Ato double labeling, hs-rho larvae (line 1B, gift from P. Marin and E. Bier, University of California, San Diego) were incubated at 37°C for 1 hr and shifted to 25°C for the time indicated, and hs-aos (four copies, gift from H. Okano, University of Tsukuba, Japan) larvae were treated similarly but dissected immediately. For Boss and Elav double labeling, the larvae were treated for five heat pulses with 1-hr interval and dissected after 18 hr at 25°C.
sca-GAL4 Expression.
For expression of DER, DN-DER (gifts from M. Freeman, Medical Research Council, United Kingdom), or constitutive active Draf (Bloomington Stock Center) with sca-GAL4 (17), third-instar larvae were shifted from 18°C to 30°C for 4 hr before immunostaining.
Elp Mutants.
Elp is a gain-of-function allele of DER (18). In this study, both ElpB1/ElpB1 and Elp1/ElpB1 mutants, and Elp1/+; l(3)2B10/Df(3R)p14 eye discs were labeled. l(3)2B10 is a hypomorphic allele of d14–3-3ɛ and Df(3R)p14 deletes d14–3-3ɛ locus (19).
RESULTS
Coexpression of Ato and dp-ERK in Proneural Clusters at the Late Stages of the Restriction Process.
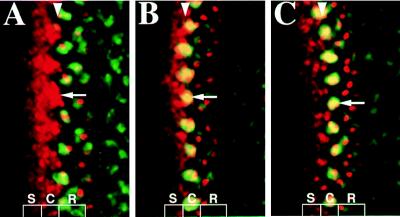
MAPK activation in the eye discs has been extensively examined (10, 14). Clusters of dp-ERK expression are colocalized with Ato proneural clusters (14). We have independently examined the colocalization of Ato and dp-ERK clusters. These dp-ERK clusters corresponded to Ato-positive clusters during the late restriction process (Fig. 1). When Ato-positive cells first emerged from the anterior stripe as 7- to 9-cell clusters, no MAPK activation was detected within individual clusters (stage I, Fig. 1A). However, when these clusters were restricted to include fewer cells, i.e., 5–7 cells, the MAPK was activated (stage II, Fig. 1B). The activation was maintained with further restriction of these clusters to fewer than three cells per cluster (stage III, Fig. 1C), while a new wave of clusters was emerging. The coexpression of Ato and dp-ERK in proneural clusters suggests a possible regulatory pathway between Ato expression and MAPK signaling.
Figure 1.
Three stages of proneural clusters that express Ato and dp-ERK. Confocal images of the eye discs showing Ato (red) and dp-ERK expression (green) at stage I (A), II (B), and III (C). In this and the following figures, anterior is left, the position of the MF is indicated by a white arrowhead (as revealed by phalloidin-FITC labeling, not shown). Arrows indicate proneural clusters, and S indicates Ato stripe expression, C, Ato cluster expression, and R, R8 precursors.
ato and da Are Required to Maintain MAPK Activation in Proneural Clusters.
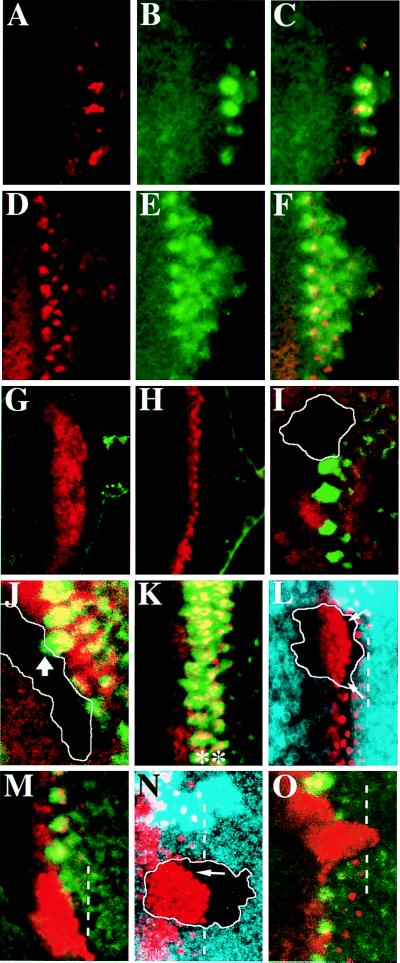
Because dp-ERK expression was localized in Ato proneural clusters at the late stages of the restriction process, the requirement of ato for MAPK activation was evaluated. During MF initiation, a few clusters of cells expressing both Ato and dp-ERK appeared near the posterior margin of the eye discs (Fig. 2 A–C). An increasing number of clusters aligned dorsoventrally in a row during anterior progression of the MF (Fig. 2 D–F). This Ato pattern in early third-instar larvae is similar to that described for late third-instar larvae (Fig. 1), except that expression in the initial stripe is missing (Fig. 2 A and D). In ato mutants, the initiation of the MF is not disturbed (4, 5). The furrow progresses to a certain distance before it completely stops, suggesting that ato is required for furrow progression. In the ato1/ato1 eye discs of early third-instar larvae, the nonfunctional, Ato mutant protein was detected as a broad stripe in the MF (Fig. 2 G and H). However, dp-ERK expression in regular clusters was abolished completely (Fig. 2 G and H). Because ato also is required for furrow progression, Ato expression gradually diminished and the furrow stopped midway across the eye disc (not shown) (5).
Figure 2.
The requirement of ato and da in MAPK activation. Early third-instar larval eye discs of +/+ (A–F), and ato1/ato1 (G and H) showing Ato (red) and dp-ERK (green) expression. Mutant clone of ato1 (I) or dakx136 (J) across the MF lacking dp-ERK expression (green). The clone boundary (marked by white line) is revealed by the lack of β-galactosidase (I, red) or Da (J, red) expression. The thick arrow in J indicates a dp-ERK cluster that overlaps the clone boundary. (K) Nts eye disc incubated at restricted temperature showing two rows of Ato clusters (red and indicated by ∗) expressing dp-ERK (green). ato1 (L and M) or dakx136 (N and O) clones showing expansion of Ato expression (red in L–O) and lacking dp-ERK expression (green in M and O). The clone boundary (marked by white line) was revealed by lack of myc staining (blue in L and N). Within the clones, expansion of Ato-positive cells reaches the most posterior row of R8 precursors (marked by dotted white lines in L–O), except that near the margin Ato is still repressed in some cases (arrows in L and N).
To eliminate the possibility that defects in dp-ERK expression were the result of the failure of furrow progression, we examined the ato mutant clones. Within the ato1/ato1 clones (n = 9) across the MF, furrow progression was slightly delayed, as revealed by phalloidin staining (not shown). The expression of dp-ERK was abolished completely (Fig. 2 I and M). da encodes an ubiquitously expressed basic helix–loop–helix protein, which forms heterodimer with Ato for transcription regulation (20). Da protein is expressed at an enhanced level in cells expressing Ato (21). In the da mutant clones (n = 14) that overlap the MF, dp-ERK expression also was abolished (Fig. 2 J and O). Taken together, these results suggest that both ato and da are required for MAPK activation in proneural clusters.
Occasionally in the ato or da clones, dp-ERK clusters overlapped the borders of the mutant clones (arrowhead in Fig. 2J). However, no clusters of dp-ERK-expressing cells located exclusively within the mutant clones, suggesting that MAPK activation in these mutant cells required the neighboring Ato- or Da-positive cells.
Notch (N) is involved in the process of lateral inhibition that restricts Ato proneural clusters into single R8 precursors (7). In the Nts discs at the restrictive temperature, failure of lateral inhibition resulted in an ectopic row of Ato-positive clusters in place of the first row of single R8 cells (Fig. 2K) (7). These ectopic Ato-positive clusters expressed dp-ERK (Fig. 2K), indicating that Ato expression maintained MAPK activation in proneural clusters.
Expansion of Ato Expression in ato and da Mutants.
In the ato1/ato1 eye discs of early third-instar larvae, Ato protein was expressed as a broad stripe (Fig. 2 G and H), wider than the first row of Ato clusters in the wild-type eye discs (compare Fig. 2 G and H to Fig. 2 A and D, respectively). To further confirm that the number of Ato-expressing cells was increased in ato1 mutants, we examined the expression of Ato mutant protein in the mutant clones. In the ato1/ato1 clones (n = 10) overlapping the MF (Fig. 2 L and M), Ato was expressed in all cells in the MF; the expression persisted to the position equivalent to the most posterior row of R8 precursors (dotted lines in Fig. 2 L and M) in neighboring ato1/+ tissue. When both the wild-type and the mutant forms of Ato protein were present in ato1/+ cells, the formation of proneural clusters was normal (Fig. 2 L and M), suggesting that expansion in the Ato expression was not caused by changes in the property of Ato mutant protein.
Ato repression in the MF also depended on the da activity. In the da clones (n = 7) that overlapped the MF, Ato repression was relieved in cells between proneural clusters (Fig. 2 N and O) (21); the extent of Ato expansion was similar to that observed in ato1 mutant clones.
However, not all cells within the ato or da clones expressed Ato. In some cases, ato or da mutant cells located near the posterior or the lateral margin of these clones exhibited Ato repression (arrows in Fig. 2 L and N). Because mutant cells near the border activated MAPK (Fig. 2J, arrow), Ato repression near the margin is likely a nonautonomous effect from the neighboring ato/+ or da/+ cells (also see Discussion).
Taken together, these results indicate that the Ato repression in cells between proneural clusters requires the functions of ato and da. Failure of this repression resulted in persistent expression of Ato in all cells after the initial stripe, thus eliminating the spacing between proneural clusters. The process of lateral inhibition that restricts proneural clusters to single R8 precursors also was blocked in the ato and da clones (Fig. 2 L–O), because some components of the N signaling pathway are transcriptionally regulated by ato and da. For example, expression of E(spl)-C in the MF requires ato activity (6).
MAPK Activation Negatively Regulates Ato Expression.
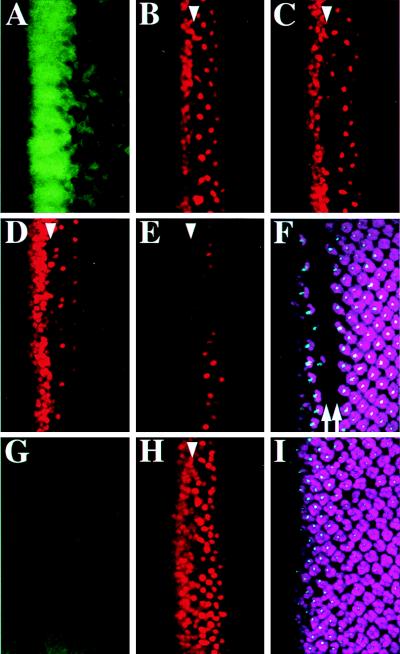
ato and da are required for MAPK activation within proneural clusters and Ato repression in cells between proneural clusters. Therefore, one possible mechanism for Ato repression may involve a nonautonomous effect exerted by the MAPK signaling in proneural clusters. We manipulated the level of MAPK activation with induced expression of a positive regulator rhomboid (rho) (22, 23) and a negative regulator argos (aos) (24, 25) of the Drosophila Egfr signaling pathway (26), and observed immediate effects on Ato expression. After heat-treating the hs-rho eye discs for 30 min, the MAPK was hyperactivated (Fig. 3A), forming a stripe-like pattern overlapping the Ato stripe and proneural clusters (data not shown). The initial effect observed was repression of Ato in the stripe region 2 hr after 1-hr heat pulse (Fig. 3B). The effect of Ato repression shifted posteriorly to the region of proneural clusters 4 hr after heat treatment (Fig. 3C), and to the region of R8 precursors 6 hr after heat treatment (Fig. 3D). Continuous heat pulses (see Materials and Methods) led to extensive loss of Ato-positive cells (Fig. 3E), failure of photoreceptor differentiation, as revealed by the lack of Boss and Elav staining (Fig. 3F), and a dorsoventral stripe of scar in the adult compound eye (not shown). As a control, the same heat treatment fail to cause any significant changes on Ato expression in the wild-type eye discs and the adult compound eyes.
Figure 3.
Down-regulation of Ato by the MAPK signaling. The hs-rho eye disc was heat-treated for 30 min (A) showing increase of MAPK activation, or for 1 hr preceded by a 2-hr rest showing Ato repression in the stripe (B), by a 4-hr rest showing repression in proneural clusters (C), or by a 6-hr rest showing repression in R8 precursors (D). (E and F) Multiple heat pulses (see Materials and Methods) result in great reduction of Ato-positive cells (E), and Boss and Elav (F) staining. The double arrows indicate the missing rows of Elav and Boss expression (F). The hs-aos disc was heat-treated for 1 hr, showing repression of dp-ERK (G) and activation of Ato (H). Prolonged heat treatment did not affect R8 specification as shown in I. (A–I) Ato is red, dp-ERK is green, Elav is purple, and Boss is blue.
In contrast, when the MAPK was inactivated by the induction of aos (Fig. 3G), numerous Ato-positive cells appeared after the MF (Fig 3H), which is consistent with the interpretation that MAPK activation down-regulates Ato expression. The appearance of these ectopic, isolated Ato-positive cells in the hs-aos discs implied that lateral inhibition was not affected through MAPK inactivation. However, even when the discs were treated with several heat pulses, induction of aos failed to increase the number of mature R8 cells (Fig. 3I). Previous results indicate that inactivation of Egfr only affects MAPK activation for a short period of time and the MAPK activity is quickly complemented later by another pathway (10). Likely, inactivation of Egfr by misexpression of aos may affect MAPK activation for a short period of time. However, our results suggest that changes in the level of MAPK activation have immediate negative effects on Ato expression.
MAPK Activation Leads to Nonautonomous Ato Repression.
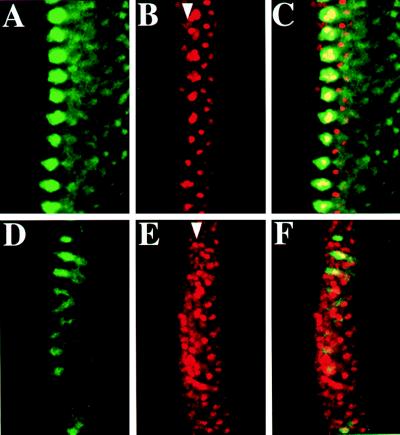
MAPK activation in proneural clusters could repress Ato expression autonomously in cells within the clusters, nonautonomously in cells outside the clusters, or both. We used scabrous-GAL4 (sca-GAL4) (17, 27) for target gene expression in the proneural clusters. After the induced expression (see Materials and Methods) of wild-type DER (9) or constitutively active Draf (28), the level of activated MAPK in the proneural clusters (Fig. 4A and data not shown) was increased and Ato expression in the stripe and proneural clusters was reduced (Fig. 4B and data not shown), indicating that MAPK activity affected Ato expression autonomously and nonautonomously. Because sca-GAL4 activates gene expression in the central and peripheral nervous system as well, induced expression of the dominant negative form of DER (DN-DER) (9) resulted in lethality at the embryonic stage except that a few survived to the larval stage. We observed the effect caused by DN-DER expression in proneural clusters at the early third-instar stage. The level of MAPK activation was reduced and many Ato-positive cells appeared in regions between the MAPK-activated clusters (Fig. 4 D and E). Taken together, these results suggest that activated MAPK represses Ato nonautonomously in cells anterior to and in cells between proneural clusters.
Figure 4.
Nonautonomous Ato repression by MAPK activation. Confocal images of a sca-GAL4;UAS-DER eye disc (A–C) showing increase of dp-ERK (A) and decrease of Ato expression in the stripe and proneural clusters (B). Each cluster of Ato-positive cells in the MF is roughly the size of the stage III clusters (Fig. 1C) in the wild-type disc. However, dp-ERK expression (A) is stronger than in Fig. 1C. (D–F) A sca-GAL4;UAS-DN-DER eye disc of an early third-instar larva. Notice the increase of Ato (E) and decrease of dp-ERK (D) expression. This figure can be compared with the wild-type disc in Fig. 2 D–F. (A–F) Ato is red and dp-ERK is green.
Hyperactivation of the MAPK Prevents Ato Proneural Cluster Formation.
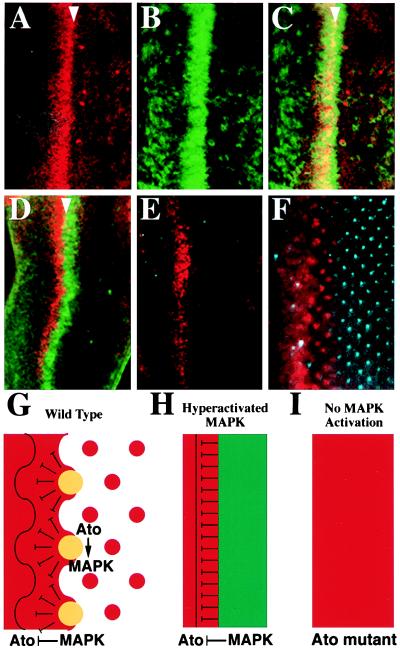
Elp is a gain-of-function allele of Egfr (18) and constitutively activates the Egfr signaling pathway. In Elp/Elp eye discs, the MAPK was hyperactivated as a dorsoventral stripe overlapping the Ato stripe (Fig. 5 A–C). Concomitantly, there were few Ato proneural clusters (Fig. 5A) posterior to the Ato/MAPK stripe. However, some R8 precursors escaped the repression and continued to form ommatidium clusters (Fig. 5A) (29), and MAPK activation was associated with these R8 precursors (Fig. 5C). In the eye discs of another mutant, Elp/+; l(3)2B10/Df(3R)p14 (see Materials and Methods), the MAPK also was highly activated in a stripe abutting the posterior margin of the Ato stripe (Fig. 5D). This stripe of MAPK expression also prevented the formation of Ato proneural clusters and R8 photoreceptors (Fig. 5E). In this mutant, the MAPK signaling induced repression in the anterior direction, resulting in failure of proneural cluster formation after the sweep of the Ato stripe.
Figure 5.
Stripe expression of activated MAPK prevents Ato cluster formation. Ato is red, dp-ERK is green, and Boss is blue. (A–C) Confocal images of ElpB1/ElpB1 eye disc showing overlap of Ato stripe (A) and dp-ERK stripe (B). Confocal image of Elp1/+; l(3)2B10/Df(3R)p14 eye disc (D) showing the anterior Ato stripe and the posterior dp-ERK stripe. (E and F) The expression pattern of Ato and the more mature R8 marker Boss in Elp1/+; l(3)2B10/Df(3R)p14 (E) and +/+ eye disc (F). Models explaining the generation of Ato clusters in wild-type (G), the failure of cluster formation in Elp/+; 2B10/Df(3R)p14 mutant (H), and expansion of Ato expression in ato1/ato1 mutant (I) eye discs. (G-I) Ato is red, dp-ERK is green, and the overlapping expression of both proteins in the clusters is yellow.
DISCUSSION
We examined the relationship between Ato expression and MAPK activation in the MF of the eye imaginal discs. Our results showed that ato and da are required for MAPK activation in proneural clusters and for Ato repression in gaps of proneural clusters. Furthermore, the Ato repression effect is likely mediated by MAPK activation in proneural clusters. These data explain how ato is required for Ato repression in cells between proneural clusters and suggest that the negative regulation of Ato is involved in patterning Ato-positive proneural clusters.
The Roles of Ato and Da in MAPK Activation.
Ato has been shown to form a heterodimer complex with Da when bound to the target DNA sequence. During eye development both ato and da are required for R8 formation (4, 21). Da protein is expressed ubiquitously, but at a high level in cells that express Ato (21). All evidence suggests that these two proteins function together in vivo. The requirement of ato and da in MAPK activation in proneural clusters suggests that a positive regulator of the MAPK signaling is transcriptionally controlled by the Ato and Da heterodimers.
Formation of chordotonal organ clusters in embryos requires the Egfr pathway (30, 31). Primary sensory organ precursor (sop) cells initially are specified by the proneural gene ato. Soon after specification, the primary sop cells induce neighboring cells to adopt the same fates through the Egfr pathway, thus forming clusters of chordotonal organs. We showed that ato also is required for MAPK activation in proneural clusters of the eye discs (Fig. 2 I and M). MAPK activation is not seen during the initial formation of proneural clusters (Fig. 1A), but during the late restriction process (Fig. 1 B and C). A time lag is required for expressing the putative positive regulators of the MAPK signaling pathway. In the border of the ato and da clones, we sometimes found that dp-ERK clusters encompasses both homozygous mutant cells and heterozygous wild-type cells (arrow in Fig. 2J), suggesting that the putative regulator activated in the wild-type tissue is secreted to the neighboring mutant cells for MAPK activation.
The Positive and Negative Regulation of ato in the MF.
During sensory organ development, the proneural genes initially are expressed in clusters of ectodermal cells (32). Proneural gene expression is quickly intensified in selected sensory organ precursor cells and diminished in the neighboring cells through lateral inhibition. The enhanced expression requires autoactivation of the proneural genes (33). In the eye discs, autoactivation of ato in proneural clusters depends on a 5′ enhancer (34). This autoactivation is prevented in cells between proneural clusters. Our results support a possible explanation to this inhibition mechanism. Because ato autoactivation requires the Ato and Da heterodimer, depletion or modification of the Ato protein would disrupt this autoactivation process. This model suggests that Ato repression occurs before autoactivation. Considering the nonautonomous effect of MAPK activation in the proneural clusters, Ato repression may occur in the more anterior stripe region. Sun et al. (34) showed that the Ato stripe expression is activated through a 3′ enhancer that confers a patterning event independent of ato autoactivation (34). It will be interesting to examine whether this patterning activity depends on the MAPK signaling.
Alternatively, Ato repression may occur at the autoactivation process. A downstream target of MAPK signaling can interfere with ato autoactivation by affecting the ato 5′ enhancer activity. This process can be achieved by modifying the transcriptional factors that bind to the 5′ enhancer or by direct binding of a transcriptional repressor to the 5′ enhancer.
The Effect of MAPK Signaling on Ato Expression.
The requirement of ato on Ato repression in the gaps between proneural clusters is likely mediated by activated MAPK in the proneural clusters. Target molecules of the MAPK signaling can transduce the signal for Ato repression in the neighboring cells. Aos, the negative regulator of Egfr and positively regulated by the Egfr pathway (26), is not likely to be the primary repressor. In the aos weak mutants, aosw11/aosw11 and aosw11/aoslΔ7, MAPK activation in proneural clusters is slightly elevated and results in slight increase in the size of the gaps between proneural clusters (data not shown), a phenotype in contrast to the role for Ato repression. Another candidate to mediate the Ato repression effect is Sca protein. Loss of sca activity in the MF interferes with the spacing between proneural clusters (35), suggesting that sca is involved in the spacing process. Gain of sca activity results in a similar phenotype of disturbing the proneural cluster spacing (36). These results leads to the suggestion that the relative level of Sca inside and outside the proneural clusters is important in regulating the spacing of proneural clusters (36). Being a secreted protein, Sca is proposed to be involved in a loop that negatively regulates ato expression for the proneural cluster formation (35).
Mutant clones of Egfr have a weak effect on the spacing of R8 precursors (11), suggesting that Egfr may contribute to MAPK activation in the MF. However, there has been no direct measurement of MAPK activation in the Egfr clones (12). MAPK activation can be mediated by two or more redundant pathways. Examining the loss-of-function mutations of the components in the MAPK signaling will provide the definitive answer.
The Generation of Evenly Spaced Proneural Clusters in the MF.
Our study suggests that Ato repression is necessary for the formation of proneural clusters. During MF progression, the Ato stripe expression is induced through a 3′ enhancer (34) which is likely activated by the Hedgehog and Decapentaplegic signaling pathways (2). Autoactivation continues ato expression in the proneural clusters in which the MAPK is activated. The activated MAPK acts nonautonomously to inhibit neighboring cells from Ato expression, including cells in the anterior stripe. Cells further away from the clusters receive the least inhibition effect and emerge as the next wave of Ato-positive clusters, forming out of phase in respect to the previous wave (Fig. 5G). In the case of MAPK hyperactivation that forms a stripe overlapping or posterior to the Ato stripe (Fig. 5 B and D), MAPK signaling exerts a nonautonomous repression effect on all cells in the Ato stripe, subsequently abolishing the proneural cluster formation (Fig. 5H). In contrast, failure of MAPK activation in ato or da mutants results in removing the repression effect, leading to Ato expression in all cells in the MF (Fig. 5I).
We speculate that at the very beginning of furrow initiation, Hedgehog and Decapentaplegic pathways activate Ato expression (37) in a small cluster of cells at the most posterior tip of the eye disc, and the MAPK subsequently is activated in the same cluster. The nonautonomous repression effect of the activated MAPK bisects the next round of Ato expression into two clusters, each in turn activating the MAPK. Each cycle of the negative loop generates one more Ato-positive cluster. Gradually the balance between MAPK activation and Ato repression is achieved and results in equally spaced Ato clusters during MF progression (Fig. 2 A–F).
Acknowledgments
We thank M. Freeman (Medical Research Council, United Kingdom), J.-C. Hsu (National Tsing Hua University, Taiwan), Y. N. Jan, (University of California, San Francisco), P. Marin and E. Bier (University of California, San Diego), H. Okano (University of Tsukuba, Japan), Y. H. Sun (Institute of Molecular Biology, Academia Sinica, Taiwan), Y. Sun (University of California, San Francisco), S. L. Zipursky (University of California, Los Angeles), and Bloomington Stock Center for antibodies and fly stocks; J.-C. Hsu, H.-M. Li (Institute of Molecular Biology, Academia Sinica, Taiwan), and Y. H. Sun for comments on the manuscript; and Y. H. Sun for discussion. This work was supported by Grant NSC-87-2311-B001-094 from the National Science Council of Taiwan and a grant from Academia Sinica, Taiwan.
ABBREVIATIONS
- MF
morphogenetic furrow
- MAPK
mitogen-activated protein kinase
- Ato
Atonal
- Da
Daughterless
- Egfr
epidermal growth factor receptor
- rho
rhomboid
- aos
argos
- N
Notch
- sca
scabrous
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Wolff T, Ready D F. In: The Development of Drosophila melanogaster. Bate M, Martinez-Arias A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 1277–1326. [Google Scholar]
- 2.Heberlein U, Moses K. Cell. 1995;81:987–990. doi: 10.1016/s0092-8674(05)80003-0. [DOI] [PubMed] [Google Scholar]
- 3.Freeman M. Development (Cambridge, UK) 1997;124:261–270. doi: 10.1242/dev.124.2.261. [DOI] [PubMed] [Google Scholar]
- 4.Jarman A P, Grell E H, Ackerman L, Jan L Y, Jan Y N. Nature (London) 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- 5.Jarman A P, Sun Y, Jan L Y, Jan Y N. Development (Cambridge, UK) 1995;121:2019–2030. doi: 10.1242/dev.121.7.2019. [DOI] [PubMed] [Google Scholar]
- 6.Dokucu M E, Zipursky S L, Cagan R L. Development (Cambridge, UK) 1996;122:4139–4147. doi: 10.1242/dev.122.12.4139. [DOI] [PubMed] [Google Scholar]
- 7.Baker N E, Yu S, Han D. Curr Biol. 1996;6:1290–1301. doi: 10.1016/s0960-9822(02)70715-x. [DOI] [PubMed] [Google Scholar]
- 8.Ligoxygakis P, Yu S-Y, Delidakis C, Baker N E. Development (Cambridge, UK) 1998;125:2893–2900. doi: 10.1242/dev.125.15.2893. [DOI] [PubMed] [Google Scholar]
- 9.Freeman M. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- 10.Kumar J P, Tio M, Hsiung F, Akopyan S, Gabay L, Seger R, Shilo B-Z, Moses K. Development (Cambridge, UK) 1998;125:3875–3885. doi: 10.1242/dev.125.19.3875. [DOI] [PubMed] [Google Scholar]
- 11.Dominguez M, Wasserman J D, Freeman M. Curr Biol. 1998;8:1039–1048. doi: 10.1016/s0960-9822(98)70441-5. [DOI] [PubMed] [Google Scholar]
- 12.Gabay L, Seger R, Shilo B-Z. Science. 1997;277:1103–1106. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- 13.Yung Y, Dolginov Y, Yao Z, Rubinfeld H, Michael D, Hanoch T, Roubini E, Lando Z, Zharhary D, Seger R. FEBS Lett. 1997;408:292–296. doi: 10.1016/s0014-5793(97)00442-0. [DOI] [PubMed] [Google Scholar]
- 14.Spencer S A, Powell P A, Miller D T, Cagan R L. Development (Cambridge, UK) 1998;125:4777–4790. doi: 10.1242/dev.125.23.4777. [DOI] [PubMed] [Google Scholar]
- 15.Xu T, Rubin G M. Development (Cambridge, UK) 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- 16.Tio M, Moses K. Development (Cambridge, UK) 1997;124:343–351. doi: 10.1242/dev.124.2.343. [DOI] [PubMed] [Google Scholar]
- 17.Nakao K, Campos-Ortega J A. Neuron. 1996;16:275–286. doi: 10.1016/s0896-6273(00)80046-x. [DOI] [PubMed] [Google Scholar]
- 18.Baker N E, Rubin G M. Nature (London) 1989;340:150–153. doi: 10.1038/340150a0. [DOI] [PubMed] [Google Scholar]
- 19.Chang H C, Rubin G M. Genes Dev. 1997;11:1132–1139. doi: 10.1101/gad.11.9.1132. [DOI] [PubMed] [Google Scholar]
- 20.Jarman A P, Grau Y, Jan L Y, Jan Y N. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- 21.Brown N L, Paddock S W, Sattler C A, Cronmiller C, Thomas B J, Carroll S B. Dev Biol. 1996;179:65–78. doi: 10.1006/dbio.1996.0241. [DOI] [PubMed] [Google Scholar]
- 22.Sturtevant M A, Roark M, Bier E. Genes Dev. 1993;7:961–973. doi: 10.1101/gad.7.6.961. [DOI] [PubMed] [Google Scholar]
- 23.Bier E, Jan L Y, Jan Y N. Genes Dev. 1990;4:190–203. doi: 10.1101/gad.4.2.190. [DOI] [PubMed] [Google Scholar]
- 24.Schweitzer R, Howes R, Smith R, Shilo B-Z, Freeman M. Nature (London) 1995;376:699–702. doi: 10.1038/376699a0. [DOI] [PubMed] [Google Scholar]
- 25.Sawamoto K, Okano H, Kobayakawa Y, Hayashi S, Mikoshiba K, Tanimura T. Dev Biol. 1994;164:267–276. doi: 10.1006/dbio.1994.1197. [DOI] [PubMed] [Google Scholar]
- 26.Schweitzer R, Shilo B-Z. Trend Genet. 1997;13:191–196. doi: 10.1016/s0168-9525(97)01091-3. [DOI] [PubMed] [Google Scholar]
- 27.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 28.Brand A H, Perrimon N. Genes Dev. 1994;8:629–639. doi: 10.1101/gad.8.5.629. [DOI] [PubMed] [Google Scholar]
- 29.Baker N, Rubin G. Dev Biol. 1992;150:381–396. doi: 10.1016/0012-1606(92)90250-k. [DOI] [PubMed] [Google Scholar]
- 30.Lage P Z, Jan Y N, Jarman A P. Curr Biol. 1997;7:166–175. doi: 10.1016/s0960-9822(97)70087-3. [DOI] [PubMed] [Google Scholar]
- 31.Okabe M, Sawamoto K, Okano H. Dev Biol. 1996;175:37–49. doi: 10.1006/dbio.1996.0093. [DOI] [PubMed] [Google Scholar]
- 32.Jan Y N, Jan L Y. In: The Development of Drosophila melanogaster. Bate M, Martinez-Arias A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 1207–1244. [Google Scholar]
- 33.Culi J, Modolell J. Genes Dev. 1998;12:2036–2047. doi: 10.1101/gad.12.13.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Y, Jan L Y, Jan Y N. Development (Cambridge, UK) 1998;125:3731–3740. doi: 10.1242/dev.125.18.3731. [DOI] [PubMed] [Google Scholar]
- 35.Lee E-C, Hu X, Yu S-Y, Baker N E. Mol Cell Biol. 1996;16:1179–1188. doi: 10.1128/mcb.16.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellis M C, Weber U, Wierdorff V, Mlodzik M. Development (Cambridge, UK) 1994;120:1959–1969. doi: 10.1242/dev.120.7.1959. [DOI] [PubMed] [Google Scholar]
- 37.Borod E R, Heberlein U. Dev Biol. 1998;197:187–197. doi: 10.1006/dbio.1998.8888. [DOI] [PubMed] [Google Scholar]