Abstract
Genes encoding homeodomain-containing proteins potentially involved in endocrine pancreas development were isolated by combined in silico and nested-PCR approaches. One such transcription factor, Arx, exhibits Ngn3-dependent expression throughout endocrine pancreas development in α, β-precursor, and δ cells. We have used gene targeting in mouse embryonic stem cells to generate Arx loss-of-function mice. Arx-deficient animals are born at the expected Mendelian frequency, but develop early-onset hypoglycemia, dehydration, and weakness, and die 2 d after birth. Immunohistological analysis of pancreas from Arx mutants reveals an early-onset loss of mature endocrine α cells with a concomitant increase in β-and δ-cell numbers, whereas islet morphology remains intact. Our study indicates a requirement of Arx for α-cell fate acquisition and a repressive action on β-and δ-cell destiny, which is exactly the opposite of the action of Pax4 in endocrine commitment. Using multiplex reverse transcriptase PCR (RT-PCR), we demonstrate an accumulation of Pax4 and Arx transcripts in Arx and Pax4 mutant mice, respectively. We propose that the antagonistic functions of Arx and Pax4 for proper islet cell specification are related to the pancreatic levels of the respective transcripts.
Keywords: Endocrine pancreas development, Arx, Pax4, mouse, hypoglycemia, fate specification
The pancreas plays a major role in nutritional homeostasis through synthesis and secretion of hormones and enzymes. This organ includes endocrine, acinar, and ductal cell types, all deriving from a common set of epithelial cells that originate in the early gut endoderm. The exocrine pancreas develops to form acinar cells and a highly branched ductal epithelium, whereas endocrine cells aggregate into islets of Langerhans. The latter are specialized micro-organs composed of four different cell types, α, β, δ, and PP cells, which produce the hormones glucagon, insulin, somatostatin, and PP (pancreatic polypeptide), respectively. Insulin and glucagon function coordinately to control glucose homeostasis, whereas somatostatin and PP regulate the secretion of other hormones and of exocrine enzymes (Adrian et al. 1978; Roncoroni et al. 1983; Csaba and Dournaud 2001). The first morphological signs of pancreas development appear at embryonic day 9.5 (E9.5) as dorsal and ventral protrusions of the primitive gut epithelium that will later fuse to form the definitive pancreas (for review, see Edlund 2002). Concomitantly, the glucagon-producing α cells are the first endocrine cells detected (Herrera et al. 1991; Teitelman et al. 1993; Upchurch et al. 1994). One day later (E10.5), a few insulin-producing cells appear, often coexpressing glucagon (Teitelman et al. 1993). At E14, the secondary transition, as defined by Pictet and Rutter (Pictet et al. 1972), leads to the generation of numerous fully differentiated β cells. Endocrine cells start to become organized in small aggregates, and within the next 24 h, the first somatostatin-producing δ cells emerge (Pictet et al. 1972). Finally, at E18, shortly before birth, PP-producing cells differentiate, whereas endocrine cells begin to form well-organized islets of Langerhans.
Until recently, the mechanisms underlying the generation of these different endocrine cell types was not well understood, but lineage studies and the generation of mice deficient for a number of pancreatic transcription factors have provided major insights into the control of islet development. One such transcription factor is Neurogenin3 (Ngn3), a basic helix-loop-helix (bHLH) protein-encoding gene member of the neurogenin/neuroD family, which is first detected in the pancreatic epithelium at E9 (Sommer et al. 1996; Apelqvist et al. 1999; Jensen et al. 2000; Schwitzgebel et al. 2000). Ngn3 loss-of-function mutant mice fail to develop endocrine cells (Gradwohl et al. 2000), whereas ectopic expression of Ngn3 under the control of the pdx1 promoter leads to the differentiation of the entire pancreas into endocrine cells (Apelqvist et al. 1999; Schwitzgebel et al. 2000). The proendocrine function for Ngn3 suggested by these data was further demonstrated by lineage studies using the Cre/LoxP system (Gu et al. 2002). Hence, whereas Ngn3 activation in pancreatic progenitor cells promotes an endocrine commitment, the specification of the four different islet subtypes appears to be controlled by other genes. A number of homeodomain-containing transcription factors have been characterized for their implication in each of the pathways leading to differentiated endocrine cell subtypes using loss-of-function mutant mice. Two groups can be distinguished on the basis of their colocalization with Ngn3; the early factors found in Ngn3-expressing cells (Nkx2.2, Nkx6.1, Pdx1, and Pax4) and the late factors (Brn4, Isl1, and Pax6) present in mature endocrine cells that are devoid of Ngn3 expression (Jensen et al. 2000; Schwitzgebel et al. 2000).
The paired-box-containing factor Pax4 is selectively expressed in the developing islets of Langerhans and is later restricted to mature endocrine cells (Smith et al. 1999; Dohrmann et al. 2000). Pax4-deficient mice develop severe diabetes at birth as a result of altered islet cell development characterized by a deficiency of β and δ cells, together with an increase of the α-cell population (Sosa-Pineda et al. 1997). Similarly, the NK-homeodomain protein-encoding genes, Nkx6.1 and Nkx2.2, are required for endocrine specification. Mice homozygous for a targeted deletion of either gene have a reduced number of β cells. In addition, a decrease in the number of α and PP cells is found in Nkx2.2 mutant mice. Nkx6.1 expression is abolished in these animals, suggesting that Nkx2.2 acts upstream Nkx6.1 (Sussel et al. 1998; Sander et al. 2000).
In contrast, the late factors Brain-4, Isl1, and Pax6 act on the development and the maintenance of differentiated islet cells. Targeted disruption of the paired-box-containing gene Pax6 or of the LIM homeodomain protein-encoding gene Isl1 leads to a drastic reduction of all endocrine subtypes (Ahlgren et al. 1997; Sander et al. 1997; St-Onge et al. 1997). Brain-4 (Brn-4), a POU-homeodomain-containing protein, is expressed in the pancreatic anlage of the mouse foregut at E10 in glucagon-producing cells and seems to transactivate glucagon gene expression (Hussain et al. 1997). However, loss-of-function mutant mice do not exhibit any defect in α-cell formation (R.S. Heller and P. Serup, in prep.), although ectopic expression of Brn-4 targeted to β cells leads to the coexpression of insulin and glucagon in this cell type (Hussain et al. 2002). These data suggest a role of Brn4 in the advanced islet to promote hormone expression. Similarly, Pdx1, a homeodomain-containing protein involved in pancreatic bud formation (Jonsson et al. 1994; Guz et al. 1995; Offield et al. 1996), plays an additional role in maintaining the β-cell phenotype. Using the Cre/Lox system, Ahlgren at al. (1998) demonstrated that β-cell-targeted deletion of Pdx1 leads to the loss of this cell type. Taken together, these data suggest that islet progenitors are progressively committed to an islet fate, specified to a particular endocrine cell subtype and finally maintained in this commitment.
In this study, we describe the functional characterization of Arx in the endocrine pancreas. The homeobox-containing gene, Arx, which is localized on the X chromosome (Blair et al. 2002), was first reported to be expressed in the mouse central nervous system (CNS) by Miura et al. (1997). Recently, Arx dysfunction has been associated with diverse forms of X-linked mental retardation and epilepsy in humans (discussed in Hirose and Mitsudome 2003). Interestingly, by means of in situ hybridization, we observed a previously unnoticed expression in the pancreas, at the onset of pancreatic bud evagination, which later becomes restricted to the islets of Langerhans. To investigate the role of this gene in pancreas development, we generated loss-of-function mutant mice. Arx-deficient animals die 2 d after birth, after having developed severe hypoglycemia. Immunohistochemical analysis of Arx mutant pancreas revealed an absence of α cells and an increased number of β and δ cells. However, the total number of endocrine cells is unaffected, suggesting a role for Arx in promoting α-cell fate. This is opposite to the phenotype observed in Pax4-deficient mice, and we demonstrate that a mutual inhibition of Arx and Pax4 during early stages of endocrine development is required to generate normal islets of Langerhans.
Results
Arx is expressed in endocrine pancreas progenitors
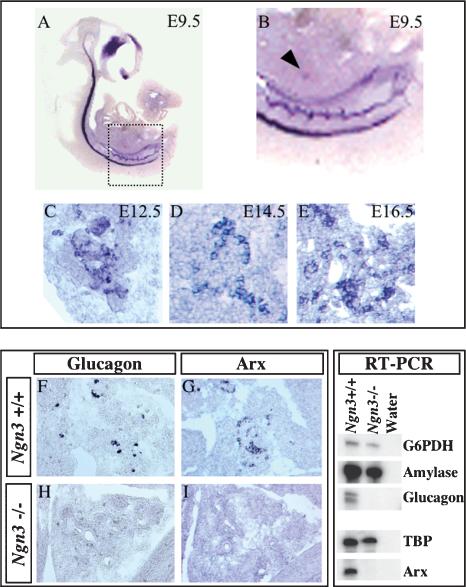
Aiming to identify members of the homeobox-containing gene family with a potential role in islet of Langerhans genesis, we undertook a search combining in silico and nested-PCR, using a mouse cDNA pancreas library as a template (P. Collombat, A. Mansouri, and P. Gruss, in prep.). Several genes with previously unnoticed expression in this organ were isolated, including Arx, a gene encoding a homeodomain-containing protein. Arx expression was described previously in the mouse central nervous system, starting from E9.5 (Miura et al. 1997). Importantly, at this particular stage, we detected expression of Arx in the pancreas anlage (Fig. 1A, arrowhead in B). Arx-expressing cells were found during the genesis of this organ, in the proliferating epithelium (Fig. 1C), the differentiating pancreatic precursors (Fig. 1D), and subsequently, in the forming islets of Langerhans (Fig. 1E).
Figure 1.
Arx is expressed in normal mouse pancreas, but not in that of Ngn3-deficient animals. (A,B) Whole-mount in situ hybridization of an E9.5 mouse embryo. Sagittal section (A) and enlarged view of the pancreatic area (B), revealing Arx transcripts in the pancreas anlage (arrowhead in B). (C-E) When analyzed by in situ hybridization, pancreas sections display widespread Arx expression at E12.5 (C), E14.5 (D), and E16.5 (E). (F-I) Detection of glucagon and Arx expression by in situ hybridization in E14.5 wild-type (F,G) and Ngn3 homozygous mutant animals (H,I). Ngn3 mutant pancreas lacks Arx expression. (RT-PCR) RT-PCR analysis performed on mRNA from E14.5 wild-type or Ngn3 mutant animals (left and middle lanes, respectively). RT-PCR controls were performed with primers designed from the glucose-6-phosphate dehydrogenase (G6PDH) and the TATA-box-binding protein (TBP) reference genes; positive controls with primers from the amylase gene and negative controls were assayed with water instead of DNA template (right lane). RT-PCR results confirm a lack of glucagon and Arx expression in Ngn3 mutant mice.
To determine whether Arx played a role in endocrine cell development, Arx expression was studied in the pancreas of mice mutant for the pro-endocrine Ngn3 gene. At E14.5, just before the peak of endocrine cell production, examination of the pancreas of Ngn3 mutant embryos revealed a complete loss of Arx expression (Fig. 1G,I). This result was further confirmed by RT-PCR performed in wild-type and Ngn3 mutant pancreas (Fig. 1, RT-PCR panel). Taken together, our data indicate new expression domains of Arx in the developing pancreas. Notably, the lack of Arx transcripts in the pancreas of Ngn3-deficient mice suggests a potential role of Arx downstream of Ngn3 in islet of Langerhans developmental processes.
Generation of Arx loss-of-function mutant mice
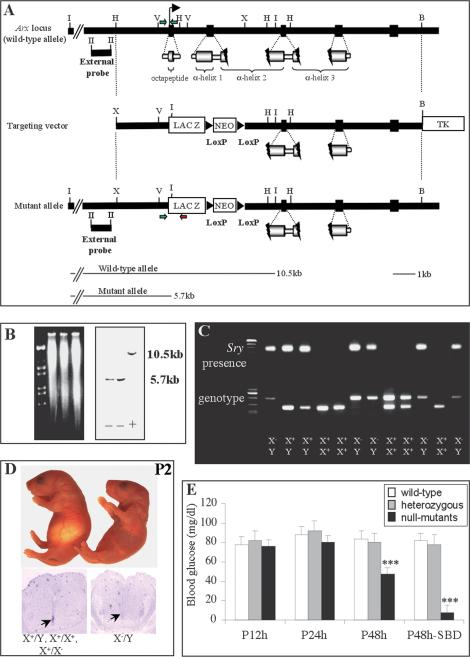
To gain insight into the potential function of Arx during pancreas development, we generated loss-of-function mutant mice by homologous recombination in embryonic stem (ES) cells. A targeting vector was designed to delete the two first exons corresponding to the N-terminal 360 amino acids (Fig. 2A). These two exons, which encode the octapeptide (Eh1 domain), α-helix 1 and approximately half α-helix 2 of the homeodomain, were replaced by the β-galactosidase gene and the Neomycin resistance gene flanked by LoxP sites. Of the 202 stably transfected ES clones obtained after antibiotic selection, two were homologous recombinants as identified by Southern analysis (Fig. 2B). Because the Arx gene is localized on the X chromosome (Blair et al. 2002), and ES cells were derived from male blastocysts, we therefore detected either the wild-type (designated as X+) or the mutated (designated as X-) allele. Chimeras obtained from the targeted ES cell clones were bred to NMRI mice with successful germ-line transmission. Heterozygous X+/X- females of the mixed background (129Sv × NMRI) were crossed with wild-type males (X+/Y), and both sex and genotype of the offspring were determined by PCR (Fig. 2C). In situ hybridization demonstrated that our targeting approach deleted the Arx gene, as its expression was undetectable using a full-length cDNA-derived probe (Fig. 2D, bottom). The following allelic frequencies were obtained: 52.3% wild-type (X+/X+ females and X+/Y males), 24.2% heterozygous (X+/X- females), and 23.5% null (X-/Y males; n = 189); such a Mendelian ratio indicates that the deletion of the Arx gene did not result in embryonic lethality. No obvious difference was observed between heterozygous and wild-type animals. We therefore used mice of both genotypes as controls throughout this study.
Figure 2.
Disruption of the Arx gene by homologous recombination and genotype analysis of heterozygous intercrosses. (A) Maps of the wild-type Arx locus, targeting vector, and disrupted Arx allele. Exons are indicated as black rectangles with encoded protein motifs underneath. The 1.3-kb 5′ external probe used for Southern blotting is shown at left. (B) BamHI, (H) HindIII, (I) EcoRI, (II) HincII, (V) EcoRV, (X) XhoI, (TK) Herpes simplex virus thymidine kinase gene. (B) Southern analysis of EcoRI-digested DNA from three male ES clones, Arx being located on the X chromosome, either the 10.5-kb wild-type allele (+) or the 5.7-kb mutated allele (-) is detected. (C) Genotyping and sex characterization by PCR of a litter from an intercross of heterozygous mice. (Top gel) Sex determination using Sry gene-derived primers producing a 350-bp band in the case of male genomic DNA. (Bottom gel) Three genotyping primers (represented by green and red arrows in A) were used to generate the wild-type (258-bp) and mutant (470-bp) allele bands. (D) Photograph of 2-day-old wild-type (left) and Arx mutants (right) with the corresponding transverse sections of neural tubes assayed by in situ hybridization with an Arx-cDNA-derived-probe (bottom). Arx-deficient animals are smaller and dehydrated and die at this stage. The loss of Arx expression in mutant mice demonstrates that Arx is inactivated. (E) Blood glucose analysis in the offspring of F1 hybrids 12 h (P12h), 24 h (P24h), and 48 h (P48h) after birth indicates that Arx-disrupted animals exhibit severe hypoglycemia that plunges shortly before death (p48h-SBD). Values are means ± S.E. of the mean (S.E.M.) and are representative of at least 20 animals. Differences between glucose levels were statistically significant by Student's t-test (***) at P48h between wild-type/heterozygous and mutant animals (p < 0.001).
Arx-deficient mice exhibit severe hypoglycemia prior to death
Arx loss-of-function mutant mice have been described previously (Kitamura et al. 2002). They are born at E19.5 and die within 12 h. Compared with wild-type animals, they have a small brain, reduced olfactory bulbs, and altered testes. However, the precise cause of death was not ascertained.
The Arx-deficient mice generated in the present study (X-/Y males) are indistinguishable from wild-type or heterozygous littermates at birth. Phenotypic discrepancies appear within the first day postpartum; despite normal feeding, as evidenced by the presence of milk in the stomach, Arx mutants display retarded growth, are dehydrated, and die at postnatal day 2 (P2; Fig. 2D, top). The different phenotypic alterations exhibited by the Arx mutant mice generated by Kitamura et al. (2002), such as early birth and shortened life-span, most probably originate in the different targeting strategies; thus, Kitamura et al. (2002) inserted a STOP-IRES-LacZ-Neomycin resistance cassette downstream of the second exon of the Arx gene. Consequently, it cannot be excluded that a gain-of-function mutation was generated. In contrast, our knockout construct was generated by specifically deleting the two first exons. We further demonstrate the absence of full-length Arx transcripts by in situ hybridization and RT-PCR analysis (Fig. 2D, bottom; data not shown).
To determine whether the observed lethality was related to anomalous endocrine pancreatic function, blood glucose levels were measured. Twelve hours after birth, glycemia appeared normal in all littermates (Fig. 2E). The first differences were observed 1 d postpartum; whereas wild-type and heterozygous animals displayed a normal blood glucose level, that of the mutants was slightly decreased. However, 2 d after birth, the mice lacking a functional Arx gene exhibited a severe hypoglycemia (46.72 ± 14 mg/dl, n = 36), which dropped sharply shortly before death (<10 mg/dl, n = 26), in contrast to age-matched wild-type (85.67 ± 12 mg/dl, n = 26) and heterozygous (80.72 ± 11 mg/dl, n = 32) animals.
Arx mutant mice display a loss of mature α cells concomitant with an increase in β- and δ-cell numbers
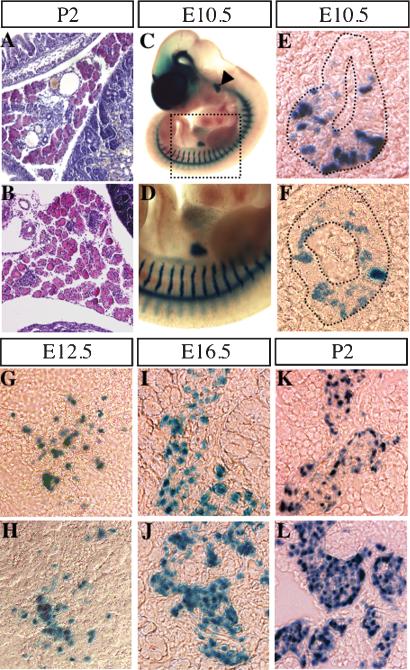
To decipher the role of Arx in endocrine pancreatic differentiation, histological analysis of pancreas from 2-day-old control and Arx mutant mice was performed. The general morphology of the pancreas and the islets of Langerhans appeared normal (Fig. 3A,B). Similarly, a kinetic analysis in islets, using X-Gal staining, recapitulated the previously observed Arx expression pattern, both in heterozygous and mutant animals (Miura et al. 1997; data not shown). More specifically, X-Gal-stained cells were detected in the pancreatic epithelium as early as E10.5 (Fig. 3C-F). An expansion of pancreatic β-galactosidase-labeled cells was seen between this stage and E16.5, together with endocrine cell development (Fig. 3G,J). The first discrepancies between heterozygous and mutant Arx mice arise at this stage and persist after birth with an increase in the number of X-Gal-positive cells in mutant animals (Fig. 3I-L).
Figure 3.
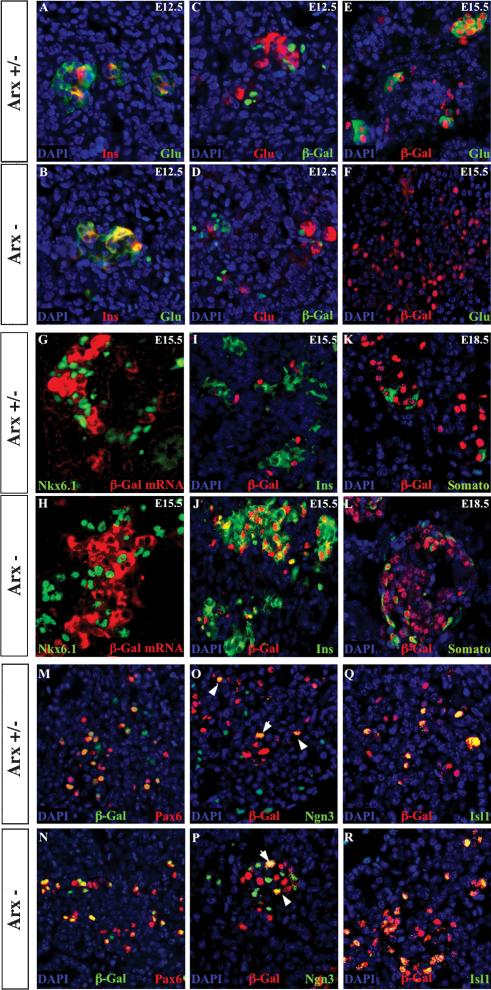
Arx expression assessed by LacZ staining demonstrates an increase in the number of β-galactosidase-producing cells within the islet of Langerhans in Arx-deficient mice. (A,B) Hematoxylin-eosin staining of 2-day-old pancreas sections reveals no morphological discrepancy between heterozygous (A) and mutant (B) Arx mice. (C,D) Whole-mount LacZ staining of Arx-deficient embryos (C) and enlarged view (D) reveals β-galactosidase expression in the pancreas as early as E10.5, as well as in additional hitherto unmentioned domains. Pancreas sections of heterozygous (E,G,I,K) and mutant (F,H,J,L) Arx animals were examined for β-galactosidase activity by X-Gal staining at different developmental stages as follows: E10,5 (E,F, outlined), E12.5 (G,H), E16.5 (I,J), and P2 (K,L). β-galactosidase-positive cells present a similar distribution at early embryonic stages in wild-type and mutant pancreas. After E16.5, the content of β-galactosidase-producing cells is increased in mutant animals as compared with littermates. Each picture is representative of at least eight animals from different litters.
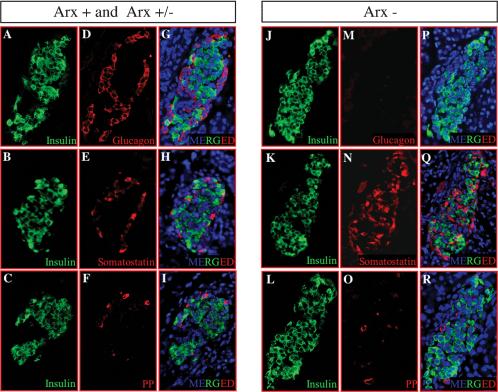
Islets were further subjected to a molecular phenotype analysis using immunohistochemistry to detect the presence of endocrine cell-subtypes producing insulin, glucagon, somatostatin, or PP. Strikingly, P2 Arx-deficient animals were completely lacking glucagon-producing α cells in the islets of Langerhans (Fig. 4D,M). We were not able to detect any glucagon-producing cells, even using higher antibody concentrations or different glucagon antibodies (data not shown). Concomitantly, there was a dramatic increase in the somatostatin-producing cell population of Arx mutant animals (Fig. 4E,N). Colocalization experiments with β cells allowed us to clearly demonstrate that δ cells also exhibited an abnormal distribution within the islet of Langerhans. In normal pancreas, at approximately E18, endocrine cell aggregates begin to develop the architecture of mature islets (Slack 1995), characterized by a core of β cells surrounded by a mantle of α, δ, and PP cells (Fig. 4G-I). In Arx mutants, δ cells were no longer localized to the mantle zone, but could be seen scattered in the core of the islet (Fig. 4E,H,N,Q). In addition, we observed a slight increase of insulin-producing β-cell number within the islets of Langerhans (Fig. 4A-C,J-L), whereas the number of PP-producing cells appeared normal (Fig. 4F,O). To confirm and quantify these differences, we serially sectioned whole P2-pancreas and determined the number of insulin-, glucagon-, somatostatin-, and PP-positive cells in every tenth section. The data from this analysis confirm the complete loss of glucagon-producing cells in mutant mice, whereas the mean number of somatostatin- and insulin-expressing cells per section was increased, 2-fold and 1.3-fold, respectively, in Arx mutants as compared with their heterozygous or wild-type littermates (Table 1). The average PP-cell number per section was not modified significantly. Collectively, our data indicate that Arx deficiency does not alter the general morphology of the pancreas and suggest that Arx function is specifically required for the proper apportionment of α-, β-, and δ-cell numbers, as well as appropriate location of the latter.
Figure 4.
Increased δ- and β-cell populations at the expense of α cells in Arx-deficient mice. Single sections of 2-day-old mice were examined for the presence of pancreatic hormone in control (A-I) and Arx mutant (J-R) animals by coimmunofluorescence. (A-C,J-L) Insulin-expressing cells were localized in well-defined islets using 488-ALEXA secondary antibody. Codetection experiments were performed with 594-ALEXA secondary antibody to visualize either glucagon-producing cells (D,M), somatostatin-producing cells (E,N), or PP-producing cells (F,O), and sections were counter-stained with DAPI (G-I,P-R, in blue). (G-I,P-R) A picture combining codetection of β cells with other endocrine cells and counter-staining is provided in each case. Note the slight increase of the insulin-producing cell population (cf. A-C and J-L), the loss of glucagon-secreting cells (cf. D and M), and the increase in the number of somatostatin-expressing cells (cf. E and N) in Arx mutant animals. Arx deletion also leads to a modification of the spatial distribution of δ cells with an additional localization within the islet core (cf. E,H and N,Q). Each picture is representative of 8-20 animals from different litters.
Table 1.
Quantification or pancreatic endocrine cell content alterations in Arx-deficient mice
| Endocrine cell distribution in Arx mutant mice
|
|||
|---|---|---|---|
| Hormone-producing cells | Wild-type, Arx heterozygotes | Arx hemizygotes | Percent difference |
| Insulin | 307 ± 35 (61%) | 404 ± 23 (74%) | +31 |
| Glucagon | 143 ± 26 (28%) | 0 | — |
| Somatostatin | 44 ± 14 (9%) | 135 ± 28 (25%) | +207 |
| Pancreatic polypeptide | 9 ± 4 (2%) | 8 ± 3 (1%) | ∼0 |
| Total cells | 503 ± 79 | 547 ± 54 | ∼0 |
Endocrine cells were counted from immunostained sections of 2-day-old pancreas estimated to be the same size. Data are shown as the mean number ± S.E. of the mean (S.E.M.) of hormone-positive cells per section (every tenth section) from at least three independent pancreata. Percentages in brackets indicate the contribution of a particular endocrine cell-subtype to the total islet cell population. Hemizygous Arx pancreas lacks α cells, whereas β- and δ-cell content are increased 1.31- and 2.07-fold, respectively. Importantly, the total endocrine cell number is unchanged following Arx distribution
Arx is differentially required for α-, β-, and δ-cell specification
To determine when the phenotypic alterations observed in Arx-deficient islets occur, we examined the pancreas of control and mutant embryos at earlier stages of development for the presence of endocrine hormones and β-galactosidase. At E10.5, the number of glucagon-expressing cells appeared normal (data not shown). Insulin-expressing cells were observed in mutant pancreas at this stage, but we were unable to determine whether their number was altered because very few cells are present at this stage, even in wild-type mice. However, at E12.5, when both insulin- and glucagon-expressing cells are readily detectable in the normal pancreas, glucagon-producing cells, often coexpressing the insulin hormone, were detected in Arx mutant pancreas in normal proportions (Fig. 5A,B). Importantly, none of these hormone-secreting cells coexpressed the β-galactosidase enzyme in the islets of Langerhans of both genotypes (Fig. 5C,D; data not shown). These results provide evidence that Arx is not implicated in the generation of early insulin/glucagon-producing cells.
Figure 5.
Alteration of mature endocrine cell-subtype specification following Arx inactivation. (A-D) Staining of E12.5 heterozygous (A,C) and mutant (B,D) Arx pancreata for insulin (A,B), glucagon (A-D), and β-galactosidase (C,D) proteins, counter-stained with DAPI (blue). β-galactosidase is not expressed in the early hormone-producing cells. There is no change in the number of insulin-expressing cells (often producing glucagon) following Arx deletion. (E-L) Sections of pancreas from E15.5 (E-J) and E18.5 (K,L) stained with anti-β-galactosidase (E,F,I-L), anti-glucagon (E,F), anti-Nkx6.1 (G,H), anti-DIG (G,H, shown in red), anti-insulin (I,J), or anti-somatostatin (K,L) antisera and counter-stained with DAPI (blue) in heterozygous (E,G,I,K) or mutant (F,H,J,L) animals except in G and H. (E,F) Glucagon-secreting cells coexpress β-galactosidase and are lacking in mutant pancreas. (G,H) β cells (stained with Nkx6.1 for technical reasons) do not express β-galactosidase in E15.5 mutant and control pancreas. (I,J) The β-cell population is increased in mutant islets. A residual presence of β-galactosidase protein is observed. (K,L) β-galactosidase staining is detected in δ cells, which are overrepresented in E18.5 Arx-deficient mice. (M-R) Endocrine differentiation appears normal following Arx inactivation. Sections of heterozygous (M,O,Q) and mutant (N,P,R) pancreas were stained with anti-Pax6 (M,N), anti-Ngn3 (O,P), or anti-Isl1 (Q,R) antisera together with anti-β-galactosidase antisera and counter-stained with DAPI (blue). (M,N) All β-galactosidase-labeled cells coexpress Pax6. Pax6-marked cells are normally represented in both genotypes. (O,P) Arrows show a few Arx- and Ngn3-labeled cells. Arx deletion does not alter the numbers of Ngn3-stained cells. (Q,R) Isl1-marked cells are exclusively found among the β-galactosidase-labeled population, but numerous cells are β-galactosidase+/Isl1-, suggesting a function of Arx in nonmature cells. Similar numbers of Isl1+ cells are observed in control and Arx mutant pancreas. Each picture is representative of at least three animals from different litters.
In contrast, when assayed at E15.5, islets of Langerhans in Arx-deficient mice were found to be completely lacking glucagon-expressing cells (Fig. 5E,F). At the same time, an increase of the insulin-producing cell population was clearly apparent (Fig. 5G-J). β-galactosidase labeling was detected in all α cells in heterozygous pancreas using protein and mRNA detection (Fig. 5E; data not shown). However, in both genotypes, β cells did not possess detectable β-galactosidase mRNA (Fig. 5G,H). Interestingly, when sections of pancreas of control and mutant mice were stained with antibodies directed against β-galactosidase and insulin proteins, we observed residual β-galactosidase proteins in insulin-producing cells of Arx-deficient embryos (Fig. 5I,J). Because the stability of the β-galactosidase protein allows the latter to be used as a short-term lineage tracer, our results suggest that β-galactosidase-expressing β cells observed in mutant animals derive from Arx-depleted cells. δ cells were also positive for β-galactosidase protein and mRNA (Fig. 5K,L; data not shown). As early as E18.5, Arx-depleted animals had developed more somatostatin/β-galactosidase-labeled cells than control mice, suggesting that Arx-deficient cells also adopt a δ-cell fate.
To further elucidate the molecular nature of the defects observed following Arx deletion, an extensive immunohistochemical analysis of the transcription factors involved in endocrine determination was performed. Sections of E14.5 pancreas were first incubated with antisera against β-galactosidase together with antisera against the Pax6, Ngn3, or Isl1 proteins (Ahlgren et al. 1997; Sander et al. 1997; Gradwohl et al. 2000). In control and Arx mutant animals, β-galactosidase staining was detected in Pax6-postive cells (Fig. 5M,N), Ngn3-labeled cells (Fig. 5O,P, arrowheads), and the vast majority of Isl1-stained cells (Fig. 5Q,R). Interestingly, the total numbers of Pax6-, Ngn3-, and Isl1-marked cells were not modified following Arx deletion. Similarly, examination of the expression of various additional transcription factors shown previously to play a role in endocrine cell specification, such as Nkx2.2, Nkx6.1, Pdx1, and HB9 (Harrison et al. 1994; Sussel et al. 1998; Li et al. 1999; Sander et al. 2000), demonstrated that they were all present and appropriately localized in heterozygous and mutant Arx animals.
Taken together, our results demonstrate the presence of two distinct populations of insulin- and glucagon-labeled cells during pancreas development. Arx does not act on the generation of the early hormone-producing cells. Instead, it is required for the specification of mature endocrine cells, as evidenced by the increased population of β and δ cells and the loss of α cells in Arx-deficient mice. In addition, similar numbers of Ngn3-stained endocrine precursors and Isl1-labeled post-mitotic endocrine cells were detected in control and mutant mice. Importantly, no alteration in the total islet cell number was detectable after birth. We therefore conclude that the lack of a functional Arx allele causes α-cell progenitors to adopt an alternative β- or δ-cell fate.
Arx and Pax4 are up-regulated in Pax4 and Arx mutant mice, respectively
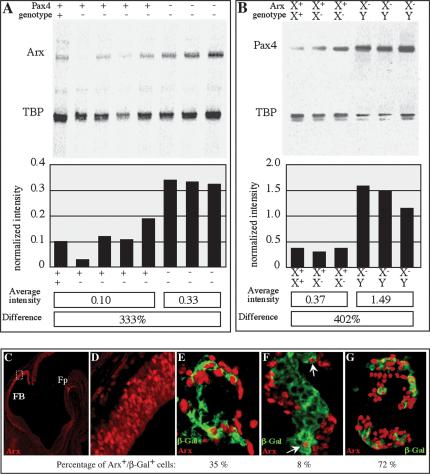
Strikingly, the deduced instructive function of Arx in specifying α-cell-subtype fate at the expense of β- and δ-cell destiny is the reverse of that attributed to the paired-box-containing factor Pax4. Mice homozygous for a mutant allele of Pax4 also exhibit an altered pancreatic endocrine cell-subtype ratio, characterized by an increased α-cell population and drastically reduced β- and δ-cell numbers (Sosa-Pineda et al. 1997). Such opposing phenotypic alterations are strongly suggestive of antagonistic functions of Pax4 and Arx during endocrine specification. To shed light on potential cross-regulatory functions of Pax4 and Arx in this phenomenon, we assayed pancreatic Pax4 and Arx mRNA content by multiplex RT-PCR in Arx and Pax4-deficient mice, respectively. The pancreas of Pax4 mutant animals contained 3.3 times more Arx mRNA than the control at E14.5 (Fig. 6A), whereas the content of Pax4 transcripts was fourfold higher in the pancreas of Arx-depleted mice (Fig. 6B). Hence, our data clearly demonstrate that Arx inactivation results in an accumulation of Pax4 transcripts associated with an increased number of β and δ cells. Conversely, Pax4 inactivation is accompanied by a rise in Arx mRNA levels, leading to an over-representation of the α cells in mature islets. We conclude that Arx and Pax4 have opposing actions on the specification of endocrine subtype destiny through an inhibitory cross-regulatory circuit that controls the transcriptional state of these two genes.
Figure 6.
Increase of Arx and Pax4 transcript levels in the pancreas of Pax4- and Arx-deficient mice, respectively. Multiplex amplification of pancreatic mRNA from Pax-4 (A) and Arx (B) mutant mice at E14.5. The genotypes of control and mutant mice are indicated above the gel scans. Measurements of the intensity of radioactive Arx and Pax4 PCR products, after normalization with the coamplified TATA-box-binding protein (TBP) amplicon, are represented as graphs. Below are indicated the average normalized intensities in control and mutant mice as well as their relative difference; the Arx transcript content is 3.3-fold higher in E14.5 Pax4 mutant pancreas compared with control animals. Arx-deficient pancreas contains four times more Pax4 mRNA at the same embryonic stage. (C,D) An anti-Arx antibody recapitulates Arx expression pattern at E11.5 (sagittal section in C and enlarged picture in D) in the forebrain (FB) and the floor plate of the diencephalon (Fp). (E-G) Analysis of Arx (red) and β-galactosidase (green) expression in Pax4 heterozygous (E,F) and homozygous (G) pancreas. The average percentage of Arx+/β-galactosidase+ cells from three serially sectioned pancreata is indicated below each picture. (E) In Pax4 heterozygous pancreas, Arx is found colocalized with the β-galactosidase protein at E13.5 in 35% of endocrine cells. (F) At E14.5, only 8% of islet cells are Arx+/β-galactosidase+. (G) At the same stage, in Pax4 homozygous pancreas, this number increases with a codetection of Arx and β-galactosidase found in 72% of the endocrine cells.
To determine whether this interaction reflects mechanisms involving individual or different cells, we analyzed Arx and Pax4 at the single-cell level. Due to the lack both of a suitable Pax4 antisense probe and of an antibody directed against Pax4, we assayed Pax4 expression using the β-galactosidase reporter gene present in heterozygous Pax4 pancreas. The recent generation of an anti-Arx antibody (recapitulating the Arx expression pattern, as demonstrated in Fig. 6C,D) allowed us to codetect Arx and Pax4 proteins in single islet cells as early as E13.5 (Fig. 6E). As the proliferation of glucagon and insulin-expressing cells proceeds, Arx and Pax4 were found to be coexpressed, but in 27% less cells (arrows in Fig. 6F), whereas the number of cells expressing either Arx or β-galactosidase increases. These data suggest that Arx and Pax4 are first found together in endocrine progenitors, however, as development continues, one of the factors prevails and the predominant protein will determine the fate of the endocrine cell. To find out whether this is achieved through direct interaction between Pax4 and Arx, we also examined Arx expression in Pax4-deficient embryos at E14.5 (Fig. 6G). We observed a 64% increase in the number of cells doubly positive for β-galactosidase and Arx proteins (Fig. 6, cf. F and G), thereby further demonstrating that Arx expression is up-regulated in the absence of functional Pax4.
Discussion
In this study, the consequences of the loss-of-function of the Arx gene on the differentiation of the endocrine pancreas are presented. In fact, although Arx has been inactivated previously in the mouse, resulting in abnormal differentiation and proliferation of the brain and testes, no expression or defect in the pancreas has been described (Kitamura et al. 2002). Our analysis clearly establishes that Arx transcripts are detectable as early as E9.5 in the pancreatic epithelium and are later found in mature α and δ cells. Mice carrying a targeted disruption of the Arx gene develop severe hypoglycemia, weakness, dehydration, and die 2 d after birth. Detailed examination of the pancreas reveals an early-onset loss of the mature α-cell population, concomitant with an augmentation of β- and δ-cell types, as well as an abnormal distribution within islets for the latter subtype.
Role of Arx in endocrine specification
By irreversibly tagging the progeny of cells via the Cre/LoxP system, Herrera (2000) clearly demonstrated that adult glucagon- and insulin-producing cells do not derive from cells that coexpressed both hormones previously, but rather emerge through distinct pathways. This study suggests that, throughout the development of the islet of Langerhans, two different populations of insulin-producing cells are generated successively, as well as two distinct types of glucagon-expressing cells. Other data have confirmed the existence of two insulin-expressing cell populations (Oster et al. 1998; Sander et al. 2000), and our results corroborate this finding. Importantly, we also clearly demonstrate the presence of two distinct glucagon-expressing cell types. In Arx mutant islets of Langerhans, the development of the early glucagon-/insulin-expressing cells remains unaffected, but mature α cells fail to appear at later stages, whereas the number of mature β and δ cells is increased. Interestingly, Arx labeling was observed at an early developmental stage, but never in hormone-expressing cells. In contrast, Arx expression was detected after E14.5, in α, β-precursors, and later in δ cells, further demonstrating that mature endocrine cells emerge from endocrine progenitors that did not express hormone previously.
Our finding that in Arx-deficient embryos α-, β-, and δ-cell content is significantly modified without alteration in shape, size, or total endocrine cell number suggests a replacement of the mature α cells by δ and β cells. Accordingly, Arx seems to determine the relative fate of these cells. This hypothesis is reinforced by the loss of Arx expression in the pancreas of Ngn3-deficient mice and the fact that pancreas from Arx mutant mice contains normal proportions of early Ngn3-labeled endocrine progenitors and Isl1-marked post-mitotic endocrine cells, as well as Nkx2.2-, Nkx6.1-, Pdx1-, and HB9-positive cells. Taken together, our data demonstrate that in Arx-deficient animals, β-, δ-, and PP-cell developmental processes are normal and lead to the generation of well-formed islets of Langerhans. However, α cells fail to appear, whereas β- and δ-cell numbers are increased. We conclude that Arx promotes the acquisition of α-cell fate by endocrine progenitors and does so by antagonizing β-and δ-cell commitment.
Arx and Pax4 act as differential regulators of endocrine cell fate
The results presented above indicate that Arx plays a key role in differential specification of islet cells. However, little is known of the processes underlying such a function; only the Pax4 gene has been shown to act differentially on endocrine cell destiny (Sosa-Pineda et al. 1997), as Pax4 deletion leads endocrine progenitors to adopt an alternative cell-subtype fate. The modifications of the islet-cell subtype content observed in these mutant mice are reminiscent of those seen in Arx loss-of-function mutant animals. Pax4- and Arx-depleted pancreas displays inversed alterations in α-, β-, and δ-cell ratio. The abnormalities seen in Arx- and Pax4-deficient mice suggest antagonistic roles for these two transcription factors. Using multiplex RT-PCR, we established that Pax4 mRNA is up-regulated in Arx mutant mice, and that the Arx transcript content is increased in Pax4-deficient pancreas. Furthermore, the recent generation of an anti-Arx antibody allowed us to demonstrate that Arx and Pax4 (the latter assayed by β-galactosidase staining in heterozygous Pax4 animals) are colocalized in early endocrine pancreas, but not at later developmental stages. Interestingly, our data suggest that in endocrine progenitors, one of these factors will ultimately predominate; if it is Pax4, β- and δ-cell fates will be specified, whereas Arx will favor α-cell commitment. Hence, we conclude that proper islet of Langerhans formation requires a tight control of the relative levels of Arx and Pax4 mRNA, probably achieved via mutual inhibition.
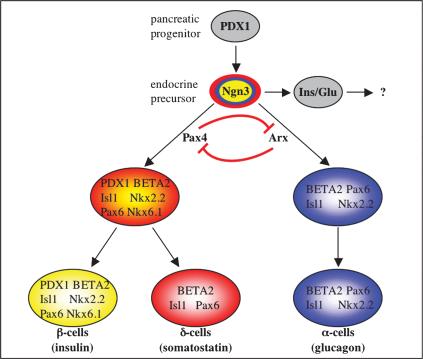
Our data, together with the results obtained by others (Sander et al. 1997; Sosa-Pineda et al. 1997; Mansouri et al. 1999; Gradwohl et al. 2000), allow us to propose the model depicted in Figure 7. In endocrine progenitors, Arx promotes α-cell fate and antagonizes β- and δ-cell destiny, whereas Pax4 favors β- and δ-cell fates at the expense of α-cell destiny. Our results suggest that these opposing activities are the consequences of a mutual cross-regulatory inhibition of these factors. The detailed mechanisms involved remain to be elucidated. Similarly, further work will be required to determine potential downstream target genes of Pax4 and Arx involved in endocrine subtype determination. In this respect, a recent study by Huotari et al. (2002) has implicated EGF signaling. Using pancreatic explant cultures, the authors have demonstrated that the EGF receptor family-binding proteins Betacellulin and Neuregulin-4 promote β- and δ-cell fates, respectively, in each case, at the expense of α-cell destiny. These results are consonant with the phenotype of Arx loss-of-function mice. To decipher the mechanisms involved in endocrine subtype commitment, a thorough examination of (1) Arx and Pax4 expression in mice with perturbed ErbB signaling and (2) EGF signaling in Arx and Pax4 mutant mice would be of particular interest. Similarly, the double inactivation of Arx and Pax4 and ectopic expression of either gene would permit us to unravel the interaction between these two factors.
Figure 7.
Schematic model representing the transcription factors implicated in the specification of the endocrine pancreas, on the basis of temporal expression and phenotypic results of specific gene deletions. Circles represent endocrine cells at particular developmental stages. The different transcription factors expressed in a particular cell type are indicated within the circles. Arrows represent different (hypothetical) endocrine differentiation steps. Loss-of-function mutant mice phenotypes demonstrate antagonistic endocrine-specifying activities for Arx and Pax4; Pax4 promotes β- and δ-cell fates, whereas Arx favors α-cell destiny. The question mark indicates a fate remaining to be elucidated. For the purpose of simplification, exocrine/ductal- and PP-cell development are not represented.
In summary, our analysis establishes a requirement of Arx for proper endocrine cell specification. We provide strong evidence that Arx and Pax4 play opposite roles in this process. Because the islet hormones, especially insulin and glucagon, have a central function in nutriment metabolism, alterations in the expression of Arx in the islets of Langerhans could modify nutriment homeostasis in the whole organism. Mutations of the human ARX gene have been associated with a particular type of X-linked lissencephaly with abnormal genitalia (Kitamura et al. 2002); it would be interesting to determine whether mutations in the human ARX gene are, additionally or independently, associated with glucose homeostasis deregulation.
Material and methods
Targeted disruption of the Arx gene
To generate Arx mutant mice, two overlapping clones containing the entire Arx gene were isolated from a 129/Sv mouse genomic library (provided by Anton Berns, Amsterdam). The gene was mapped using a combination of Southern analysis and restriction enzyme digests. The targeting construct is depicted in Figure 2A. Briefly, aiming to delete the ATG start-codon without altering any potential regulatory element, a 2.0-kb digested HindIII/EcoRV 5′ fragment was ligated to a 450-bp PCR-amplified fragment designed to encompass the region just upstream of the ATG start-codon. The 2.5-kb fragment obtained was ligated to a cassette containing a fusion of a nuclear localization signal (NLS) in-frame with the β-galactosidase gene [kindly provided by M. Buckingham (Pasteur Institute, Paris, France)]. Downstream, we inserted the Neomycin resistance gene flanked by LoxP sites. Next, the fragment encompassing the former 2.5-kb 5′ fragment, the NLS-β-galactosidase gene and the LoxP-Neomycin resistance gene was ligated upstream of an 8.1-kb XhoI/BamHI 3′ fragment. Finally, the Herpes simplex virus-thymidine kinase cassette was added in the 3′ position. The XhoI enzyme was used to linearize our construct prior to electroporation into ES cells.
Generation of mutant animals
The generation of recombinant ES clones and Arx mutant mice was performed as described by Mansouri (2001). A total of 202 stably transfected ES clones were obtained after antibiotic selection. Clones were screened for the presence of the disrupted Arx gene by Southern blotting with a 1.3-kb HincII-digested 5′ external probe (shown in Fig. 3A). Mutant ES cells were aggregated with NMRI 8-cell-stage embryos using standard techniques. Chimeric male mice were obtained and mated to NMRI females to generate heterozygous animals. Mice were geno-typed by PCR using a mixture of three primers (Fig. 2A) to identify the wild-type and the mutant alleles, generating a 0.25-kb and a 0.4-kb PCR product, respectively (P1, CGGCTCACTACACTTGTTACCGCTTGTCC; P2, AGCAGCCCTCTTCCTGGTACTGATTGCTC; and P3, TGCTGCAAGGCGATAAGTTGGGTAACGC).
β-galactosidase staining
Whole embryos or tissues were isolated and the yolk sac saved for DNA preparation and genotyping. After fixation in 4% paraformaldehyde, embryos and tissues were washed in PBS and then incubated in staining solution [4 mM K3 [Fe(CN)]6], 4 mM K4[Fe(CN)6], 0.02% NP-40, 0.01% Na-deoxycholate, 5 mM EGTA, 2 mM MgCl2, and 0.4 mg/mL 5-bromo-4-chloro-3-inodolyl-D-galactopyranoside. Prior to sectioning, tissues were dehydrated through ethanol series, cleared in toluene, and embedded in paraffin.
Hematoxylin-eosin staining
Paraformaldehyde-fixed tissues, embedded in paraffin, were cut in 6-μm sections and applied to Probe-on Plus slides (Fisher Scientific). Tissues were incubated after rehydration in hematoxylin for 2.5 min, rinsed in water, dipped quickly in 0.5% HCl/70% ethanol (v/v), and washed in water. After immersion in 0.2% NaHCO3, they were rinsed in water, dipped in 0.1% eosin for 20 sec, and washed briefly in water before dehydration and mounting.
RNA in situ hybridization
For whole-mount RNA in situ hybridization, tissues were isolated, fixed in 4% paraformaldehyde, and dehydrated through methanol series. Rehydrated embryos were bleached in 6% H2O2 for 1 h, permeabilized with 10 μg/mL proteinase K for 1-15 min, and prehybridized at 70°C for 90 min in a medium containing 50% formamide, 5× SSC at pH 4.5, 50 μg/mL yeast tRNA, 50 μg/mL heparin. Hybridization was performed overnight under the same conditions. For RNA in situ hybridization on sections, embryos were collected, fixed, incubated overnight in 30% sucrose, embedded in cryomatrix, and 10-μm sections were applied to Probe-on Plus slides (Fisher Scientific). Defrosted sections were hybridized overnight with DIG-labeled probes in a medium containing 50% formamide, 10% dextran sulphate, 1 mg/mL yeast tRNA, 0.02% BSA, 0.02% Ficoll, and 0.02% PVP.
Embryos and sectioned tissues were washed successively in 50% formamide, 1× SSC, 0.1% Tween-20 at 70°C for 70 min, and MABT at pH 7.5 (100 mM maleic acid; 150 mM NaCl; 0.1% Tween-20) at room temperature for 1 h and blocked in PBS containing 20% inactivated fetal calf serum for 90 min. Anti-DIG antibody (1:2500) was applied overnight in the same solution at room temperature. Tissues were washed thoroughly in MABT for 2 h, rinsed with NTMT (100 mM NaCl; 100 mM Tris-HCl at pH 9.5; 50 mM MgCl2; 1% Tween-20) for 1 h, stained in a solution containing 350 μg/mL NBT, 175 μg/mL BCIP, rinsed in PBS, and fixed in 4% paraformaldehyde. The Arx cRNA probe used was transcribed from a PCR-generated fragment encompassing the 8-1168-bp region of Arx full-length cDNA. The β-galactosidase cRNA PCR-generated probe contains the 675-1140-bp portion of the β-galactosidase cDNA.
Immunohistochemistry
Tissues were fixed in 4% paraformaldehyde, embedded in paraffin and 6-μm sections were applied to Probe-on Plus slides (Fisher Scientific). Deparaffinized and rehydrated sections were permeabilized in 0.2% Triton X-100 for 5 min and blocked in PBS containing 10% inactivated fetal calf serum for 90 min. Primary antibodies were diluted in the same medium, applied on sections, and incubated overnight at 4°C. Slides were washed in PBS and incubated for 90 min with the appropriate secondary antibody diluted in PBS containing 10% inactivated fetal calf serum. Slides were washed in PBS, mounted with DAPI, and viewed by fluorescent microscopy. Sections that had been subjected to in situ hybridization were washed in PBS, blocked, and incubated with primary antibodies as described above.
The primary antibodies used in these assays were as follows: mouse monoclonal anti-insulin diluted 1/1000 (Sigma); mouse monoclonal anti-glucagon diluted 1/1000 (Sigma); guinea-pig anti-insulin diluted 1/1000 (Sigma); guinea-pig anti-glucagon diluted 1/1000 (Sigma); rabbit anti-somatostatin diluted 1/600 (Dako); rabbit anti-PP diluted 1/200 (Dako); rabbit anti-β-galactosidase diluted 1/300 (Molecular Probes); mouse anti-β-galactosidase diluted 1/5000 (Promega); rabbit anti-Nkx6.1 diluted 1/3000; rabbit anti-Pdx1 diluted 1/2000 [kindly provided by C. Wright (Vanderbilt University, Nashville, TN)]; rabbit anti-Isl1 diluted 1/2000 [kindly provided by T. Jessell (Columbia University, New York)]; rabbit anti-Hb9 diluted 1/10000 (kindly provided by T. Jessell); rabbit anti-Nkx2.2 diluted 1/1000 (kindly provided by T. Jessell); rabbit anti-Pax6 diluted 1/500 [kindly provided by S. Saule (UMR 146, Orsay, France)]; rabbit anti-Ngn3 diluted 1/2000 [kindly provided by M. German (Diabetes Center, San Francisco, CA)]; and rabbit anti-Arx diluted 1/4500.
The secondary antibodies (Molecular Probes, dilution 1/1000) used for immunofluorescence were as follows: 594-alexa anti-mouse; 488-alexa anti-mouse; 594-alexa anti-rabbit; 488-alexa anti-rabbit; 594-alexa anti-guinea pig; and 488-alexa anti-guinea pig.
Glucose levels
Glucose levels were determined with the One Touch Glucose monitoring kit (Johnson & Johnson) using 15 μL of peripheral blood from 0.5-, 1-, and 2-day-old mice. Blood glucose levels are represented as an average ± standard deviation.
RNA isolation and cDNA synthesis
RNA was isolated by lysing tissues in guanidinium thiocyanate/phenol buffer (Trizol; Invitrogen) according to the manufacturer's instructions. After resuspension in RNase-free water, ethanol precipitation, and wash in 70% ethanol, the total RNA was diluted in RNase-free water to 2 μg/μL. cDNA synthesis was performed with the superscript single-stranded cDNA synthesis kit (Invitrogen) according to manufacturer's instructions. Briefly, 10 μg of total RNA were denatured at 70°C for 10 min, quickly chilled on ice, and subjected to cDNA synthesis at 45°C for 1 h.
RT-PCR and multiplex RT-PCR reactions
RT-PCR were carried out in 50-μL reactions using 0.5 μL of the diluted cDNA product as template with 47 μL of PCR mix (Genecraft); 10 pM of each primer and 2.5 U of Taq polymerase (Genecraft). Multiplex RT-PCR reactions were performed strictly following the protocol described by Jensen et al. (1996) by use of primers derived from insulin, glucagon, somatostatin, α-tubulin (data not shown), TATA-box-binding protein, Arx, and Pax4 genes. Previously unmentioned primers and multiplex RT-PCR conditions were as follows: Arx 5′Oligo (CAGCATTTGGCAGGCTCT) and 3′oligo (AGGATGTTGAGCTGCGTGAG), producing a 178-bp PCR product (26 cycles), and Pax4 5′Oligo (ACCTCATCCCAGGCCTATCT) and 3′oligo (AGGCCTCTTATGGCCAGTTT), producing a 220-bp PCR product (25 cycles). Reaction products labeled with [α-32P]dCTP (Amersham) were separated on 6% polyacrylamide gels, dried, and exposed overnight to a phosphorimage storage screen. Screens were subsequently scanned by a Molecular Dynamics PhosphorImager Series 400, and band intensities were calculated using the Imagequant software with rectangle mode/local background/volume integration. All amplicons were measured against the internal standards TATA-box-binding protein.
Preparation of polyclonal antibodies
Rabbits were injected with a mixture of three peptides corresponding to amino acids 2-16, 167-181, and 314-329 of Arx coupled to keyhole limpet hemocyanin (Eurogentec).
Acknowledgments
We are most grateful to Dr. K. Kniesel for bioinformatic assistance and Dr. X. Zhou, as well as Prof. Penicaud and Dr. F. Pituello, for helpful discussions. We thank Prof. J. Smith and Dr. G. Goudreau for careful reading of the manuscript, Dr. D. Treichel, Dr. A. Stoykova, Dr. K. Chowdhury, and our laboratory members for their constant support. We also thank S. Mahsur and U. Franke for excellent technical assistance. This work was supported by the DFG (Graduiertenkolleg 242), the Max-Planck Society, the JDRF Center for Beta Cell Therapy in Europe, and the NIH Beta Cell Biology Consortium.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.269003.
References
- Adrian T.E., Bloom, S.R., Hermansen, K., and Iversen, J. 1978. Pancreatic polypeptide, glucagon and insulin secretion from the isolated perfused canine pancreas. Diabetologia 14: 413-417. [DOI] [PubMed] [Google Scholar]
- Ahlgren U., Pfaff, S.L., Jessell, T.M., Edlund, T., and Edlund, H. 1997. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature 385: 257-260. [DOI] [PubMed] [Google Scholar]
- Ahlgren U., Jonsson, J., Jonsson, L., Simu, K., and Edlund, H. 1998. β-Cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the β-cell phenotype and maturity onset diabetes. Genes & Dev. 12: 1763-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apelqvist A., Li, H., Sommer, L., Beatus, P., Anderson, D.J., Honjo, T., Hrabe de Angelis, M., Lendahl, U., and Edlund, H. 1999. Notch signalling controls pancreatic cell differentiation. Nature 400: 877-881. [DOI] [PubMed] [Google Scholar]
- Blair H.J., Reed, V., Gormally, E., Wilson, J.B., Novak, J., McInnes, R.R., Phillips, S.J., Taylor, B.A., and Boyd, Y. 2002. Positioning of five genes (CASK, ARX, SAT, IMAGE cDNAs 248928 and 253949) from the human X chromosome short arm with respect to evolutionary breakpoints on the mouse X chromosome. Mamm. Genome 11: 710-712. [DOI] [PubMed] [Google Scholar]
- Csaba Z. and Dournaud, P. 2001. Cellular biology of somatostatin receptors. Neuropeptides 35: 1-23. [DOI] [PubMed] [Google Scholar]
- Dohrmann C., Gruss, P., and Lemaire, L. 2000. Pax genes and the differentiation of hormone-producing endocrine cells in the pancreas. Mech. Dev. 92: 47-54. [DOI] [PubMed] [Google Scholar]
- Edlund H. 2002. Pancreatic organogenesis-developmental mechanisms and implications for therapy. Nat. Rev. Genet. 3: 524-532. [DOI] [PubMed] [Google Scholar]
- Gradwohl G., Dierich, A., LeMeur, M., and Guillemot, F. 2000. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. 97: 1607-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G., Dubauskaite, J., and Melton, D.A. 2002. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 129: 2447-2457. [DOI] [PubMed] [Google Scholar]
- Guz Y., Montminy, M.R., Stein, R., Leonard, J., Gamer, L.W., Wright, C.V., and Teitelman, G. 1995. Expression of murine STF-1, a putative insulin gene transcription factor, in β cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development 121: 11-18. [DOI] [PubMed] [Google Scholar]
- Harrison K.A., Druey, K.M., Deguchi, Y., Tuscano, J.M., and Kehrl, J.H. 1994. A novel human homeobox gene distantly related to proboscipedia is expressed in lymphoid and pancreatic tissues. J. Biol. Chem. 269: 19968-19975. [PubMed] [Google Scholar]
- Herrera P.L. 2000. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development 127: 2317-2322. [DOI] [PubMed] [Google Scholar]
- Herrera P.L., Huarte, J., Sanvito, F., Meda, P., Orci, L., and Vassalli, J.D. 1991. Embryogenesis of the murine endocrine pancreas; early expression of pancreatic polypeptide gene. Development 113: 1257-1265. [DOI] [PubMed] [Google Scholar]
- Hirose S. and Mitsudome, A. 2003. X-linked mental retardation and epilepsy: Pathogenic significance of ARX mutations. Brain. Dev. 25: 161-165. [DOI] [PubMed] [Google Scholar]
- Huotari M.A., Miettinen, P.J., Palgi, J., Koivisto, T., Ustinov, J., Harari, D., Yarden, Y., and Otonkoski, T. 2002. ErbB signaling regulates lineage determination of developing pancreatic islet cells in embryonic organ culture. Endocrinology 143: 4437-4446. [DOI] [PubMed] [Google Scholar]
- Hussain M.A., Lee, J., Miller, C.P., and Habener, J.F. 1997. POU domain transcription factor brain 4 confers pancreatic α-cell-specific expression of the proglucagon gene through interaction with a novel proximal promoter G1 element. Mol. Cell. Biol. 17: 7186-7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M.A., Miller, C.P., and Habener, J.F. 2002. Brn-4 transcription factor expression targeted to the early developing mouse pancreas induces ectopic glucagon gene expression in insulin-producing β cells. J. Biol. Chem. 277: 16028-16032. [DOI] [PubMed] [Google Scholar]
- Jensen J., Serup, P., Karlsen, C., Funder-Nielsen, T., and Madsen, O.D. 1996. mRNA profiling of rat islet tumors reveals Nkx6.1 as a β-cell-specific homeodomain transcription factor. J. Biol. Chem. 271: 18749-18758. [DOI] [PubMed] [Google Scholar]
- Jensen J., Heller, R.S., Funder-Nielsen, T., Pedersen, E.E., Lind-sell, C., Weinmaster, G., Madsen, O.D., and Serup, P. 2000. Independent development of pancreatic α- and β-cells from neurogenin3-expressing precursors: A role for the notch pathway in repression of premature differentiation. Diabetes 49: 163-176. [DOI] [PubMed] [Google Scholar]
- Jonsson J., Carlsson, L., Edlund, T., and Edlund, H. 1994. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 371: 606-609. [DOI] [PubMed] [Google Scholar]
- Kitamura K., Yanazawa, M., Sugiyama, N., Miura, H., Iizuka-Kogo, A., Kusaka, M., Omichi, K., Suzuki, R., Kato-Fukui, Y., Kamiirisa, K., et al. 2002. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat. Genet. 32: 359-369. [DOI] [PubMed] [Google Scholar]
- Li H., Arber, S., Jessell, T.M., and Edlund, H. 1999. Selective agenesis of the dorsal pancreas in mice lacking homeobox gene Hlxb9. Nat. Genet. 23: 67-70. [DOI] [PubMed] [Google Scholar]
- Mansouri A. 2001. Determination of gene function by homologous recombination using embryonic stem cells and knockout mice. Methods Mol. Biol. 175: 397-413. [DOI] [PubMed] [Google Scholar]
- Mansouri A., St-Onge, L., and Gruss, P. 1999. Role of Pax genes in endoderm-derived organs. Trends Endocrinol. Metab. 10: 164-167. [DOI] [PubMed] [Google Scholar]
- Miura H., Yanazawa, M., Kato, K., and Kitamura, K. 1997. Expression of a novel aristaless related homeobox gene 'Arx' in the vertebrate telencephalon, diencephalon and floor plate. Mech. Dev. 65: 99-109. [DOI] [PubMed] [Google Scholar]
- Offield M.F., Jetton, T.L., Labosky, P.A., Ray, M., Stein, R.W., Magnuson, M.A., Hogan, B.L., and Wright, C.V. 1996. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 122: 983-995. [DOI] [PubMed] [Google Scholar]
- Oster A., Jensen, J., Serup, P., Galante, P., Madsen, O.D., and Larsson, L.I. 1998. Rat endocrine pancreatic development in relation to two homeobox gene products (Pdx-1 and Nkx 6.1). J. Histochem. Cytochem. 46: 707-715. [DOI] [PubMed] [Google Scholar]
- Pictet R.L., Clark, W.R., Williams, R.H., and Rutter, W.J. 1972. An ultrastructural analysis of the developing embryonic pancreas. Dev. Biol. 29: 436-467. [DOI] [PubMed] [Google Scholar]
- Roncoroni L., Violi, V., Montanari, M., and Muri, M. 1983. Effect of somatostatin on exocrine pancreas evaluated on a total external pancreatic fistula of neoplastic origin. Am. J. Gastroenterol. 78: 425-428. [PubMed] [Google Scholar]
- Sander M., Neubuser, A., Kalamaras, J., Ee, H.C., Martin, G.R., and German, M.S. 1997. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes & Dev. 11: 1662-1673. [DOI] [PubMed] [Google Scholar]
- Sander M., Sussel, L., Conners, J., Scheel, D., Kalamaras, J., Dela Cruz, F., Schwitzgebel, V., Hayes-Jordan, A., and German, M. 2000. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of β-cell formation in the pancreas. Development 127: 5533-5540. [DOI] [PubMed] [Google Scholar]
- Schwitzgebel V.M., Scheel, D.W., Conners, J.R., Kalamaras, J., Lee, J.E., Anderson, D.J., Sussel, L., Johnson, J.D., and German, M.S. 2000. Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development 127: 3533-3542. [DOI] [PubMed] [Google Scholar]
- Slack J.M. 1995. Developmental biology of the pancreas. Development 121: 1569-1580. [DOI] [PubMed] [Google Scholar]
- Smith S.B., Ee, H.C., Conners, J.R., and German, M.S. 1999. Paired-homeodomain transcription factor PAX4 acts as a transcriptional repressor in early pancreatic development. Mol. Cell. Biol. 19: 8272-8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer L., Ma, Q., and Anderson, D.J. 1996. neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol. Cell. Neurosci. 8: 221-241. [DOI] [PubMed] [Google Scholar]
- Sosa-Pineda B., Chowdhury, K., Torres, M., Oliver, G., and Gruss, P. 1997. The Pax4 gene is essential for differentiation of insulin-producing β cells in the mammalian pancreas. Nature 386: 399-402. [DOI] [PubMed] [Google Scholar]
- St-Onge L., Sosa-Pineda, B., Chowdhury, K., Mansouri, A., and Gruss, P. 1997. Pax6 is required for differentiation of glucagon-producing α-cells in mouse pancreas. Nature 387: 406-409. [DOI] [PubMed] [Google Scholar]
- Sussel L., Kalamaras, J., Hartigan-O'Connor, D.J., Meneses, J.J., Pedersen, R.A., Rubenstein, J.L., and German, M.S. 1998. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic β cells. Development 125: 2213-2221. [DOI] [PubMed] [Google Scholar]
- Teitelman G., Alpert, S., Polak, J.M., Martinez, A., and Hanahan, D. 1993. Precursor cells of mouse endocrine pancreas coexpress insulin, glucagon and the neuronal proteins tyrosine hydroxylase and neuropeptide Y, but not pancreatic polypeptide. Development 118: 1031-1039. [DOI] [PubMed] [Google Scholar]
- Upchurch B.H., Aponte, G.W., and Leiter, A.B. 1994. Expression of peptide YY in all four islet cell types in the developing mouse pancreas suggests a common peptide YY-producing progenitor. Development 120: 245-252. [DOI] [PubMed] [Google Scholar]