Abstract
The Drosophila gene buttonhead (btd) is required for the establishment of three embryonic head segments. It encodes a zinc-finger-type transcription factor expressed in the corresponding head segment anlagen in the blastoderm stage embryo. The DNA-binding properties of the btd protein (BTD) are indistinguishable from the human transcription factor Sp1. Furthermore, BTD and Sp1 are capable of activating transcription in transfected cultured cells through interaction with the same DNA target sites. Herein we show that BTD and Sp1 functionally interact with the same TATA box-binding protein-associated factors and support in vitro transcription activation through these contacts. Transgene expression of BTD results in the rescue of the head segments that fail to develop in btd mutant embryos, whereas Sp1 or Sp1 containing the zinc finger region of BTD rescues mandibular segment development. The results suggest that BTD contains functional domains other than an equivalent DNA-binding region and interaction sites of the TATA box-binding protein-associated factors, which are necessary to establish head segments that fail to develop in response to Sp1.
Head segments of the Drosophila embryo are generated in response to the overlapping activities of the segmentation genes orthodenticle (otd), empty spiracles (ems), and buttonhead (btd) (1). These genes are expressed in three overlapping head segment anlagen of the blastoderm embryo that fail to be established in the corresponding mutants (2–5). btd is required for antennal, intercalary, and mandibular segment formation (1). It encodes a transcription factor, BTD, with a modular design similar to the human transcription factor Sp1 (5). BTD is able to bind to the G + C-rich DNA target site of Sp1 and to activate transcription in an Sp1-like manner in Drosophila Schneider cells (5).
Transcriptional activation by human Sp1 involves the general RNA polymerase II transcription factor TFIID (6). TFIID is composed of the TATA box-binding protein (TBP) and a set of TBP-associated factors (TAFs) (for review, see refs. 7 and 8). TAFs provide interfaces for enhancer-bound transcription factors that contact the basal transcription apparatus and direct the activation of transcription. Sp1 selectively interacts with TAFII110 (9), one of the eight TAFs of Drosophila that include TAFII250, TAFII150, TAFII110, TAFII80, TAFII60, TAFII40, TAFII30α, and TAFII30β (10). Partial TFIID complexes composed of TBP, TAFII250, TAFII150, and TAFII110 are sufficient to support transcriptional activation in vitro, whereas partial complexes lacking TAFII110 or mutant Sp1 lacking the interaction surface fail to activate transcription (11, 12). Furthermore, different transcription factors, such as NTF-1, functionally interact with other TAFs (12). This suggested that the different subunits of TFIID, each with distinct structural characteristics, provide specific substrates for enhancer-bound transcriptional activators to contact the basal transcription apparatus and thereby direct the activation of gene expression.
Herein we show that BTD and Sp1 interact with the same TAF subunits and support in vitro transcription activation via these contacts. In addition, the DNA-binding regions of the two proteins are equivalent. However, transgene-dependent Sp1 expression in place of BTD rescues only some aspects of mandibular development in btd mutant embryos. The results suggest that the generation of the intercalary and the antennal segment requires functional interaction domains of BTD that are not present in Sp1.
MATERIALS AND METHODS
Drosophila Strains and Mutant Embryos.
The following fly strains were used in this work: Oregon R; y w btdXG/FM7c, ftz-lacZ; svbYP17b btdXG/FM7c (5, 13, 14); TAFII110ΔC/TM6B (15). Homozygous lines of the genomic btd (Pbtd) and Sp1 (btd>Sp1) rescue constructs (5) and the new constructs were also used. The svb btd double mutant allows the detection of btd mutant cuticles when crossing this strain to the transgene (13). The following strains were constructed: a fly strain homozygous for btd>Sp1 on the second and third chromosome and svbYP17b btdXG/FM7c; Pbtd. When crossing the latter flies to the TAF mutant, the svb locus allowed us to recognize btd mutant embryos that are rescued by one copy of Pbtd and in 50% of the cases are also heterozygous for TAFII110ΔC.
Generation and Analysis of the Rescue Construct.
Sp1BTDzf (Sp1 with the BTD zinc finger region) consists of a 1.8-kb filled-in BamHI fragment and a 670-bp filled-in BglI–HindIII fragment from pSp1–778c (16) that was ligated with a 300-bp filled-in AvaII–SfiI fragment from pKSbtd (17). After addition of XbaI linkers, the fragment was cloned into pBluescript II KS+ (Stratagene). After checking the sequence, the construct was subcloned into the P element vector pCbtdRV-2ndBΔXba, which provides the 5.2-kb btd cis-acting element (17). To generate transgenic flies, constructs were injected into w sn3 embryos by using pΔ2–3 (18) as helper (19). Transgenic progeny were balanced over CyO or TM3. At least three independent transgenic lines were analyzed.
Protein Binding and in Vitro Transcription Assay.
The baculovirus transfer vector pSLFlag (20) was used to generate flag-tag fusion constructs for overproduction of proteins. The following constructs were generated. For the full-length BTD, a 2.2-kb SmaI–SspI fragment from pKSbtd (17) was cloned into pSLFlag digested with NdeI and SmaI and blunt-ended with mung bean nuclease. For a full-length Sp1, a filled-in 2.4-kb NotI–NdeI fragment from pSp1–778c (16) was cloned into pSLFlag digested with NdeI and SmaI and blunt-ended with mung bean nuclease. For N-terminal BTD without the zinc finger region, a 1,030-bp filled-in SmaI–partial BspEI fragment from pKSbtd was cloned into pSLFlag digested with NdeI and SmaI and blunt-ended with mung bean nuclease. For the C-terminal BTD without a zinc finger region, a 880-bp blunt-ended BglI–SspI fragment from pKSbtd was cloned into pSLFlag digested with NdeI and SmaI and filled in. All constructs were checked by sequencing. Generation of recombinant baculovirus with Baculogold viral DNA (PharMingen), expression of Flag-epitope-tagged proteins, and their purification from Sf9 cells was performed as described (21, 22). Interaction studies were performed with approximately 50 ng of Flag-epitope-tagged proteins immobilized on Flag-M2 antibody resin (Eastman Kodak) and [35S]methionine-labeled proteins that were generated in a TNT-coupled in vitro transcription/translation system (Promega) (22). Protein complexes were analyzed by SDS/PAGE and visualized by autoradiography. In vitro transcription reactions (22, 23) were programmed with 10 ng of BTD or Sp1 and 50 ng of reporter gene plasmid (12). The reconstituted Drosophila transcription system is as described (23).
Histochemistry and Cuticle Preparations.
Immunological staining of whole-mount embryos was performed as described (24) with the Vectastain ABC Elite Kit (Vector, Boehringer Ingelheim) and the primary antibodies anti-β-galactosidase (Cappel), anti-En (4D9) (25), and mAb22C10 (26). Homozygous btd mutant embryos were identified with blue balancers (27). Stained embryos and cuticle preparations (13) were photographed with a Zeiss Axiophot microscope.
RESULTS AND DISCUSSION
Sp1 and BTD Interact with the Same TAFs.
Biochemical interaction assays and functional in vitro studies provided critical support that transcriptional activation by Sp1 is mediated by binding to TAFII110 (9). Protein–protein interactions (i.e., Sp1 and TAFII110) were sufficient to support transcriptional activation in vitro, whereas partial complexes lacking TAFII110 failed to do so (12). Because the C2H2 zinc finger DNA-binding motif and glutamine- and serine/threonine-rich N-terminal domains are conserved between Sp1 and BTD (16), we tested whether BTD exerts Sp1-like biochemical properties. For this, we performed binding studies involving components of the transcriptional apparatus, including TFIIA and the eight Drosophila TAFs (see Introduction; Fig. 1). To allow for identical posttranslational modifications, we produced Flag-epitope-tagged BTD and Sp1 expressed in Sf9 cells that were infected with recombinant baculovirus (Fig. 2A). The epitope-tagged proteins were immunopurified by immobilization on Flag M2 antibody resins. The protein-coated resins were incubated with [35S]methionine-labeled TFIIA or TAFs that were generated by using a coupled in vitro transcription/translation system.
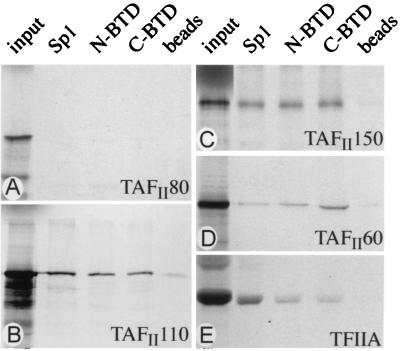
Figure 1.
Comparison of BTD and Sp1 in vitro interactions with TAFs. Flag-tagged BTD and Sp1 were expressed and purified from Sf9 cells and incubated with [35S]methionine-labeled recombinant TFIIA or TAFs. An example of noninteracting TAFs, such as TAFII80, is shown in A. C-terminal BTD (C-BTD), N-terminal BTD (N-BTD), and Sp1 interact strongly with TAFII110 (B) and TAFII150 (C) and weakly with TAFII60 (D) and TFIIA (E). Input refers to 10% of the labeled TAFs that was used for the binding reaction. Beads refers to a control incubation of unreacted resin with labeled TAFs and TFIIA.
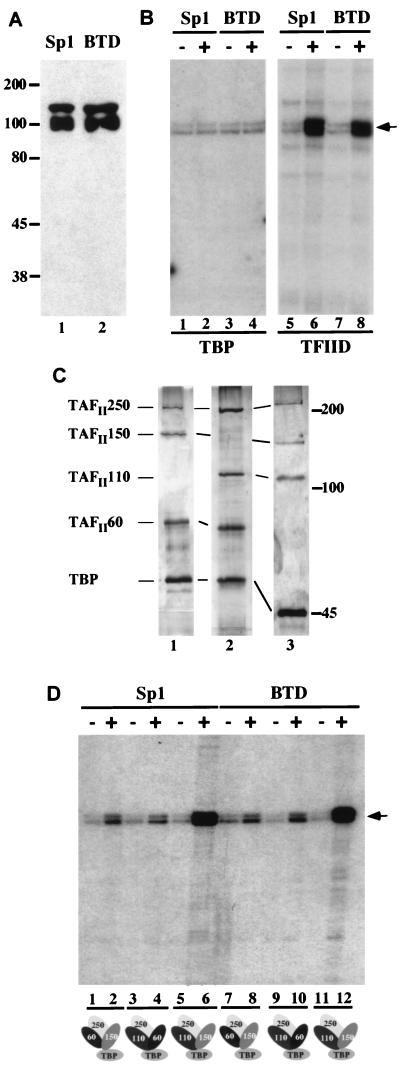
Figure 2.
Transcriptional activation by BTD and Sp1 in vitro. (A) Western Blot showing purified Sp1 (lane 1) and BTD (lane 2) produced in baculovirus-infected Sf9 cells. The purified proteins were detected by anti-Flag antibody; molecular weights (in kDa) are indicated. (B) Sp1- and BTD-dependent transcriptional activation in vitro. In vitro transcription reactions were composed of a reconstituted Drosophila transcription system containing either TBP (lanes 1–4) or TFIID (lanes 5–8), which were programmed with the reporter plasmid (GC)3BCAT (12) and either Sp1 (lanes 2 and 6) or BTD (lanes 4 and 8). TBP was unable to mediate Sp1- and BTD-dependent transcription activation (lanes 1–4), whereas TFIID strongly stimulated transcription activation (lanes 6 and 8). (C) Silver-stained SDS/polyacrylamide gel of in vitro-assembled partial TBP–TAF complexes composed of TAFII250, TAFII150, TAFII60, and TBP (lane 1); TAFII250, TAFII110, TAFII60, and TBP (lane 2); or TAFII250, TAFII150, TAFII110, and TBP (lane 3). Molecular weights (in kDa) are indicated to the right. (D) Autoradiographs of in vitro transcription reaction products, programmed with partial TBP–TAF complexes shown in C (indicated at the bottom) with or without 10 ng of purified Sp1 (lanes 1–6) or 10 ng of BTD (lanes 7–12). Transcription products were assayed by primer extension; the position of the reverse transcript is indicated by an arrow. The combination of TBP, TAFII250, TAFII110, and TAFII150 (lanes 6 and 12) causes strong and synergistic activation of transcription, whereas comparatively weak activation is observed with other partial complexes.
Affinity beads containing either the purified N-terminal or C-terminal region of BTD, excluding the zinc finger domain, or full-length purified Sp1 were able to specifically retain TAFII110 and TAFII150 (Fig. 1 B and C). In addition, weak binding was observed with TAFII60 and TFIIA (Fig. 1 D and E), whereas the other four TAFs were not retained on the BTD- or Sp1-coated affinity resins (for example, see Fig. 1A). This indicates that both BTD and Sp1 can target the same components of the TFIID complex in vitro. We next asked whether the conserved contacts of BTD also support the activation of transcription in a cell-free reaction system in an Sp1-like fashion.
Interacting TAFs Support BTD-Dependent in Vitro Transcription.
To compare the transcriptional properties of Sp1 and BTD in a cell-free transcription assay, we used a reconstituted Drosophila transcription system composed of the purified recombinant basal transcription factors TFIIA, -B, -E, and -F, purified endogenous TFIIH, RNA polymerase II, and either purified recombinant TBP or in vitro-assembled TBP–TAF complexes (Fig. 2). We programmed the cell-free reaction system with recombinant epitope-tagged BTD or Sp1 that was expressed and purified as described above (Fig. 2A). As a reporter gene, we used a plasmid construct containing five Sp1 in vitro DNA-binding sites upstream of the E1B TATA box that governs the transcription of the reporter gene encoding chloramphenicol acetyltransferase. Reporter gene transcription was monitored by primer extension analysis.
In vitro reactions containing endogenous Drosophila TFIID supported BTD-dependent activation of transcription, whereas reactions containing only TBP failed to do so (Fig. 2B). This result suggests that BTD, like Sp1, requires TAF subunits of the TFIID complex to mediate activation of transcription in vitro. To assess which TAF subunits mediate BTD-dependent activation of transcription, we used in vitro-assembled TBP–TAF complexes (Fig. 2C) instead of the endogenous TFIID, previously shown to mediate Sp1-dependent transcriptional activation (12). Reactions containing in vitro-assembled TBP–TAF complexes composed of TBP, TAFII250, and various combinations of TAFII60, TAFII150, and TAFII110 were assayed (Fig. 2D). Strongest activation of transcription in response to Sp1 and BTD was mediated by partial complexes containing TAFII150 and TAFII110 (Fig. 2D). This observation is consistent with the earlier finding that TAFII110 and TAFII150 mediate Sp1-dependent activation in a synergistic manner (12). The results suggest further that the presence of TAFII60 did not provide additional transcriptional activation when acting in a combination with TAFII110 or TAFII150, meaning that there is no synergistic interaction between TAFII110 or TAFII150 and TAFII60 (Fig. 2D, compare lanes 8, 10, and 12). These findings demonstrate indistinguishable properties of Sp1 and BTD with respect to their abilities to activate transcription in a TAF-mediated manner.
Sp1 and BTD Contact the Same Functional Targets.
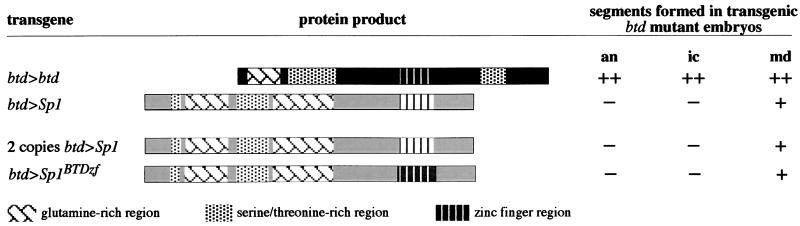
btd mutant embryos lack three adjacent head segments: the mandibular, the intercalary, and the antennal segments (1, 5). The development of these segments can be identified by a distinct set of head sensory organs in the embryo or the larval cuticle (1, 28, 29) and by the expression domains of the segment polarity gene engrailed (en) in the germ-band-extended embryo (30). btd transgene expression under the control of the btd 5.2-kb cis-acting element (btd>btd, Fig. 3) provides the proper spatial and temporal expression in the three btd-dependent head segment anlagen and rescues the btd mutant head phenotype (17). In contrast, a transgene containing the Sp1 cDNA in place of the btd cDNA (btd>Sp1, Fig. 3) provides a partial mandibular segment only (5), even when multiple copies of btd>Sp1 were supplied (Fig. 3 and data not shown). Thus, Sp1 can activate BTD target genes required for mandibular development but is unable to support intercalary and antennal development (31).
Figure 3.
Schematic representation of transgenes and their expression response in btd mutant embryos. Transgene constructs (Left) and rescue response of transgene-bearing btd mutant embryos are shown as assayed by Engrailed expression, the presence of sensory neurons in the head region, and cuticular head markers (ref. 28; for details, see Fig. 4). btd>btd and btd>Sp1 rescue is shown for comparison (5, 17). ++, a full rescue of btd mutant embryos [i.e., the corresponding set of segment markers could be scored in transgene-bearing btd mutant embryos (n ≥ 50 embryos analyzed)]; +, a partial rescue (i.e., only a subset of segment markers was found); −, no rescue. Representative examples of rescued embryos are shown in Fig. 4.
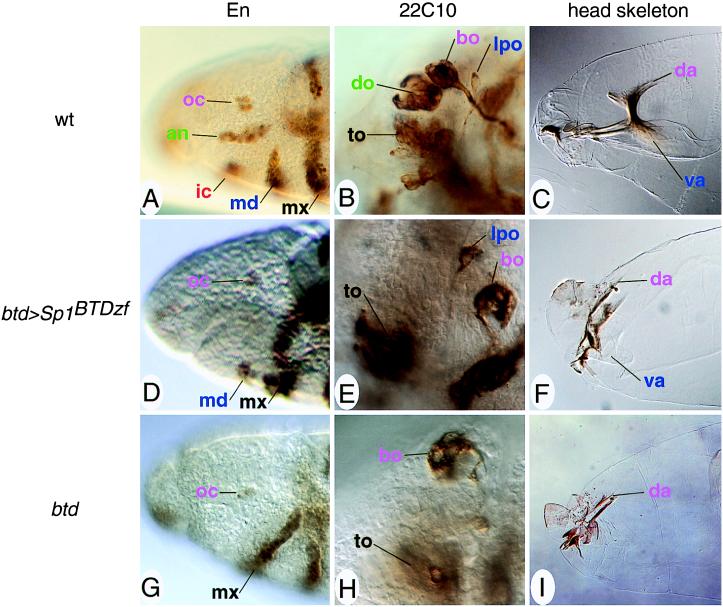
One possibility to account for the different in vivo properties of BTD and Sp1 is that they exhibit different DNA-binding characteristics that allow them to distinguish different sets of target sites. To assess this possibility directly, we generated a Sp1-derived protein that contains the BTD zinc finger DNA-binding domain (Sp1BTDzf). This hybrid gene was placed under the control of the btd enhancer sequence and expressed in transgenic btd mutant embryos. Expression of the btd>Sp1BTDzf (Figs. 3 and 4 D–F, compare with wild type in Fig. 4 A–C and the btd mutant in Fig. 4 G–I) transgene resulted in an Sp1-like rescue of the mandibular segment. Thus, differences in target site specificity and/or binding affinity of the Sp1 and BTD DNA-binding domains are not critical with respect to the biological responses caused by the two proteins. Furthermore, taking into account their identical TAF requirement in vitro, the relevant differences between Sp1 and BTD should also not be dependent on their ability to stimulate transcriptional activation of the biologically relevant target genes.
Figure 4.
Head rescue pattern of btd>Sp1BTDzf-bearing btd mutant embryos. (A–C) Head structures of wild-type embryos. (D–F) Head structures of btd>Sp1BTDzf-bearing btd mutant embryos. (G–I) Head structures of btd mutant embryos. (A, D, and G) Anti-En antibody labeling of stage 10 embryos. (Magnification, ×400.) Only the mandibular segment is partially rescued (D); antennal and intercalary segments are missing (compare with A and G). (B, E, and H) mAb22C10 staining of stage 14/15 embryos showing head sensory organs at a representative focal plane. (Magnification, ×1,000.) The lateropharyngeal organ of mandibular origin is rescued whereas the dorsal organ (an antennal organ) is not (E, compare with B and H). (C, F, and I) Head skeletons of stage 17 embryos show that intercalary and antennal cuticle structures are not rescued, whereas the ventral arm of mandibular origin is partially present in Sp1BTDzf-expressing embryos (F). (Magnification, ×400.) Embryos are dorsal up and anterior to the left and staged according to ref. 32. Abbreviations and color code: ocular (oc) segmental structures (bo, Bolwig organ; da, dorsal arms) are labeled in violet; antennal (an) structures (do, dorsal organ) are green; intercalary (ic) structures are orange red; mandibular (md) structures (lpo, lateropharyngeal organ; va, ventral arms) are blue; maxillary (mx) structures (to, terminal organ) are black (for a detailed description of the segmental identity of organs, Engrailed stripes, and cuticle structures, see ref. 28).
Activation of in Vivo BTD Target Genes Is Not TAF-Specific.
We tested this proposal by a genetic interaction assay, involving a dominant negatively acting Drosophila mutant of TAFII110 (15) and a homozygous btd mutant that was rescued by a single copy of the btd-expressing transgene. The rationale behind this mutant combination was that the btd activity in such embryos was limiting and that an interference with the TAFII110–BTD interaction should further decrease btd action and thus impair btd-dependent head development. However, trans-heterozygous TAFII110/btd mutant embryos develop a normal head segmentation pattern (data not shown). BTD must therefore be able to properly activate its target genes by means other than synergistic interactions involving TAFII110 and TAFII150 exclusively. However, lowering the level of BTD or TAFII110 further or reducing the TAFII150 level in addition to BTD and TAFII110 might result in head defects and thereby reveal the necessity of interactions between the two factors.
BTD Must Contain Information to Mediate Intercalary and Antennal Segment Development.
Our data provide evidence that BTD and human Sp1 interact in vitro with the same subset of TAFs and use the same TAFs for transcriptional activation in a reconstituted cell-free assay. In addition, Sp1 containing the BTD DNA-binding domain supports partial mandibular development but fails to provide intercalary and antennal structures. The same is true for a BTD miniprotein composed of the BTD DNA-binding domain fused to the strong transcriptional VP16 activation domain (F. Schöck, unpublished results). Because Sp1 and the VP16 activation domain contact different TAFs [i.e., TAFII40 and the basal factor TFIIB in the case of VP16 (33)], binding of the transcription factor and its contacts with basal transcription factors appear to be sufficient to establish mandibular pattern elements. This minimal requirement explains why BTD-dependent mandibular development can be achieved in response to a biologically unrelated transcription factor such as human Sp1. However, the transgene expression study also establishes that this property of BTD is not sufficient to establish segments that require ems (intercalary segment) or ems and otd (antennal segment) in addition to BTD. This suggests that BTD contains properties to interact with EMS and OTD or with their downstream factors. Furthermore, because Sp1 is not sufficient to establish all aspects of mandibular development, it also appears likely that the proper formation of this segment requires an additional BTD-specific component. We currently examine whether the unique BTD properties are due to specific interaction domains and whether such domains are required for interactions with, for example, EMS and OTD.
Acknowledgments
We thank Dr. R. Tjian (Berkeley) for providing the tools for the in vitro experiments and for hosting B.A.P. in his laboratory. We also thank him for encouragement and support. We also thank C. Klämbt for providing generous amounts of mAb22C10; H. Taubert for injections; G. Dowe for sequencing; and M. González-Gaitán, R. Rivera-Pomar, and G. Vorbrüggen for helpful discussions. The anti-En antibody (4D9) was obtained from the Developmental Studies Hybridoma Bank maintained by the University of Iowa (Iowa City). This work was supported by the Human Frontier Science Organization (H.J.) and by fellowships of the Fonds der Chemischen Industrie (F.S.) and the Alexander-von-Humboldt Stiftung (B.A.P.).
ABBREVIATIONS
- TBP
TATA box-binding protein
- TAF
TBP-associated factor
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Cohen S, Jürgens G. Nature (London) 1990;346:482–485. doi: 10.1038/346482a0. [DOI] [PubMed] [Google Scholar]
- 2.Dalton D, Chadwick R, McGinnis W. Genes Dev. 1989;3:1940–1956. doi: 10.1101/gad.3.12a.1940. [DOI] [PubMed] [Google Scholar]
- 3.Finkelstein R, Smouse D, Capaci T, Spradling A, Perrimon N. Genes Dev. 1990;4:1516–1527. doi: 10.1101/gad.4.9.1516. [DOI] [PubMed] [Google Scholar]
- 4.Walldorf U, Gehring W J. EMBO J. 1992;11:2247–2259. doi: 10.1002/j.1460-2075.1992.tb05284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wimmer E A, Jäckle H, Pfeifle C, Cohen S M. Nature (London) 1993;366:690–694. doi: 10.1038/366690a0. [DOI] [PubMed] [Google Scholar]
- 6.Smale S T, Schmidt M C, Berk A J, Baltimore D. Proc Natl Acad Sci USA. 1990;87:4509–4513. doi: 10.1073/pnas.87.12.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Struhl K, Moqtaderi Z. Cell. 1998;94:1–4. doi: 10.1016/s0092-8674(00)81213-1. [DOI] [PubMed] [Google Scholar]
- 8.Tansey W, Herr W. Cell. 1997;88:729–732. doi: 10.1016/s0092-8674(00)81916-9. [DOI] [PubMed] [Google Scholar]
- 9.Hoey T, Weinzierl R, Gill G, Chen J-L, Dynlacht B, Tjian R. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 10.Conaway R, Conaway J. In: Transcription Factors. Rickwood D, Hames B, editors. New York: Wiley; 1997. pp. 1–11. [Google Scholar]
- 11.Gill G, Pascal E, Tseng Z H, Tjian R. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 13.Wimmer E A, Frommer G, Purnell B A, Jäckle H. Mech Dev. 1996;59:53–62. doi: 10.1016/0925-4773(96)00575-8. [DOI] [PubMed] [Google Scholar]
- 14.Wieschaus E, Nüsslein-Volhard C, Jürgens G. Roux′s Arch Dev Biol. 1984;193:296–307. doi: 10.1007/BF00848158. [DOI] [PubMed] [Google Scholar]
- 15.Sauer F, Wassarman D A, Rubin G M, Tjian R. Cell. 1996;87:1271–1284. doi: 10.1016/s0092-8674(00)81822-x. [DOI] [PubMed] [Google Scholar]
- 16.Kadonaga J T, Courey A J, Ladika J, Tjian R. Science. 1988;242:1566–1570. doi: 10.1126/science.3059495. [DOI] [PubMed] [Google Scholar]
- 17.Wimmer E A, Cohen S M, Jäckle H, Desplan C. Development (Cambridge, UK) 1997;124:1509–1517. doi: 10.1242/dev.124.8.1509. [DOI] [PubMed] [Google Scholar]
- 18.Laski F A, Rio D C, Rubin G M. Cell. 1986;44:7–19. doi: 10.1016/0092-8674(86)90480-0. [DOI] [PubMed] [Google Scholar]
- 19.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 20.Lichtsteiner S, Tjian R. EMBO J. 1995;14:3937–3945. doi: 10.1002/j.1460-2075.1995.tb00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richardson C. In: Methods in Molecular Biology. Walker J, editor. Vol. 39. Totowa, NJ: Humana; 1995. [Google Scholar]
- 22.Sauer F, Hansen S K, Tjian R. Science. 1995;270:1783–1788. doi: 10.1126/science.270.5243.1783. [DOI] [PubMed] [Google Scholar]
- 23.Hansen S K, Tjian R. Cell. 1995;82:565–575. doi: 10.1016/0092-8674(95)90029-2. [DOI] [PubMed] [Google Scholar]
- 24.González-Gaitán M, Jäckle H. In: The Molecular Biology of Insect Disease Vectors. Crampton J, Beard C, Louis C, editors. London: Chapman & Hall; 1997. pp. 283–294. [Google Scholar]
- 25.Patel N H, Martin-Blanco E, Coleman K G, Poole S J, Ellis M C, Kornberg T B, Goodman C S. Cell. 1989;58:955–968. doi: 10.1016/0092-8674(89)90947-1. [DOI] [PubMed] [Google Scholar]
- 26.Zipursky S, Venkatesh T, Teplow D, Benzer S. Cell. 1984;36:15–26. doi: 10.1016/0092-8674(84)90069-2. [DOI] [PubMed] [Google Scholar]
- 27.González-Gaitán M, Jäckle H. Development (Cambridge, UK) 1995;121:2313–2325. doi: 10.1242/dev.121.8.2313. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt-Ott U, González-Gaitán M, Jäckle H, Technau G M. Proc Natl Acad Sci USA. 1994;91:8363–8367. doi: 10.1073/pnas.91.18.8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt-Ott U, González-Gaitán M, Technau G. Roux’s Arch Dev Biol. 1995;205:31–44. doi: 10.1007/BF00188841. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt-Ott U, Technau G M. Development (Cambridge, UK) 1992;116:111–125. doi: 10.1242/dev.116.1.111. [DOI] [PubMed] [Google Scholar]
- 31.Wimmer E A, Simpson-Brose M, Cohen S M, Desplan C, Jäckle H. Mech Dev. 1995;53:235–245. doi: 10.1016/0925-4773(95)00439-8. [DOI] [PubMed] [Google Scholar]
- 32.Campos-Ortega J, Hartenstein V. The Embryonic Development of Drosophila melanogaster. Berlin: Springer; 1997. [Google Scholar]
- 33.Goodrich J A, Hoey T, Thut C J, Admon A, Tjian R. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]