Abstract
Phylogenetic trees for the four extant species of African hominoids are presented, based on mtDNA control region-1 sequences from 1,158 unique haplotypes. We include 83 new haplotypes of western chimpanzees and bonobos. Phylogenetic analysis of this enlarged database, which takes intraspecific geographic variability into account, reveals different patterns of evolution among species and great heterogeneity in species-level variation. Several chimpanzee and bonobo clades (and even single social groups) have retained substantially more mitochondrial variation than is seen in the entire human species. Among the 811 human haplotypes, those that branch off early are predominantly but not exclusively African. Neighbor joining trees provide strong evidence that eastern chimpanzee and human clades have experienced reduced effective population sizes, the latter apparently since the Homo sapiens–neanderthalensis split. Application of topiary pruning resolves ambiguities in the phylogenetic tree that are attributable to homoplasies in the data set. The diverse patterns of mtDNA sequence variation seen in today’s hominoid taxa probably reflect historical differences in ecological plasticity, female-biased dispersal, range fragmentation over differing periods of time, and competition among social groups. These results are relevant to the origin of zoonotic diseases, including HIV-1, and call into question some aspects of the current taxonomic treatment and conservation management of gorillas and chimpanzees.
Keywords: human evolution, molecular evolution, Pan, Gorilla
In spite of the absence of relevant fossils, studies of genetic variation have settled the question of whether humans are more closely related to gorillas or chimpanzees in favor of the latter (1). However, most such comparative studies have been based on no more than six individuals of each taxon with the exception of humans (2–9). Furthermore, the individual apes used in these comparisons are typically of unknown geographic origin, and there has been a tendency to treat all chimpanzees as a homogeneous group. Advances in DNA sequencing technology and noninvasive genotyping (10–13) now permit a more thorough analysis of the geographic variation within and among mitochondrial DNA (mtDNA) sequences in each of the living African hominoids. The noncoding control region 1 (CR1) was selected for analysis because it is hypervariable and was thought to be relatively free from direct natural selection and therefore was thought to provide a less ambiguous record of mutational change and phylogenetic relationships (9).
Here we present a comparison of genetic variation in all nine recognized taxa of African or African-derived hominoid: (i) western lowland gorilla (Gorilla g. gorilla); (ii) eastern mountain gorilla (Gorilla g. beringei); (iii) eastern lowland gorilla (Gorilla g. graueri); (iv) humans (Homo sapiens); (v) bonobo (Pan paniscus); (vi) central chimpanzee (Pan t. troglodytes); (vii) eastern chimpanzee (Pan t. schweinfurthi); (viii) western chimpanzee (Pan t. verus); and (ix) the newly recognized Nigerian chimpanzee (tentatively referred to as Pan t. vellerosus) (14). We also include three other groups in our analyses: a Neandertal (Homo s. neanderthalensis) and Bornean and Sumatran orangutans (Pongo p. pygmaeus and Pongo p. abelii, respectively). Using common names except when ambiguous, we show that these taxa have very different amounts and patterns of genetic variation, with humans being the least variable. Strong evidence is presented for a relatively recent demographic bottleneck, selective sweep, or population expansion in two clades, humans and eastern chimpanzees. We also show that topiary pruning (15) resolves ambiguities in the phylogenetic tree that are caused by homoplasies in the data set.
MATERIALS AND METHODS
Data.
We examined part of hypervariable CR1, corresponding to the human mtDNA nucleotide positions 16,053–16,465 (16), from 1,158 unique haplotypes, almost all (,1000) of known geographic provenance. These include 83 previously unpublished haplotypes representing 70 chimpanzees and 13 bonobos and 1,070 published sequences from GenBank (811 humans from around the world, 26 gorillas, 11 bonobos, and 222 chimpanzees). In addition, we included one Neanderthal (17) and one human nuclear sequence of mitochondrial origin (numt) involving a CR1 sequence that became inserted into chromosome 11 sometime in the past (18). Haplotypes compared ranged in length from 300–415 bp except for three comprising 270–298 bp that were included to capture the full extent of known variability. Three published orangutan sequences representing both subspecies were used as an outgroup for analytical purposes (19, 20). GenBank accession nos. of all sequences used, with their 35 specific source references and our alignments, are available from P.G.
The new chimpanzee samples from 12 geographically separate natural communities across West Africa characterize a previously poorly documented region: Bafing, western Mali; Tenkere, northwestern Sierra Leone; eastern Liberia; and Côte d’Ivoire (nine sites). Chimpanzee DNA was extracted from single shed hairs collected in 137 abandoned night nests constructed and occupied once by single individuals or mother–offspring pairs. We used the methods of Morin et al. (21) for PCR amplification and direct double-stranded 35S sequencing and used an ABI377 (Applied Biosystems) automated sequencer. The total length of the amplified fragment was 448 bp, and the primers used were L16041 (21), H16498 (9). Sixty bonobo fecal samples were collected at Eyengo, Lomako, Democratic Republic of Congo, and DNA was amplified by using primers L15997 5′-CACCATTAGCACCCAAAGCT and H16498 (9), was cycle-sequenced, and was run on an ABI377 sequencer.
Phylogenetic Reconstruction.
Sequences were aligned manually; three to four indels were required for alignment, and all gorillas showed an 80-bp gap described previously (22). The adenosine insert of the Neandertal sequence at position 16,264 was included for the alignment. Phylogenetic trees were generated with the paup program (test version 4.0d61a) written by David L. Swofford (Smithsonian Institution, Washington, DC) (ties broken randomly, ignoring sites with missing data in pairwise comparisons, among-site variation in mutation rate). Pairwise distance matrices for neighbor-joining analysis (23) were calculated by using several different distance measures and resulted in very similar topologies. A tree based on Kimura’s two-parameter model distances (24) is presented for the unpruned data, and uncorrected P values were used for the pruned data because we assumed that substitutions at hypermutable sites had been removed. Both data sets gave very similar topologies by using other commonly used genetic distances. Bootstrap values are based on 1,000 resamplings. Using different transition/transversion ratios also had very little effect on the tree topology. The large size of the entire data set precluded maximum likelihood analyses or parsimony, which, with far more taxa than informative sites, tends to be untrustworthy.
Topiary Pruning.
This method (15) was used to resolve ambiguities in the phylogenetic tree caused by homoplasies. It is based on the assumption that homoplastic substitutions are over-represented in sequences differing by only a few substitutions because such pairs of sequences are likely to differ at highly mutable sites, which will give rise to homoplasies. In this method, a series of numbered copies of the original data set are made. The original sequences then are compared in all possible pairwise combinations. Sequences that differ from each other by PL substitutions are compared with a consensus sequence. PL = number of substitutions by which sequences differ from each other. Bases that differ from the consensus sequence are deleted from copy number PL and all higher-numbered copies. The method produces a series of progressively more “pruned” data sets, labeled PL = 1, 2,… n, in which homoplasies resulting from high rates of mutation at specific sites are preferentially removed. During the pruning process, genetically similar haplotypes collapse into a single sequence containing characteristics of each. The consensus sequence used here was that for the entire data set (including Pongo). Homoplasies will evidently affect the nature of the consensus sequence. To examine the impact of homoplasies on the consensus sequence, we subdivided the taxa into subgroups by using a consensus sequence for each of the five main taxa. No significant differences in the results were seen when different consensus sequences were employed, using the commonest base at each site for five subsets of the data: humans, chimpanzees, bonobos, gorillas, and orangutans (data not shown).
RESULTS
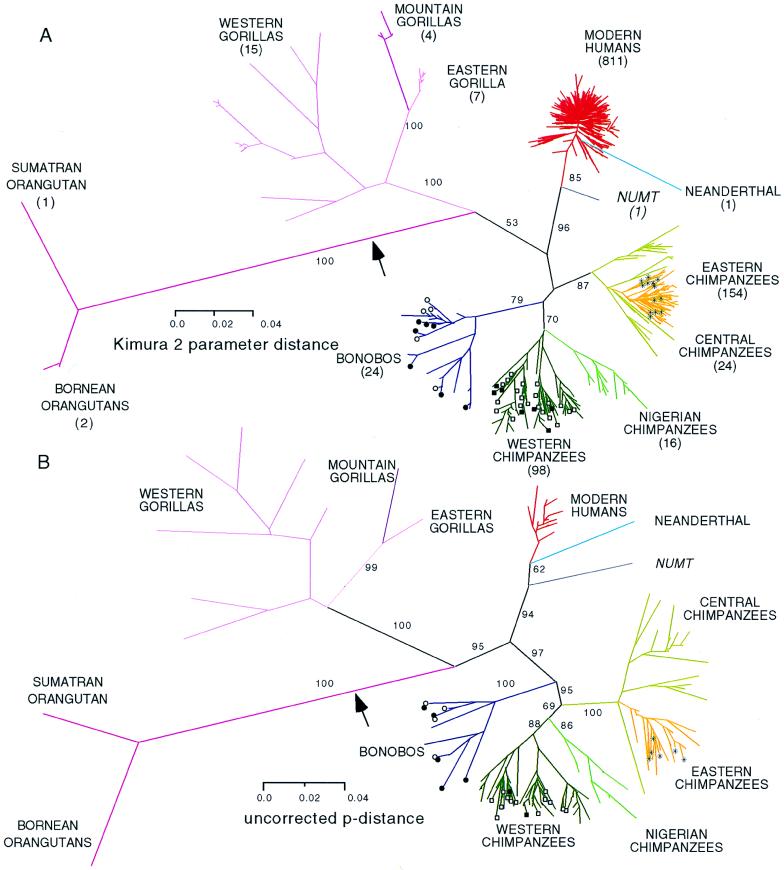
Fig. 1A shows the neighbor-joining tree. Recognized taxa are monophyletic. However, it is not possible to determine the branching order of major groups within Pan because of low bootstrap values of the internodes connecting them. Low bootstrap values also were obtained when only transversions were used (data not shown).
Figure 1.
Unrooted phylogram of the neighbor-joining tree of 1,158 different CRI sequences before (A) and after (B) after topiary pruning to level PL = 8 to remove homoplasies. Bootstrap values ≥50% for the primary internodes are shown. Position of the midpoint root is indicated by arrow. Different colors indicate species (humans, bonobos, and gorillas) and subspecies (chimpanzees). Symbols indicate individuals belonging to the same social group: □, Taï Forest, Côte d’Ivoire; ■, Solo, Mali; ○, Eyengo, Lomako, Democratic Republic of Congo; ●, E-group, Wamba, Democratic Republic of Congo; ∗, Kasakela, Gombe, Tanzania.
Remarkably, within single social groups (communities) in both western chimpanzees and bonobos, we find apes belonging to two or three deep subclades. The most striking example is from the Taï forest, where the 19 haplotypes show greater diversity than the entire human clade, even though they occur in a single breeding group. The phylogenetic position of the human numt, the CR1 sequence that became inserted in the nuclear genome after the Homo–Pan split, is the same as previously reported (18). The Neanderthal roots within the extant humans, but this result is an artifact for reasons discussed below.
Fig. 1B shows the same data after removal of homoplasies by topiary pruning to level PL = 8. Many of the original 1,158 haplotyes collapse as a result of pruning, and the dataset is reduced to 148 taxa. Internodes linking humans, bonobos, and chimpanzees are now all supported by bootstrap values of ≥94. These high values confirm and add probabilistic weight to the order of branching reported previously (1) by using much smaller sample sizes and individuals of unknown geographic origin. Branch length indicates orangutans separated from all other taxa first, followed by gorillas, then by the Homo–Pan clade, and finally chimpanzees from bonobos. Although their branching order is ambiguous, there is a clear separation among P. t. verus, Nigerian chimpanzees, and P. t. troglodytes (with P. t. schweinfurthi), supported by bootstrap values ≥86.
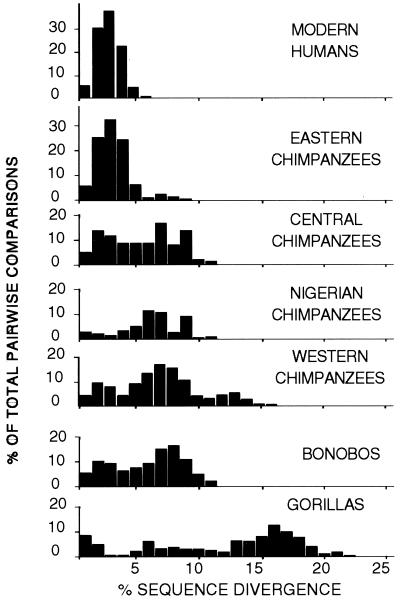
Fig. 2 shows the distribution of percent pairwise sequence divergences of each taxon normalized to the maximum sequence length. Humans and eastern chimpanzees show the smallest divergence, and even that is exaggerated because many of the differences in these taxa occurred at hypervariable sites (8, 15, 25). The narrow unimodal mismatch distributions of humans and eastern chimpanzees are typical of rapidly expanding populations. It has been suggested that the western chimpanzees may be the oldest of the chimpanzee subspecies (21, 26), and these data support this hypothesis.
Figure 2.
Pairwise sequence divergence in humans and other African hominoids.
Average transversional differences within clades were determined and normalized to the maximum sequence length. Unlike the 811 human sequences, which average only 0.41 transversional differences, the African great ape sequences show larger differences: eastern chimpanzees (1.14), central chimpanzees (2.36), western chimpanzees (3.40), bonobos (3.43), Nigerian chimpanzees (6.47), and gorillas (12.37) (data in refs. 14, 21, 22, and 27–29). Assuming comparable mutation rates, this indicates that all great ape clades are older than that of humans.
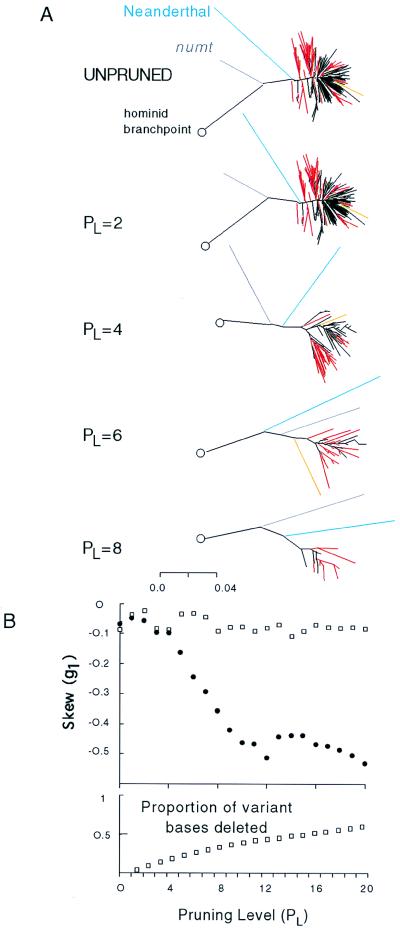
Fig. 3A illustrates the progressive effects of topiary pruning on the hominid clade. The deeply rooted all-African branches of the clade persist throughout the pruning process, though the vast majority of the non-African branches collapse into a single shallower branch. By level PL = 4, the Neanderthal sequence roots basal to all modern humans as expected, but, at earlier pruning levels, it sometimes alternates in position with the numt. By level PL = 8, the order numt followed by Neanderthal is weakly supported by a bootstrap value of 62. This suggests that the branching order and relative ages of these two “taxa” cannot be determined without additional data. Fig. 3A also shows, that although the majority of deep human branches are entirely African, several deep branches involving non-Africans persist throughout the pruning process. Some of these sequence groups move away from the base of the clade during pruning, and some move toward the base. These latter consist of an Indian Brahmin (30) and collapsed haplotypes made up of sequences from Africans, Sardinians, Melanesians, and East Asians.
Figure 3.
Effects of topiary pruning. (A) Progressive effect of pruning of the whole data set on the hominid clade from PL = 0–8. Exclusively African (red), non-African (black), one Indian (orange), neanderthal (blue), and numt (gray) haplotypes are shown. (B) Pruning results in an increase in phylogenetic information, measured as change in negative skew of the length distribution of 10,000 random trees at each pruning level (●), relative to the random removal of the same number of bases (□).
DISCUSSION
Distinguishing mtDNA CR1 Sequences from Those That Have Moved to the Nucleus.
The inadvertent inclusion of numt sequences in the data set would result in artifactual deep branches in the trees presented here. This is of concern because numts have been found in hominoids. Zischler et al. (18) found an invariant insert (Fig. 1A) in some humans but, despite repeated attempts, did not find it in chimpanzees or gorillas; most known numt sequences represent molecular “fossils” and have remained relatively unchanged since insertion (34, 35). Recently, another insert was found in all four hominoid genera (36), but it is very different from all known hominoid mtDNA sequences. We have three reasons for believing that our new sequences and most or all of the others analyzed are mitochondrial rather than nuclear: (i) mtDNA amplifies preferentially from sources (hair, feces) in which it is 1,000× more abundant than nuclear DNA (37); (ii) mother–offspring pairs were examined in western chimpanzees (12 cases) and bonobos (10 cases) and, in each case, identical sequences were found; and (iii) some of the western chimpanzee mother–offspring pairs have haplotypes found on different deep branches (Fig. 1A); if one of these deep subclades represented a numt, then one would expect the same pattern to appear in the other clades as well, and this was not observed.
Effects of Topiary Pruning.
During the early stages of topiary pruning, the amount of phylogenetic information in the data set increases. Fig. 3B shows that pruning the entire data set from PL = 1 to 10 progressively increases the negative skew of the length distribution of 10,000 random trees produced with paup; by level PL = 6, the skew has become highly significant, and further pruning after PL = 10 has little effect. In contrast, pruning the data set by random removal of the same number of variant bases as were deleted during topiary pruning results in no increase in the skew or information content (31).
Although topiary pruning as a method has yet to be widely examined, its application here resolves a number of inconsistencies that arise when the entire data set is used. These include the distinction between chimpanzees and bonobos (which is supported by a bootstrap value of 95 at level PL = 8) and the inconsistent positioning of the Neanderthal sequence. Alternative approaches to phylogenetic signal enhancement, including removal of indels and/or truncation of the sequences to remove length inconsistencies, had little influence on the topology of the tree (data not shown).
Topiary pruning produces different results in the different clades. In the human and eastern chimpanzee clades, most haplotypes have collapsed by level PL = 10 (data not shown). Even after pruning to this level, however, the clades of bonobos, western gorillas, and western chimpanzees retain very deep subclades. As noted above, although the vast majority of the non-African branches of the human clade collapse into a single shallower branch (Fig. 3A), some deep non-African branches persist. It is not possible to rule out a geographically diverse origin for our species (32, 33).
Evidence for Demographic Bottlenecks and Range Expansions.
When compared with African great apes, the lineage leading to humans must have experienced a lower genetic effective population size (Ne) (38). Whether this was attributable to a single chance demographic bottleneck (39), repeated bottlenecks, a selective sweep(s), a population expansion, or some combination of all of these remains unresolved. Evidence for departure from selective neutrality in a mitochondrial gene in humans (but not in chimpanzees) has been presented recently (40). Although mtDNA diversity in our own species is limited at the present time, the long Neandertal and numt branches indicate that our ancestral hominids harbored much greater diversity before this demographic event. In contrast to humans, the other great apes still exhibit deep branches.
Although it has been argued that our species has not gone through a reduction in Ne (39, 42), counterarguments have been proposed (43, 44). The fact that some strongly selected polymorphic nuclear loci, e.g., HLA, show no evidence of a bottleneck indicates that the history of these loci may have been very different from that of CR1. Wise et al. (45), who also noted lower mtDNA diversity in humans compared with chimpanzees, found the reverse situation at >33 nuclear markers. This paradox suggested to them that selection may have acted differentially in these two clades or that nuclear and mitochondrial genomes have evolved differently because of interspecific differences in mating patterns and natural history. The possible effect of ascertainment bias on their result should be examined because the loci used to compare chimpanzees with humans were selected for their known variability in humans only (46, 47). Furthermore, because their measure for nuclear variability, heterozygosity (H), does not differentiate between ancient and new alleles, it does not permit us to distinguish a population expansion with many new alleles from the persistence of many old alleles in a population of constant size.
Eastern chimpanzees also appear to have experienced a low Ne, as most of the branches of this clade collapsed during pruning, though less rapidly than those of the human clade. Relatively little divergence is seen in the pairwise sequence comparison (Fig. 2). This supports the conclusion that the eastern chimpanzees have expanded their range recently (between 20,000 and 61,000 years ago) from central Africa (28). Judging from the trees in Fig. 1, the question arises regarding whether eastern chimpanzees should not be regarded as a subset of a much more variable central/eastern subclade.
Hominoid Relationships.
The diverse patterns of mtDNA sequence variation seen in today’s hominoid populations probably reflect historical differences in ecological plasticity, sex-biased dispersal, range fragmentation over differing periods of time, and competition among social groups. The shapes of the Pan and Gorilla clades are consistent with very different evolutionary histories from those of humans. Both Gorilla and P. troglodytes show deep subclades, but those of the chimpanzees are “bushier,” and several different haplotypes are found in the same social groups. This difference is unlikely to be an artifact of smaller sample size in the gorilla because the 26 gorilla haplotypes were drawn from 64 individuals, many with identical sequences (22). Gorillas are confined to rain forests and live in far smaller groups (10 vs. as many as 100 in P. troglodytes) (48, 49). It seems likely that gorillas or their ancestors were subdivided into small populations that followed divergent evolutionary paths as a result of habitat fragmentation associated with contraction of the rain forest into refugia during drier Pliocene-Pleistocene hypothermal periods (50, 51). In contrast, P. troglodytes is known for its ability to survive in a variety of different habitat types (ranging from dry, sparsely wooded savanna to dense, moist forest) across tropical Africa (52). During drier periods, barriers that isolated different subpopulations can be expected to have arisen more readily for gorillas than for chimpanzees.
The three highly variable, deeply branched subclades in the western chimpanzees are not simply the result of high mutation rates at some nucleotide positions that give the appearance of deep clades because these would have been removed by topiary pruning. It is likely that these three haplotype groups originated in allopatry and subsequently became widely sympatric. Alternatively, the pattern could be caused by the persistence of ancestral haplotypes in continuously large populations. The high degree of diversity seen in relatively small and scattered social groups of chimpanzees and bonobos may be, in part, a function of high female dispersal rates in Pan (53); haplotypes are shared over up to 583 km in eastern chimpanzees (28) and up to 1,000 km in western chimpanzee (54).
The extensive genetic differentiation in chimpanzees has not been accompanied by concordant morphological and behavioral differentiation in the different clades. The three geographically defined subspecies of chimpanzees are morphologically indistinguishable except at the level of minor dental/craniometric features (55–57). Although regional behavioral differences have been found, these do not follow current subspecies divisions but, rather, reflect probable adaptations to ecological conditions or cultural differences (58, 59). All three subspecies will interbreed in captivity, but estimates of hybrid inviability (if any) are not available. Most captive chimpanzees are of unknown geographic provenance, and subspecies identity and the effects of hybrid genetic inviability cannot be separated from the high background neonate mortality observed in captivity (60).
Our results confirm the existence of a previously unrecognized subclade of chimpanzees in Nigeria and northern Cameroon, recently described (14) on the basis of 11 sequences from Nigeria. In addition, 4 of 54 apes of Wise et al. (45) captured randomly in the wild and 3 captive apes we examined (in Cameroon and Italy) are referable to this subclade of chimpanzees. However, assigning a taxonomic status to subclades should be delayed until appropriate geographical sampling is completed.
The divergence patterns in our results are in general agreement with current hominoid divergence time estimates (in millions of years ago) that are based on numerous sequences, methods, and assumptions, with the caveat that the ages of the major chimpanzee clades have been underappreciated: orangutans, 12.5 million years ago (range: 8–15); gorillas, 7.7 (6.6–10.0); hominids, 4.7 (4.2–5.2); bonobos, 2.5 (2.0–3.0); eastern and western gorillas (>2.5); the two subspecies of orangutan, 1.7 (0.6–3.4); and the two major clades of chimpanzees (western, and central and eastern), 1.6 (refs. 1, 6, 7, 21, and 61).
There are currently 15 recognized species of extinct hominids, and at least half of these have been suggested to lie between H. sapiens and the Homo–Pan ancestor 5 million years ago (62). Several of these taxa persisted for ≥1 million years (longer than H. sapiens), and, if Ne were on the order of 104 (1), each of these species had sufficient time to evolve mtDNA diversity as great as that seen in the other hominoids. Because this was not observed, something unusual happened in the early history of our species; these maternally inherited sequences show that the descent of (wo)man was very different from the pattern seen in other living hominoids.
The hominoid results have four implications: (i) Although some of the evolutionarily significant units defined on the basis of these mtDNA sequences are not concordant with current taxonomic practice, formal taxonomic revision should await clarification of their degree of genetic isolation with nuclear markers and a thorough review of the morphological and ecobehavioral evidence; (ii) biomedical research results that were based on samples of chimpanzees of unknown provenance should be reexamined because comparisons involving mixed species groups and hybrids may be compromised; (iii) when elucidating the history of zoonotic diseases like HIV-1, intraspecific variability must be taken into account (63); and (iv) future conservation efforts should be directed toward saving as much of the genetic diversity in each of these newly recognized evolutionarily significant units as possible.
Acknowledgments
We thank H. Boesch, B. Jetzer, G. Nohon, D. Lia, M. Douosson, H. Kpazahi, R. Alp, J. Moore, K. Amman, and G. Bandini for help in obtaining chimpanzee hair samples; T. Goldberg, T. Disotell, K. Saltonstall for preprints and sequences; M. Krings for sharing his database; and T. Price, A. Richman, and K. Garner for comments. Research by the authors was authorized by the relevant governments in Africa, and hair was imported under Convention on International Trade in Endangered Species permits. This work was supported by the Swiss National Foundation (P.G. and C.B.), Roche Foundation and Freiwillige Akademische Gesellschaft (P.G.), Deutsche Forschungsgemeinschaft (U.G.), Max-Planck-Society (B.F. and G.H.), and the U.S. National Science Foundation (C.W. and D.S.W.), Department of Energy (C.W.), and National Institutes of Health (C.W. and D.S.W.).
ABBREVIATIONS
- CR1
mtDNA control region-1
- numt
nuclear sequence of mitochondrial origin
Footnotes
References
- 1.Ruvolo M. Mol Biol Evol. 1997;14:248–265. doi: 10.1093/oxfordjournals.molbev.a025761. [DOI] [PubMed] [Google Scholar]
- 2.Sibley C G, Ahlquist J E. J Mol Evol. 1987;26:99–121. doi: 10.1007/BF02111285. [DOI] [PubMed] [Google Scholar]
- 3.Miyamoto M M, Slightom J L, Goodman M. Science. 1987;238:369–373. doi: 10.1126/science.3116671. [DOI] [PubMed] [Google Scholar]
- 4.Caccone A, Powell J R. Evolution (Lawrence, Kans) 1989;43:925–942. doi: 10.1111/j.1558-5646.1989.tb02540.x. [DOI] [PubMed] [Google Scholar]
- 5.Saitou N. Am J Phys Anthropol. 1991;84:75–85. doi: 10.1002/ajpa.1330840107. [DOI] [PubMed] [Google Scholar]
- 6.Horai S, Hayasaki K, Kondo R, Tsugane K, Takahata N. Proc Natl Acad Sci USA. 1995;92:532–536. doi: 10.1073/pnas.92.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruvolo M, Zehr S, von Dornum M, Pan D, Chang B, Lin J. Mol Biol Evol. 1993;10:1115–1135. doi: 10.1093/oxfordjournals.molbev.a040068. [DOI] [PubMed] [Google Scholar]
- 8.Tamura K, Nei M. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 9.Kocher T D, Wilson A C. In: Evolution of Life: Fossils Nucleotides and Culture. Osawa S, Honjo T, editors. Tokyo: Springer; 1991. pp. 391–413. [Google Scholar]
- 10.Higuchi R, von Beroldingen C H, Sensabaugh G F, Erlich H A. Nature (London) 1988;332:543–546. doi: 10.1038/332543a0. [DOI] [PubMed] [Google Scholar]
- 11.Gerloff U, Sclötterer C, Rassmann K, Rambold I, Hohmann G, Fruth B, Tautz D. Mol Ecol. 1995;4:515–518. [Google Scholar]
- 12.Woodruff D S. Primates. 1993;34:333–346. [Google Scholar]
- 13.Morin P A, Woodruff D S. In: Molecular Genetic Approaches in Conservation. Smith T B, Wayne R K, editors. Oxford: Oxford Univ. Press; 1996. pp. 298–313. [Google Scholar]
- 14.Gonder M K, Oates J F, Disotell T R, Forstner M R J, Morales J C, Melnick D J. Nature (London) 1997;388:337. doi: 10.1038/41005. [DOI] [PubMed] [Google Scholar]
- 15.Wills C. Evolution (Lawrence, Kans) 1996;50:977–989. doi: 10.1111/j.1558-5646.1996.tb02340.x. [DOI] [PubMed] [Google Scholar]
- 16.Anderson S, Bankier A T, Barrell B G, de Bruijn M H, Coulson A R, Drouin J, Eperon I C, Nierlich D P, Roe B A, Sanger F, et al. Nature (London) 1981;290:457–464. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 17.Krings M, Stone A, Schmitz R W, Kranitzki H, Stoneking M, Pääbo S. Cell. 1997;90:19–30. doi: 10.1016/s0092-8674(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 18.Zischler H, Geisert H, von Haessler A, Pääbo S. Nature (London) 1995;378:489–492. doi: 10.1038/378489a0. [DOI] [PubMed] [Google Scholar]
- 19.Xu X, Arnason U. J Mol Evol. 1996;43:431–437. doi: 10.1007/BF02337514. [DOI] [PubMed] [Google Scholar]
- 20.Horai S, Satta Y, Hayasaka K, Kondo R, Inoue T, Ishida T, Hayashi S. J Mol Evol. 1992;35:32–43. doi: 10.1007/BF00160258. [DOI] [PubMed] [Google Scholar]
- 21.Morin P A, Moore J J, Chakraborty R, Jin L, Goodall J, Woodruff D S. Science. 1994;265:1193–1201. doi: 10.1126/science.7915048. [DOI] [PubMed] [Google Scholar]
- 22.Garner K J, Ryder O A. Mol Phylogenet Evol. 1996;6:39–48. doi: 10.1006/mpev.1996.0056. [DOI] [PubMed] [Google Scholar]
- 23.Saitou M, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 24.Kimura M. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 25.Wills C. Evolution (Lawrence, Kans) 1995;49:593–607. doi: 10.1111/j.1558-5646.1995.tb02296.x. [DOI] [PubMed] [Google Scholar]
- 26.Kortland A. New Perspectives in Ape and Human Evolution. Amsterdam: Stichting voor Psychobiologie; 1972. [Google Scholar]
- 27.Goldberg T L, Ruvolo M. Biol J Linn Soc. 1997;61:301–324. [Google Scholar]
- 28.Goldberg T L, Ruvolo M. Mol Biol Evol. 1997;14:976–984. doi: 10.1093/oxfordjournals.molbev.a025841. [DOI] [PubMed] [Google Scholar]
- 29.Hashimoto C, Furuichi T, Takenaka O. Primates. 1996;37:305–318. [Google Scholar]
- 30.Mountain J L, Hebert J M, Bhattacharya S, Underhill P A, Ottolenghi C, Gadgil M, Cavalli Sforza L L. Am J Hum Genet. 1995;56:979–992. [PMC free article] [PubMed] [Google Scholar]
- 31.Hillis D M, Huelsenbeck J P, Cunningham C W. Science. 1994;264:671–677. doi: 10.1126/science.8171318. [DOI] [PubMed] [Google Scholar]
- 32.Altheide T K, Hammer M F. Am J Hum Genet. 1997;61:462–466. doi: 10.1016/S0002-9297(07)64077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harding R M, Fullerton S M, Griffiths R C, Bond J, Cox M J, Schneider J A, Moulin D S, Clegg J B. Am J Hum Genet. 1997;60:772–789. [PMC free article] [PubMed] [Google Scholar]
- 34.Ward R H, Frazier B H, Dew-Jager S, Pääbo S. Proc Natl Acad Sci USA. 1991;88:8720–8724. doi: 10.1073/pnas.88.19.8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lundstrom R, Tavaré S, Ward R H. Proc Natl Acad Sci USA. 1992;89:5961–5965. doi: 10.1073/pnas.89.13.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zischler H, Geisert H, Castresana J. Mol Biol Evol. 1998;15:463–469. doi: 10.1093/oxfordjournals.molbev.a025943. [DOI] [PubMed] [Google Scholar]
- 37.Michaels G S, Hauswirth W W, Laipis P J. Dev Biol. 1982;94:246–251. doi: 10.1016/0012-1606(82)90088-4. [DOI] [PubMed] [Google Scholar]
- 38.Nei M, Maruyama T, Chakraborty R. Evolution (Lawrence, Kans) 1975;29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- 39.Wills C. Nature (London) 1990;348:398. doi: 10.1038/348398a0. [DOI] [PubMed] [Google Scholar]
- 40.Wise C, Sraml M, Easteal S. Genetics. 1998;148:409–421. doi: 10.1093/genetics/148.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ayala F J. Science. 1995;270:1930–1936. doi: 10.1126/science.270.5244.1930. [DOI] [PubMed] [Google Scholar]
- 42.Hey J. Mol Biol Evol. 1997;14:166–172. doi: 10.1093/oxfordjournals.molbev.a025749. [DOI] [PubMed] [Google Scholar]
- 43.Harpending H. Hum Biol. 1994;66:591–600. [PubMed] [Google Scholar]
- 44.Rogers A R. Evolution (Lawrence, Kans) 1995;49:608–615. doi: 10.1111/j.1558-5646.1995.tb02297.x. [DOI] [PubMed] [Google Scholar]
- 45.Wise C A, Sraml M, Rubinsztein D C, Easteal S. Mol Biol Evol. 1997;14:707–716. doi: 10.1093/oxfordjournals.molbev.a025810. [DOI] [PubMed] [Google Scholar]
- 46.Ellegren H, Primmer C R, Sheldon B C. Nat Genet. 1995;11:360–362. doi: 10.1038/ng1295-360. [DOI] [PubMed] [Google Scholar]
- 47.Forbes S H, Hogg J T, Buchanan F C, Crawford A M, Allendorf F W. Mol Biol Evol. 1995;12:1106–1113. doi: 10.1093/oxfordjournals.molbev.a040284. [DOI] [PubMed] [Google Scholar]
- 48.Goodall J. The Chimpanzees of Gombe: Patterns of Behavior. Cambridge, MA: Harvard Univ. Press; 1986. [Google Scholar]
- 49.Watts D P. In: Great Ape Societies. McGrew W C, Marchant L F, Nishida T, editors. Cambridge, U.K.: Cambridge Univ. Press; 1996. pp. 16–28. [Google Scholar]
- 50.de Menocal P B. Science. 1995;270:53–59. [Google Scholar]
- 51.Vrba G H, Denton G H, Partridge T C, Burckle L H, editors. Paleoclimate and Evolution, with Emphasis on Human Origins. New Haven, CT: Yale Univ. Press; 1995. [Google Scholar]
- 52.Kortlandt A. J Hum Evol. 1983;12:231–278. [Google Scholar]
- 53.Pusey A E, Packer C. In: Primate Societies. Cheney D L, Seyfarth R M, Smuts B B, Wrangham R W, Struhsaker T T, editors. Chicago: Univ. Chicago Press; 1987. pp. 250–266. [Google Scholar]
- 54.Gagneux P. Ph. D. thesis. Basel: Basel University; 1998. [Google Scholar]
- 55.Albrecht G H, Miller J M A. In: Species, Species Concepts and Primate Evolution. Kimbel W H, Martin L B, editors. New York: Plenum; 1993. pp. 123–161. [Google Scholar]
- 56.Shea B T, Leigh S R, Groves C P. In: Species, Species Concepts and Primate Evolution. Kimbel W H, Martin L B, editors. New York: Plenum; 1993. pp. 265–296. [Google Scholar]
- 57.Uchida A. Bull Peabody Museum. 1996;4:1–184. [Google Scholar]
- 58.McGrew W C. Chimpanzee Material Culture: Implications for Human Evolution. Cambridge, U.K.: Cambridge Univ. Press; 1992. [Google Scholar]
- 59.Boesch C. In: Evolution of Social Behavior Patterns in Primates and Man. Runciman W G, Maynard Smith J, Dunbar R I M, editors. Oxford: Oxford Univ. Press; 1997. pp. 251–268. [Google Scholar]
- 60.Morin P A, Moore J J, Woodruff D S. Proc R Soc Lond Ser B. 1992;249:293–297. doi: 10.1098/rspb.1992.0117. [DOI] [PubMed] [Google Scholar]
- 61.Zhi L, Karesh W B, Janczewski D N, Frazier-Taylor H, Sajuthi D, Gombeck F, Andau M, Martenson J S, O’Brien J O. Curr Biol. 1996;6:1326–1336. doi: 10.1016/s0960-9822(02)70719-7. [DOI] [PubMed] [Google Scholar]
- 62.Lewin R. Human Evolution. Cambridge, MA: Harvard Univ. Press; 1997. [Google Scholar]
- 63.Gao F, Bailes E, Robertson D L, Chen Y, Rodenburg C M, Michael S F, Cummins L B, Arthur L O, Peeters M, Shaw G M, et al. Nature (London) 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]