Abstract
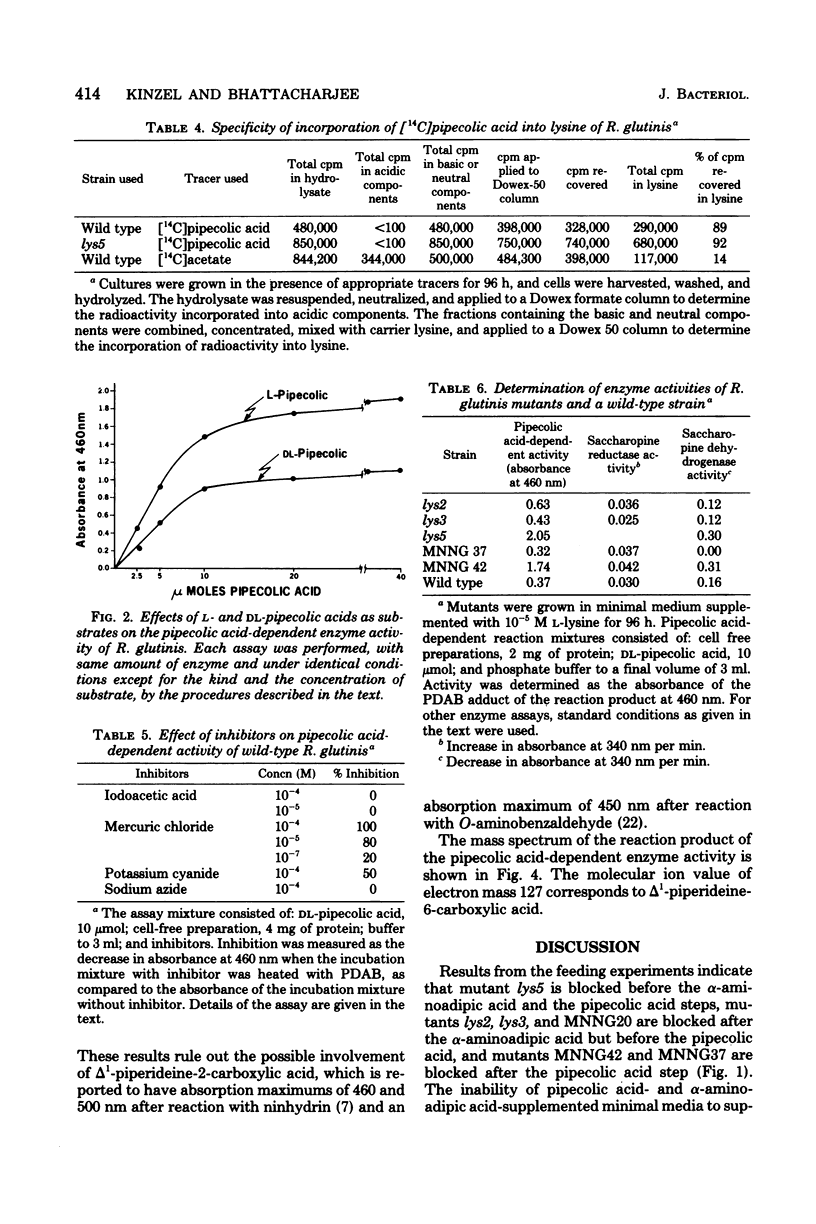
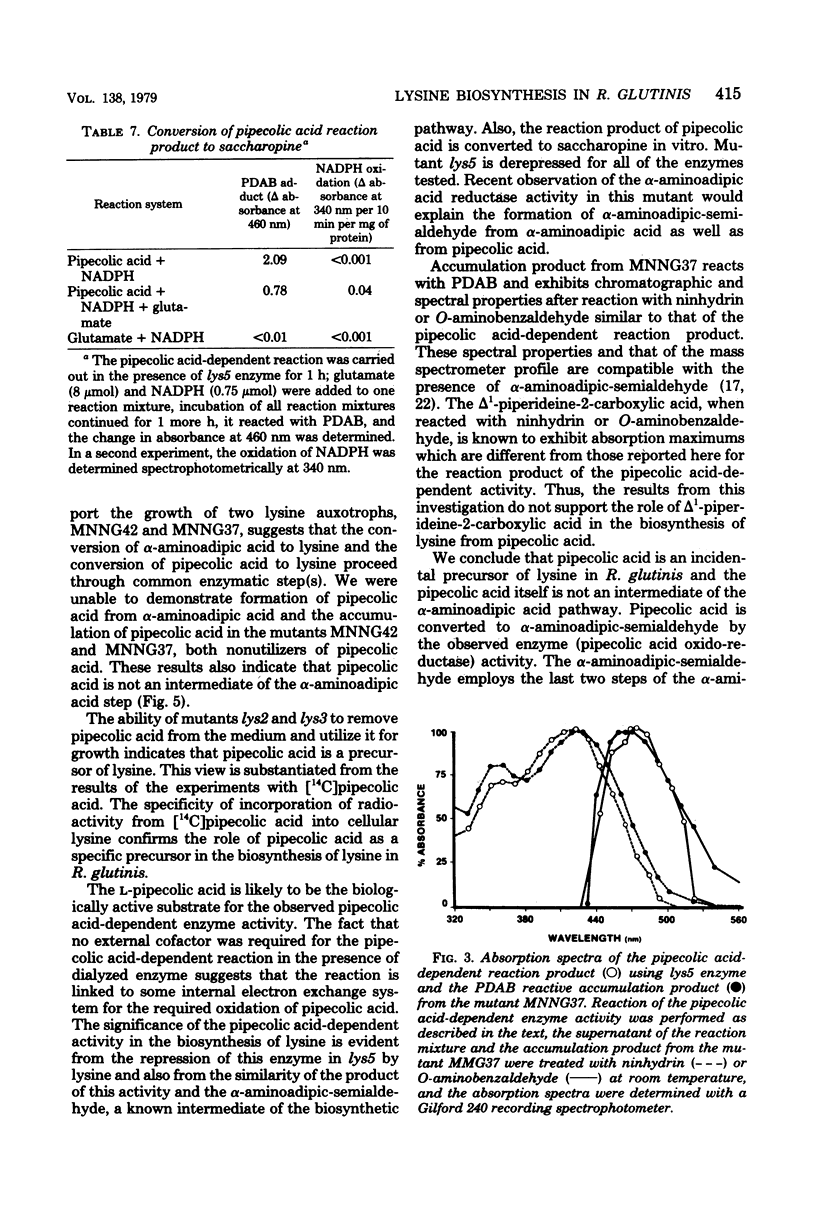
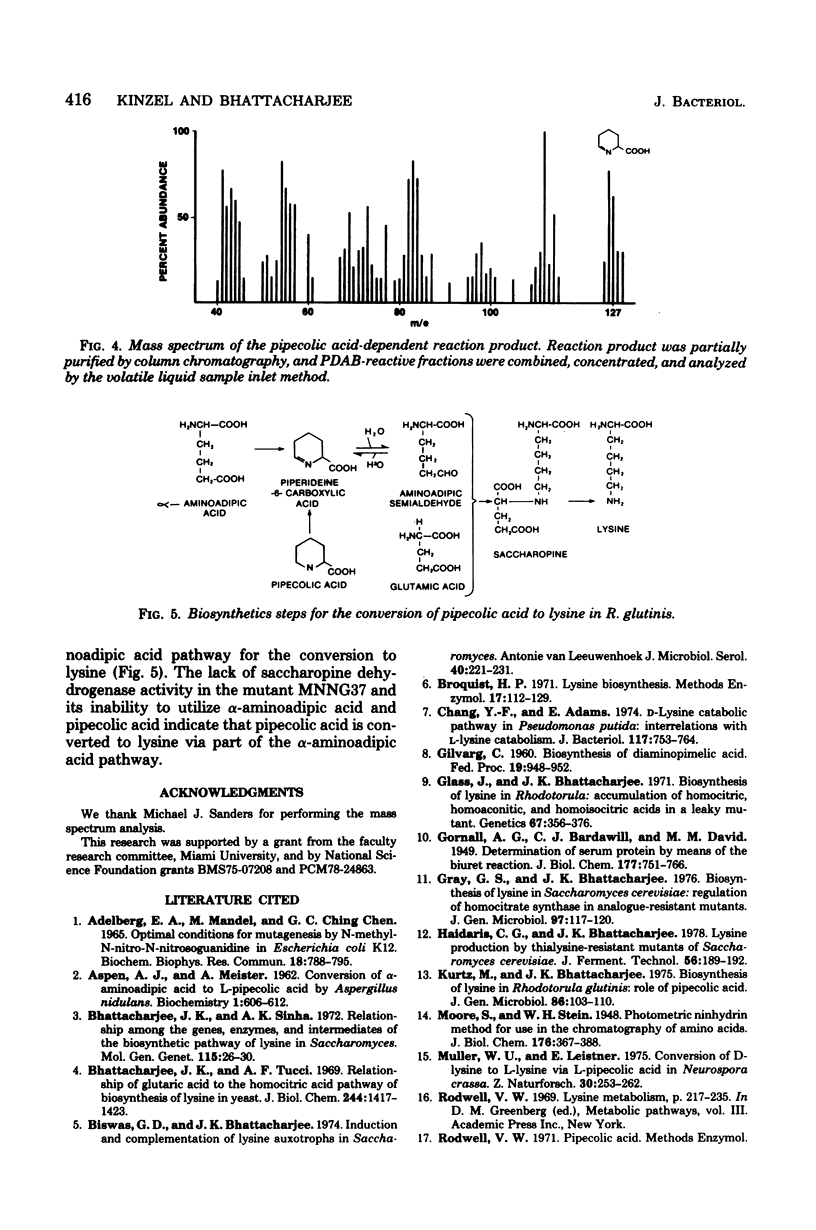
The role of pipecolic acid in the biosynthesis of lysine was investigated in Rhodotorula glutinis, an aerobic red yeast. Supplementation of pipecolic acid in the minimal medium supported the growth of mutants lys2, lys3, and lys5; α-aminoadipic acid supported the growth of lys5; but neither α-aminoadipic acid nor pipecolic acid supported the growth of mutants MNNG42 and MNNG37. During the growth of the appropriate mutants, pipecolic acid was removed from the growth medium and the intracellular pool. In tracer experiments, radioactivity from [14C]pipecolic acid was selectively incorporated into the cellular lysine of lys5 and the wild-type strain. l-Pipecolic acid-dependent enzyme activity did not require any cofactor and was inhibited by mercuric chloride and potassium cyanide. This activity was present in the wild-type strain and all of the mutants tested and was repressed in mutant lys5 when grown in the presence of higher concentration of lysine. The reaction product of pipecolic acid was converted to saccharopine by lys5 enzyme in the presence of glutamate and reduced nicotin-amide adenine dinucleotide phosphate. Mutant MNNG37 lacked the saccharopine dehydrogenase activity, indicating that this step is involved in the conversion of α-aminoadipic acid and pipecolic acid to lysine. Mutants MNNG37 and MNNG42 accumulated a p-dimethylaminobenzaldehyde-reacting product in the culture supernatant and in the intracellular pool. Chromatographic properties of the p-dimethylaminobenzaldehyde adduct and that of the pipecolic acid-dependent reaction product were similar. The reaction product and the accumulation product were characterized on the basis of mass and absorption spectra as α-aminoadipic-semialdehyde, which in solution remains in equilibrium with Δ1-piperideine-6-carboxylic acid. Since α-aminoadipic-semialdehyde is a known intermediate of the α-aminoadipic acid pathway for the biosynthesis of lysine, it is concluded that pipecolic acid is converted to lysine in R. glutinis via α-aminoadipic-semialdehyde and saccharopine.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ASPEN A. J., MEISTER A. Conversion of alpha-aminoadipic acid to L-pipecolic acid by Aspergillus nidulans. Biochemistry. 1962 Jul;1:606–612. doi: 10.1021/bi00910a010. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee J. K., Sinha A. K. Relationship among the genes, enzymes, and intermediates of the biosynthetic pathway of lysine in Saccharomyces. Mol Gen Genet. 1972;115(1):26–30. doi: 10.1007/BF00272214. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee J. K., Tucci A. F. Relationship of glutaric acid to the homocitric acid pathway of biosynthesis of lysine in yeast. J Biol Chem. 1969 Mar 25;244(6):1417–1423. [PubMed] [Google Scholar]
- Biswas G. D., Bhattacharjee J. K. Induction and complementation of lysine auxotrophs in Saccharomyces. Antonie Van Leeuwenhoek. 1974;40(2):221–231. doi: 10.1007/BF00394380. [DOI] [PubMed] [Google Scholar]
- Chang Y. F., Adams E. D-lysine catabolic pathway in Pseudomonas putida: interrelations with L-lysine catabolism. J Bacteriol. 1974 Feb;117(2):753–764. doi: 10.1128/jb.117.2.753-764.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GILVARG C. Biosynthesis of diaminopimelic acid. Fed Proc. 1960 Dec;19:948–952. [PubMed] [Google Scholar]
- Glass J., Bhattacharjee J. K. Biosynthesis of lysine in Rhodotorula: accumulation of homocitric, homoaconitic, and homoisocitric acids in a leaky mutant. Genetics. 1971 Mar;67(3):365–376. doi: 10.1093/genetics/67.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray G. S., Bhattacharjee J. K. Biosynthesis of lysine in Saccharomyces cerevisiae: regulation of homocitrate synthase in analogue-resistant mutants. J Gen Microbiol. 1976 Nov;97(1):117–120. doi: 10.1099/00221287-97-1-117. [DOI] [PubMed] [Google Scholar]
- Kurtz M., Bhattacharjee J. K. Biosynthesis of lysine in Rhodotorula glutinis: role of pipecolic acid. J Gen Microbiol. 1975 Jan;86(1):103–110. doi: 10.1099/00221287-86-1-103. [DOI] [PubMed] [Google Scholar]
- Müller W. U., Leistner E. Conversion of D-lysine via L-pepecolic acid in Neurospora crassa. Z Naturforsch C. 1975 Mar-Apr;30(2):253–262. doi: 10.1515/znc-1975-3-419. [DOI] [PubMed] [Google Scholar]
- SCHWEET R. S., HOLDEN J. T., LOWY P. H. The metabolism of lysine in Neurospora. J Biol Chem. 1954 Dec;211(2):517–529. [PubMed] [Google Scholar]
- Sinha A. K., Bhattacharjee J. K. Control of a lysine-biosynthetic step by two unlinked genes of Saccharomyces. Biochem Biophys Res Commun. 1970;39(6):1205–1210. doi: 10.1016/0006-291x(70)90689-3. [DOI] [PubMed] [Google Scholar]
- Sinha A. K., Kurtz M., Bhattacharjee J. K. Effect of hydroxylysine on the biosynthesis of lysine in saccharomyces. J Bacteriol. 1971 Nov;108(2):715–719. doi: 10.1128/jb.108.2.715-719.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soda K., Misono H., Yamamoto T. L-Lysine:alpha-ketoglutarate aminotransferase. I. Identification of a product, delta-1-piperideine-6-carboxylic acid. Biochemistry. 1968 Nov;7(11):4102–4109. doi: 10.1021/bi00851a045. [DOI] [PubMed] [Google Scholar]
- Umbarger H. E. Amino acid biosynthesis and its regulation. Annu Rev Biochem. 1978;47:532–606. doi: 10.1146/annurev.bi.47.070178.002533. [DOI] [PubMed] [Google Scholar]



